Optimization of Electrically Active Magnetic Nanoparticles as Accurate and Efficient Microbial Extraction Tools
Abstract
:1. Introduction
2. Experimental Section
- (a)
- The addition of sodium chloride to a concentration of about 0.14 M during conjugation of antibodies onto MNPs.
- (b)
- The concentration of antibodies present during conjugation of antibodies onto MNPs.
- (c)
- The concentration of Mab-EAMNPs present during IMS.
- (d)
- The number of days elapsed since conjugation of antibodies onto EAMNPs, which affect the capture evaluation by culture after IMS.
3. Materials and Methods
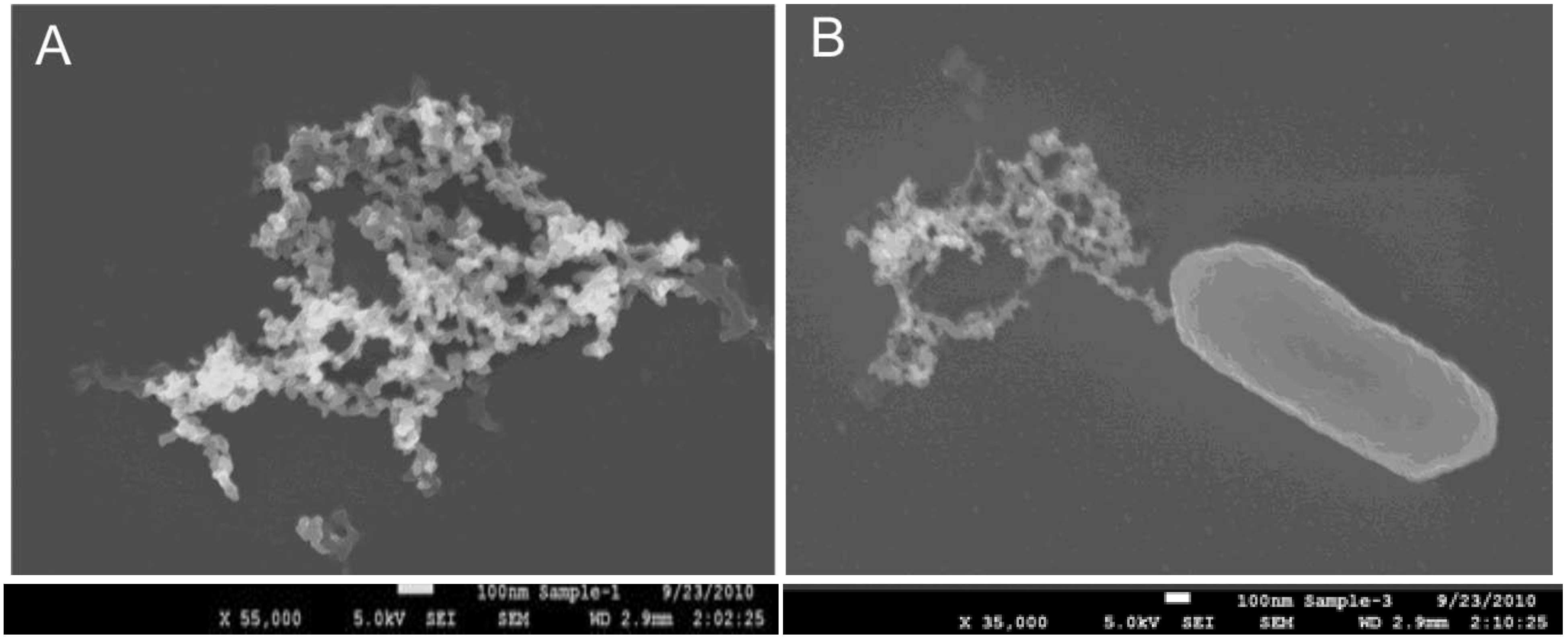
3.1. EAMNP Production
3.2. EAMNP Antibody Conjugation
3.3. Immuno-Magnetic Separation (IMS) and Plating of Bacteria
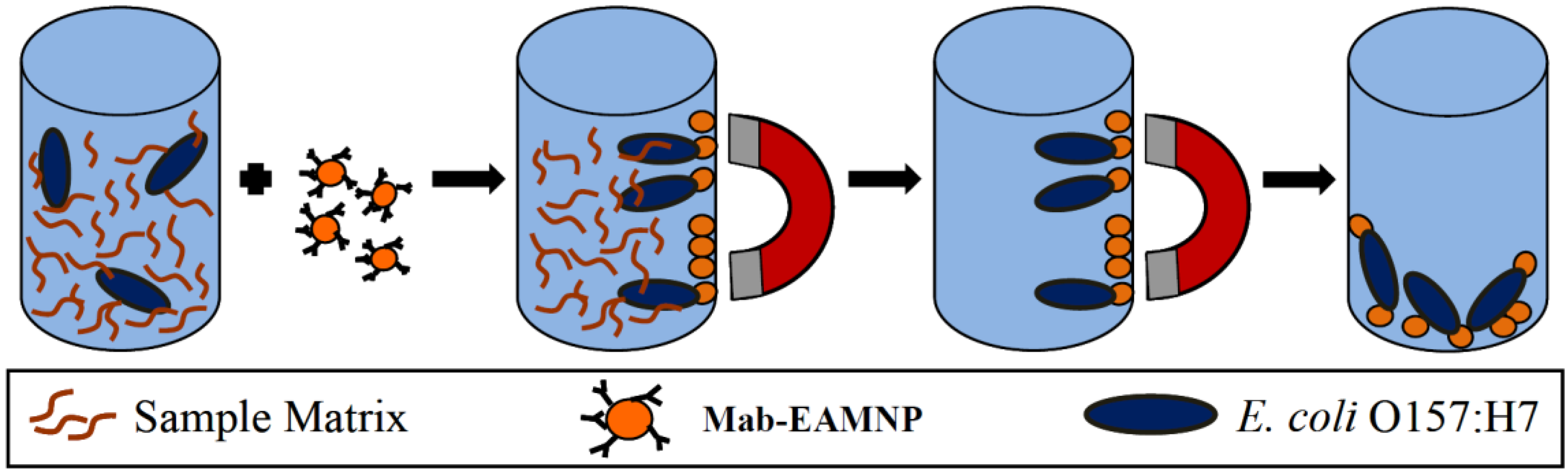
3.4. Statistical Analysis
4. Results and Discussion
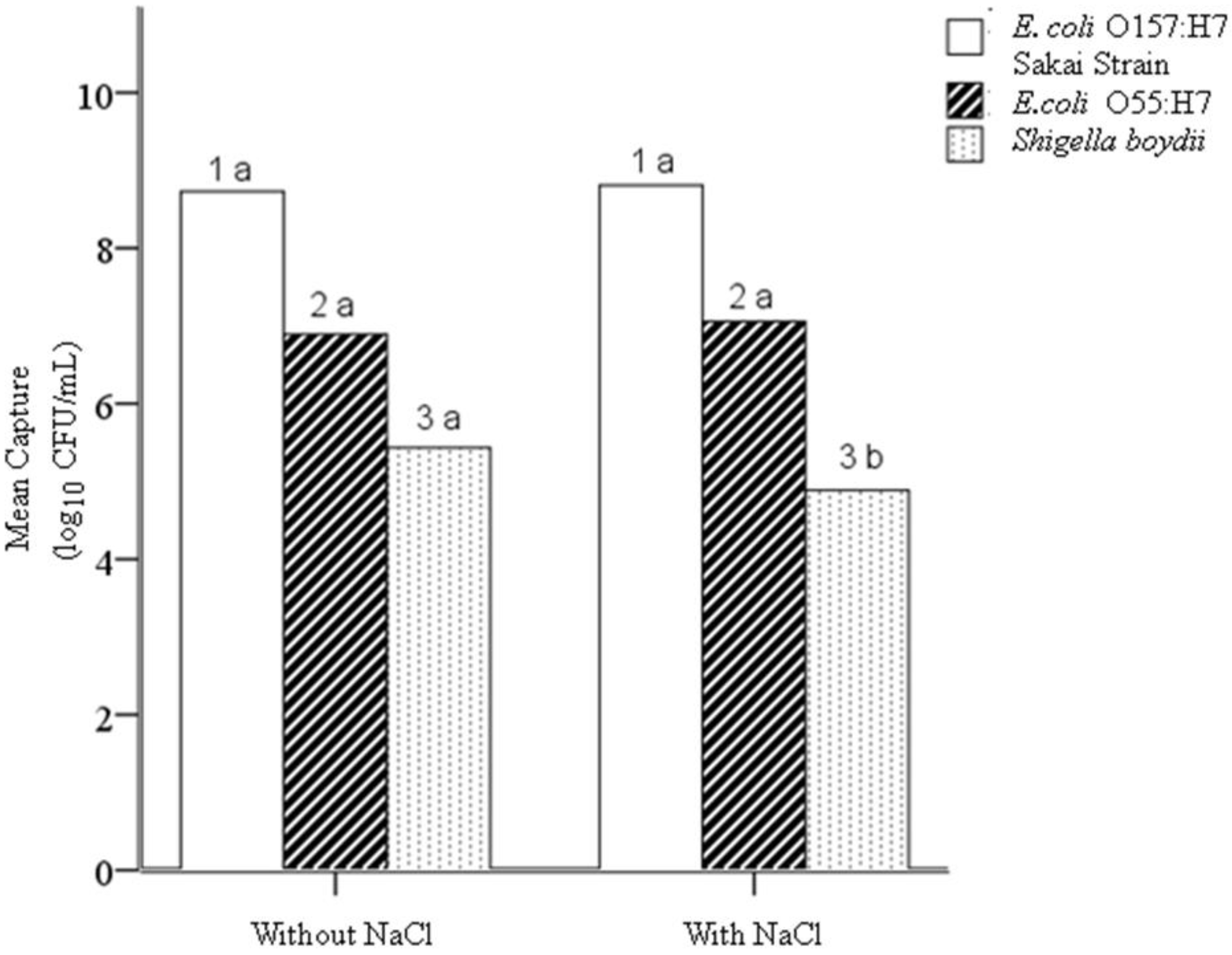
4.1. Hypothesis 1a: Effect of Sodium Chloride Addition during Conjugation
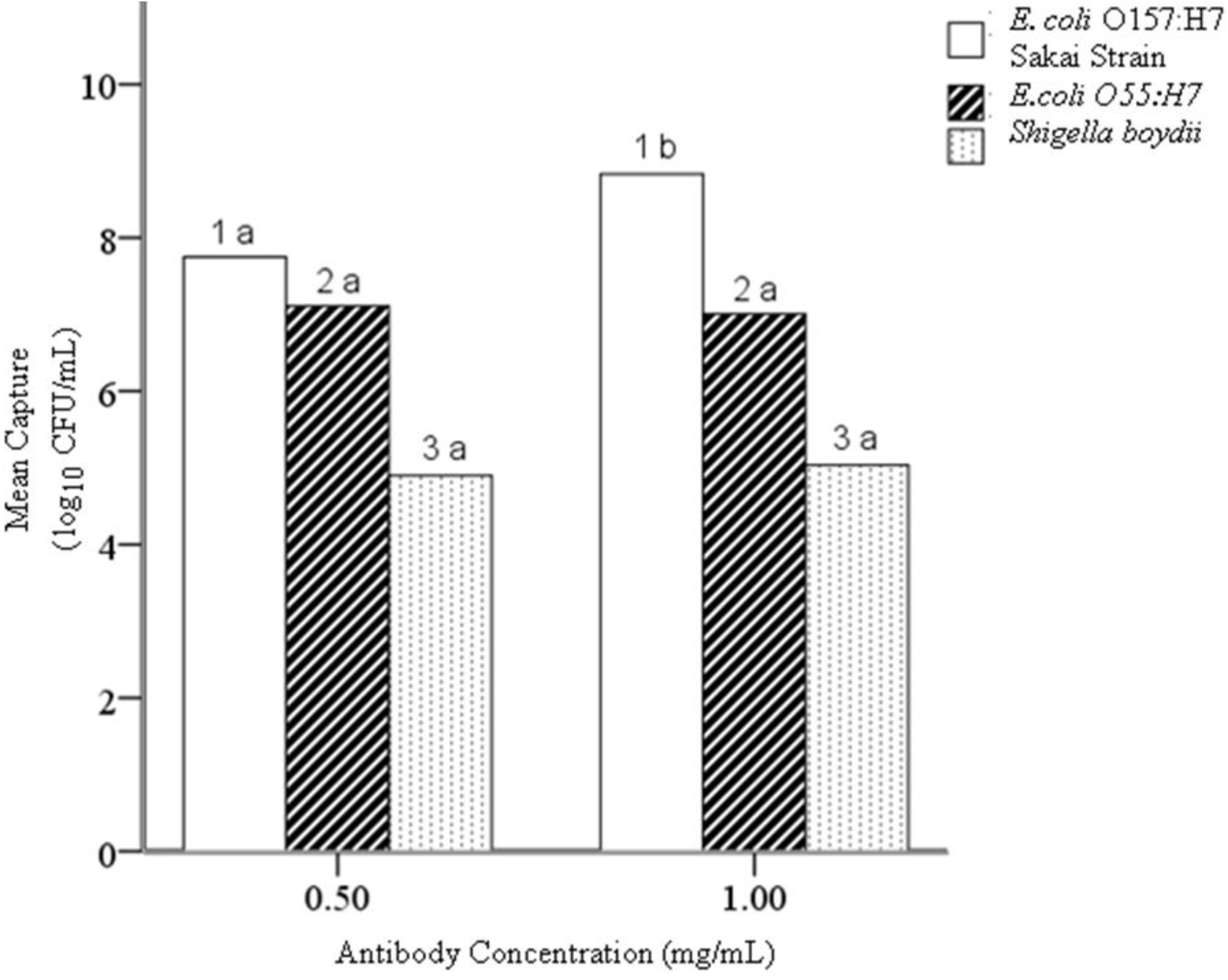
4.2. Hypothesis 1b: Effect of Antibody Concentration during Conjugation
4.3. Hypothesis 1c: Effect of Mab-EAMNP Concentration during IMS
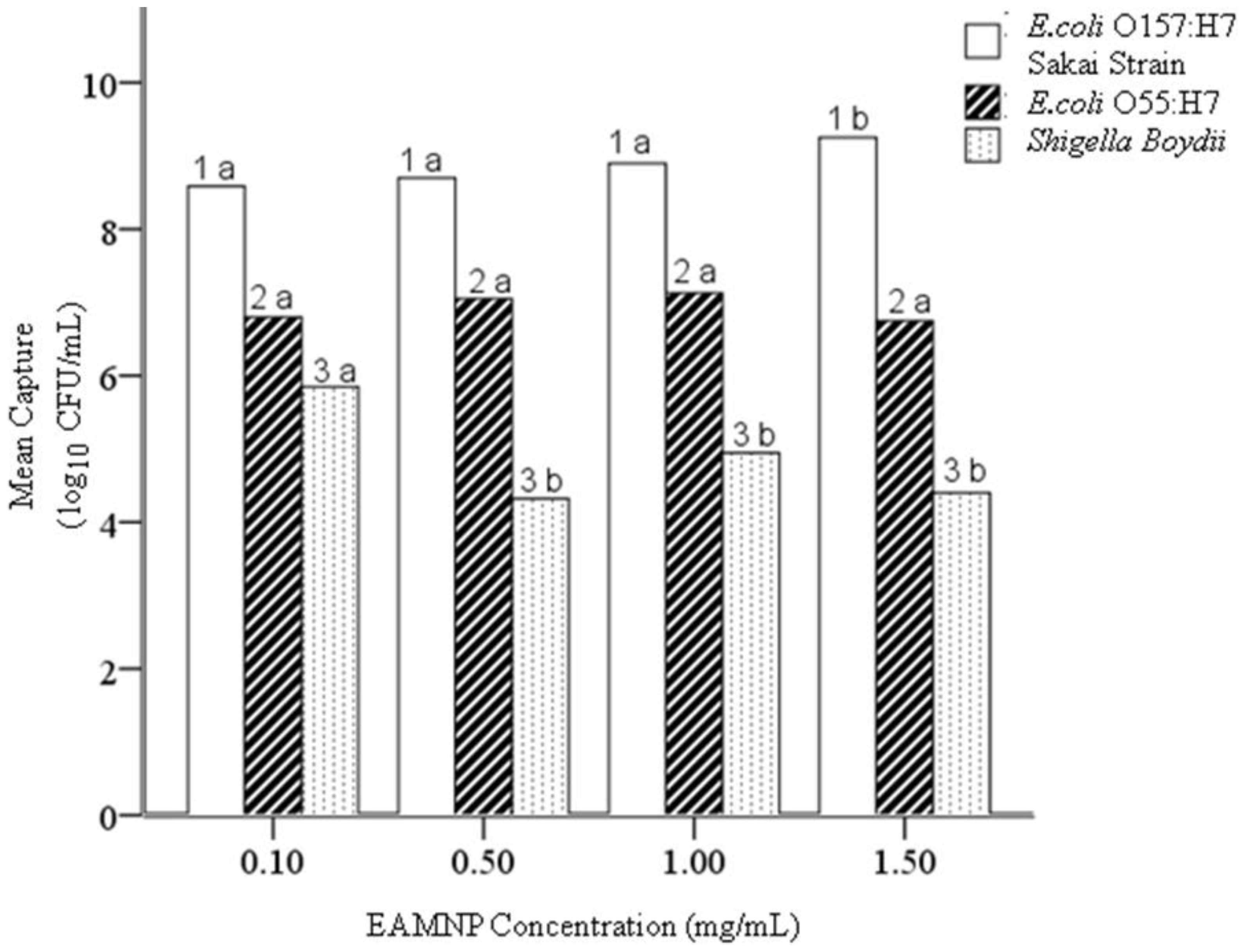
4.4. Hypothesis 1d: Effect of Age of Mab-EAMNP
Solution during IMS

5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Spink, J. Defining Food Fraud and the Chemistry of the Crime. In Proceedings of the FDA Open Meeting, Economically Motivated Adulteration, College Park, MD, USA, 1 May 2009.
- Varshney, M.; Yang, L.; Su, X.L.; Li, Y. Magnetic nanoparticle-antibody conjugates for the separation of Escherichia coli O157:H7 in ground beef. J. Food Prot. 2005, 68, 1804–1811. [Google Scholar] [PubMed]
- Center for Science in the Public Interest (CSPI). Keep America’s Food Safe, The Case for Increased Funding at FDA. 2007. Available online: http://www.cspinet.org/foodsafety/fdafunding.html (accessed on 20 January 2012).
- Olsson, F.; Weeda, T.; Bode, M. PC Attorneys at Law, 2010. The FDA Food Safety Modernization Act. A Summary Published by Institute of Food Technologists (IFT), Chicago. Available online: http://www.ift.org/public-policy-andregulations/recent-news/2011/january/w/media/PublicPolicy/PolicyDevelopment/FDAFoodSafetyModernizationAct_summary.pdf (accessed on 17 April 2012).
- Sjerven, J. FDA Issues Criteria for Determining Frequency of Inspections. Agency Outlines Method for Determining Whether a Food Plant Should be Designated “High Risk” or “Non-High Risk”. Food Business News.net. Available online: http://www.foodbusinessnews.net/News/News%20Home/Food%20Safety%20News/2012/3/FDA%20issues%20criteria%20for%20determining%20frequency%20of%20inspections.aspx (accessed on 27 March 2012).
- Ge, B.; Meng, J. Advanced Technologies for Pathogen and Toxin Detection in Foods: Current Applications and Future Directions. J. Assoc. Lab. Autom. 2009, 14, 235–241. [Google Scholar] [CrossRef]
- Cheng, Y.; Liu, Y.; Huang, J.; Li, K.; Zhang, W.; Xian, Y.; Jin, L. Nanoscale assembly of amine-functinalized colloidal iron oxide. J. Magn. Magn. Mater. 2009, 321, 1529–1532. [Google Scholar] [CrossRef] [PubMed]
- Gehring, A.G.; Brewster, J.D.; Irwin, P.L.; Tu, S.I.; van Houten, L.J. 1-Naphthylphosphate as an enzymatic substrate for enzyme-linked immune-magnetic electrochemistry. J. Electroanal. Chem. 1999, 469, 27–33. [Google Scholar] [CrossRef]
- Gehring, A.G.; Tu, S.I. Enzyme-linked immune-magnetic electrochemical detection of live Escherichia coli O157:H7 in Apple Juice. J. Food Prot. 2005, 68, 146–149. [Google Scholar] [PubMed]
- Jaffrezic-Renault, N.; Martelett, C.; Chevolot, Y.; Cloarec, J.P. Biosensors and biobarcode assays based on biofunctionalized magnetic microbeads. Sensors 2007, 7, 589–614. [Google Scholar] [CrossRef]
- Maalouf, R.; Hassen, W.; Fournier-Wirth, C.; Coste, J.; Jaffrezic-Renault, N. Comparison of two innovatives approaches for bacterial detection: Paramagnetic nanoparticles and self-assembled multilayer processes. Microchim. Acta 2008, 163, 157–161. [Google Scholar] [CrossRef]
- Perez, F.G.; Mascini, M.; Tothill, I.E.; Turner, A.P.F. Immuno-magnetic separation with mediated flow injection analysis amperometric detection of viable Escherichia coli O157. Anal. Chem. 1998, 70, 2380–2386. [Google Scholar] [CrossRef] [PubMed]
- Ruan, C.; Wang, H.; Li, Y. A bienzyme electrochemical biosensor coupled with immuno-magnetic separation for rapid detection of Escherichia coli O157:H7 in food samples. Trans. ASAE 2002, 45, 249–255. [Google Scholar] [CrossRef]
- Tu, S.I.; Golden, M.; Cooke, P.; Paoli, G.; Gehring, A. Detection of Escherichia coli O157:H7 through the formation of sandiwiched complexes with immuno-magnetic and fluorescent beads. J. Rapid Methods Autom. Microbiol. 2005, 13, 269–282. [Google Scholar] [CrossRef]
- Varshney, M.; Li, Y. Interdigitated array microelectrode based impedance biosensor coupled with magnetic nanoparticle-antibody conjugates for detection of Escherichia coli O157:H7 in food samples. Biosens. Bioelectron. 2007, 22, 2408–2414. [Google Scholar] [CrossRef] [PubMed]
- Varshney, M.; Li, Y.; Srinivasan, B.; Tung, S. A label-free, microfluidics and interdigitated array microelectrode-based impedance biosensor in combination with nanoparticles immunoseparation for detection of Escherichia coli O157:H7 in food samples. Sens. Actuators B Chem. 2007, 128, 99–107. [Google Scholar] [CrossRef]
- Yang, L.; Li, Y. Detection of viable Salmonella using microelectrode-based capacitance measurement coupled with immuno-magnetic separation. J. Microbiol. Methods 2006, 64, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Wang, R.; Wang, Y.; Su, X.; Ying, Y.; Wang, J.; Li, Y. Evaluation of different micro/nanobeads used as amplifiers in QCM immunosensor for more sensitive detection of E. coli O157:H7. Biosens. Bioelectron. 2011, 29, 23–28. [Google Scholar] [CrossRef] [PubMed]
- FSIS. Risk Assessment of the Public Health Impact of Escherichia coli O157:H7 in Ground Beef, 2001. Available online: http://www.fsis.usda.gov/science/risk_assessments/index.asp (accessed on 25 April 2012).
- FDA. Bacteriological Analytical Manual (BAM); U.S. Food & Drug Administration: Silver Springs, MD, USA, 2009. [Google Scholar]
- Pal, S.; Setterington, E.B.; Alocilja, E.C. Electrically active magnetic nanoparticles for concentrating and detecting Bacillus anthracis spores in a direct-charge transfer biosensor. Sens. J. IEEE 2008, 8, 647–654. [Google Scholar] [CrossRef]
- Pal, S.; Alocilja, E.C. Electrically active polyaniline coated magnetic (EAPM) nanoparticle as novel transducer in biosensor for detection of Bacillus anthracis spores in food samples. Biosens. Bioelectron. 2009, 24, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Setterington, E.; Cloutier, B.; Ochoa, J.; Cloutier, A.; Alocilja, E. Rapid, sensitive, and specific immunomagnetic separation of foodborne pathogens. Int. J. Food Saf. Nutr. Public Health 2011, 4, 83–100. [Google Scholar] [CrossRef]
- AOAC. Final Report and Executive Summaries from the AOAC International Presidential Task Force on Best Practices in Microbiological Methodology; AOAC International: Rockville, MD, USA, 2006. [Google Scholar]
- Adams, M.R.; Moss, M.O. Food Microbiology, 3rd ed.; RSC Publishing: Cambridge, UK, 2008. [Google Scholar]
- Barick, K.C.; Aslam, M.; Prasad, P.V.; Dravid, V.P.; Bahadur, D. Nanoscale assembly of amine-functionalized colloidal iron oxide. J. Magn. Magn. Mater. 2009, 321, 1529–1532. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, C.; McCarthy, J.D. Modifications to methods for the enumeration and detection of injured Escherichia coli O157:H7 in foods. Int. J. Food Microbiol. 2000, 55, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Durso, L.M.; Keen, J.E. Shiga-toxigenic Escherichia coli O157 and non-Shiga-toxigenic E. coli O157 respond differently to culture and isolation from naturally contaminated bovine faeces. J. Appl. Microbiol. 2007, 103, 2457–2464. [Google Scholar] [CrossRef] [PubMed]
- Fitzmaurice, J.; Duffy, G.; Kilbride, B.; Sheridan, J.J.; Carroll, C.; Maher, M. Comparison of a membrane surface adhesion recovery method with an IMS method for use in a polymerase chain reaction method to detect Escherichia coli O157:H7 in minced beef. J. Microbiol. Methods 2004, 59, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Fung, D. Rapid methods and automation in food microbiology: 25 years of development and predictions. In IUFeST World Congress Book: Global Issues in Food Science and Technolgy; Academic Press: Salt Lake City, UT, USA, 2008. [Google Scholar]
- Hennekens, C.; Buring, J. Screening. In Epidemiology in Medicine; Mayrent, S., Ed.; Little Brown and Company: Boston, MA, USA; Toronto, ON, Canada, 1987; pp. 327–347. [Google Scholar]
- Mettler, D.; Tholen, D. G108—Guidelines for Estimating Uncertainty for Microbiological Counting Methods, 2007. Available online: http://www.a2la.org/guidance/MU_for_Micro_Labs.pdf (accessed on 26 April 2012).
- AOAC International. The AOAC Research Institute Performance Tested Methods Program Workshop: Validation of Rapid Microbiological Methods, 2009. Available online: http://www.aoac.org/testkits/PTM_MicroWorkshop_8-12-09.pdf (accessed on 26 April 2011).
- Bangs Laboratories, Inc. TechNote 204: Adsorption to Microspheres. Available online: http://www.bangslabs.com/sites/default/files/imce/docs/TechNote%20204%20Web.pdf (accessed on 5 November 2014).
- Bangs Laboratories, Inc. TechNote 201: Working with Microspheres, 2008. Available online: http://www.bangslabs.com/sites/default/files/imce/docs/TechNote%20201%20Web.pdf (accessed on 5 November 2014).
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cloutier, B.C.; Cloutier, A.K.; Alocilja, E.C. Optimization of Electrically Active Magnetic Nanoparticles as Accurate and Efficient Microbial Extraction Tools. Biosensors 2015, 5, 69-84. https://doi.org/10.3390/bios5010069
Cloutier BC, Cloutier AK, Alocilja EC. Optimization of Electrically Active Magnetic Nanoparticles as Accurate and Efficient Microbial Extraction Tools. Biosensors. 2015; 5(1):69-84. https://doi.org/10.3390/bios5010069
Chicago/Turabian StyleCloutier, Barbara C., Ashley K. Cloutier, and Evangelyn C. Alocilja. 2015. "Optimization of Electrically Active Magnetic Nanoparticles as Accurate and Efficient Microbial Extraction Tools" Biosensors 5, no. 1: 69-84. https://doi.org/10.3390/bios5010069
APA StyleCloutier, B. C., Cloutier, A. K., & Alocilja, E. C. (2015). Optimization of Electrically Active Magnetic Nanoparticles as Accurate and Efficient Microbial Extraction Tools. Biosensors, 5(1), 69-84. https://doi.org/10.3390/bios5010069





