Polymerization Parameters Influencing the QCM Response Characteristics of BSA MIP
Abstract
:1. Introduction

2. Experimental Section
2.1. Materials and Devices
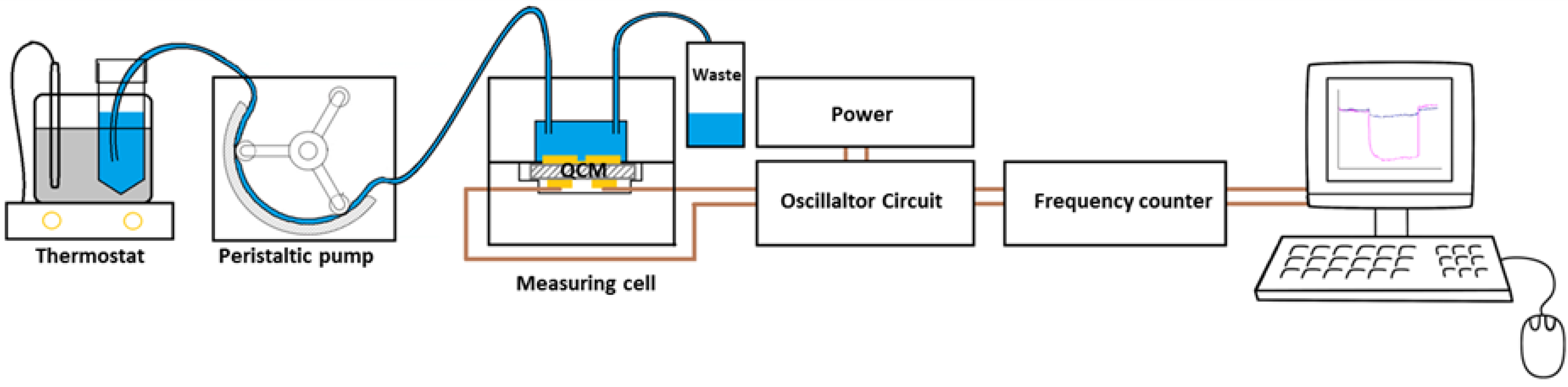
2.2. Experimental
3. Results and Discussion
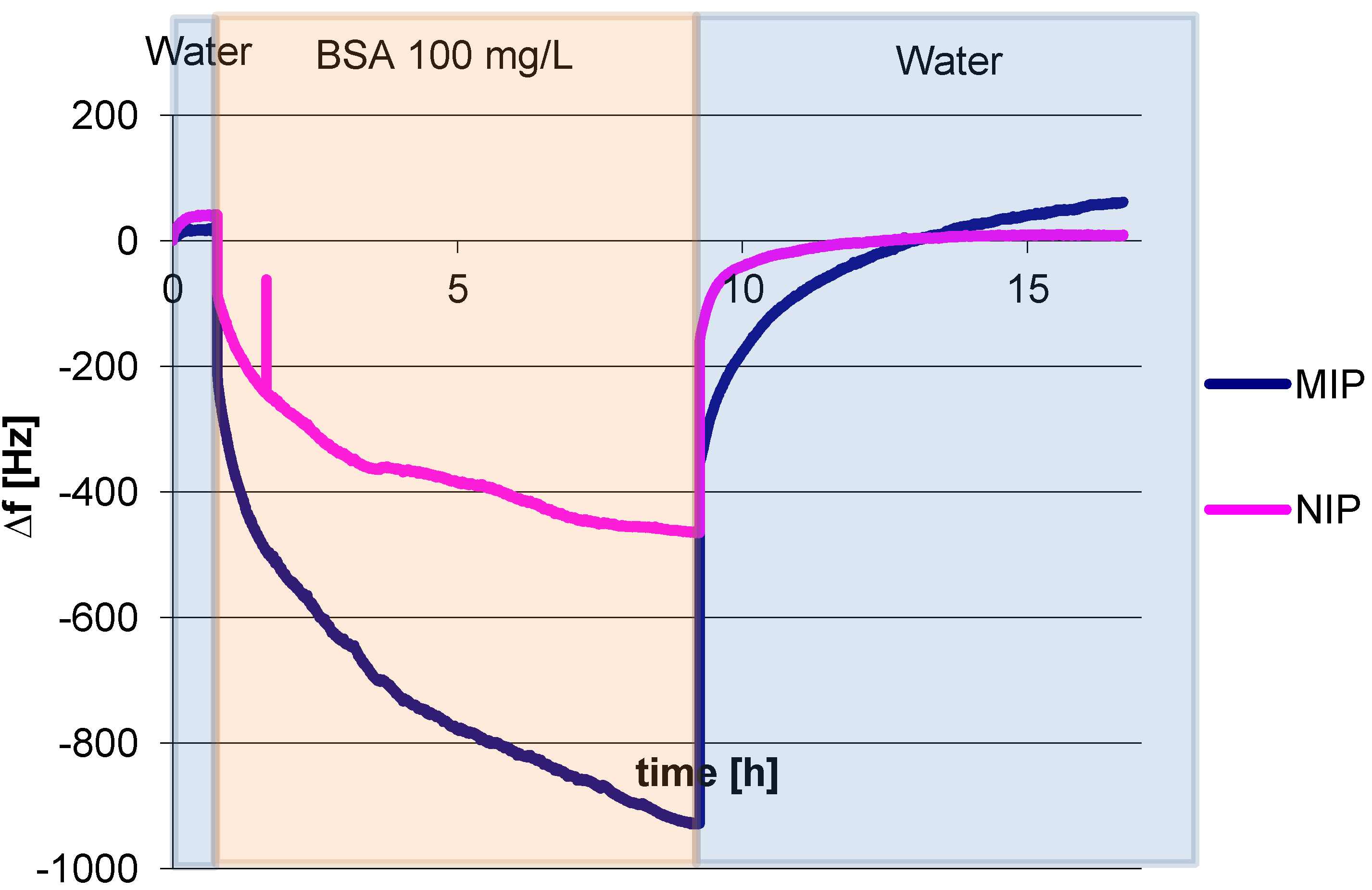
3.1. Polymer Optimization: Cross Linker
| Cross-linker (%) | 30% | 40% | 50% | 60% | 70% |
|---|---|---|---|---|---|
| mg | 15 | 15 | 25 | 30 | 35 |
| Monomers (mg) | 35 | 17.5 | 25 | 20 | 15 |

3.2. Polymer Optimization: Functional Monomer
| Name | cross linker | functional monomer 1 | functional monomer 2 | functional monomer ratio w/w | |
|---|---|---|---|---|---|
| ratio (w/w) | - | 70% | 30% | - | |
| 35 mg | |||||
| MA 1 | DHEBA | methacrylic acid | vinylpyrrolidone | 5:2 | |
| AA 1 | DHEBA | acrylic acid | vinylpyrrolidone | 5:2 | |
| AA 2 | DHEBA | acrylic acid | vinylpyrrolidone | 1:1 | |
| AA 3 | DHEBA | acrylic acid | vinylpyrrolidone | 2:3 | |
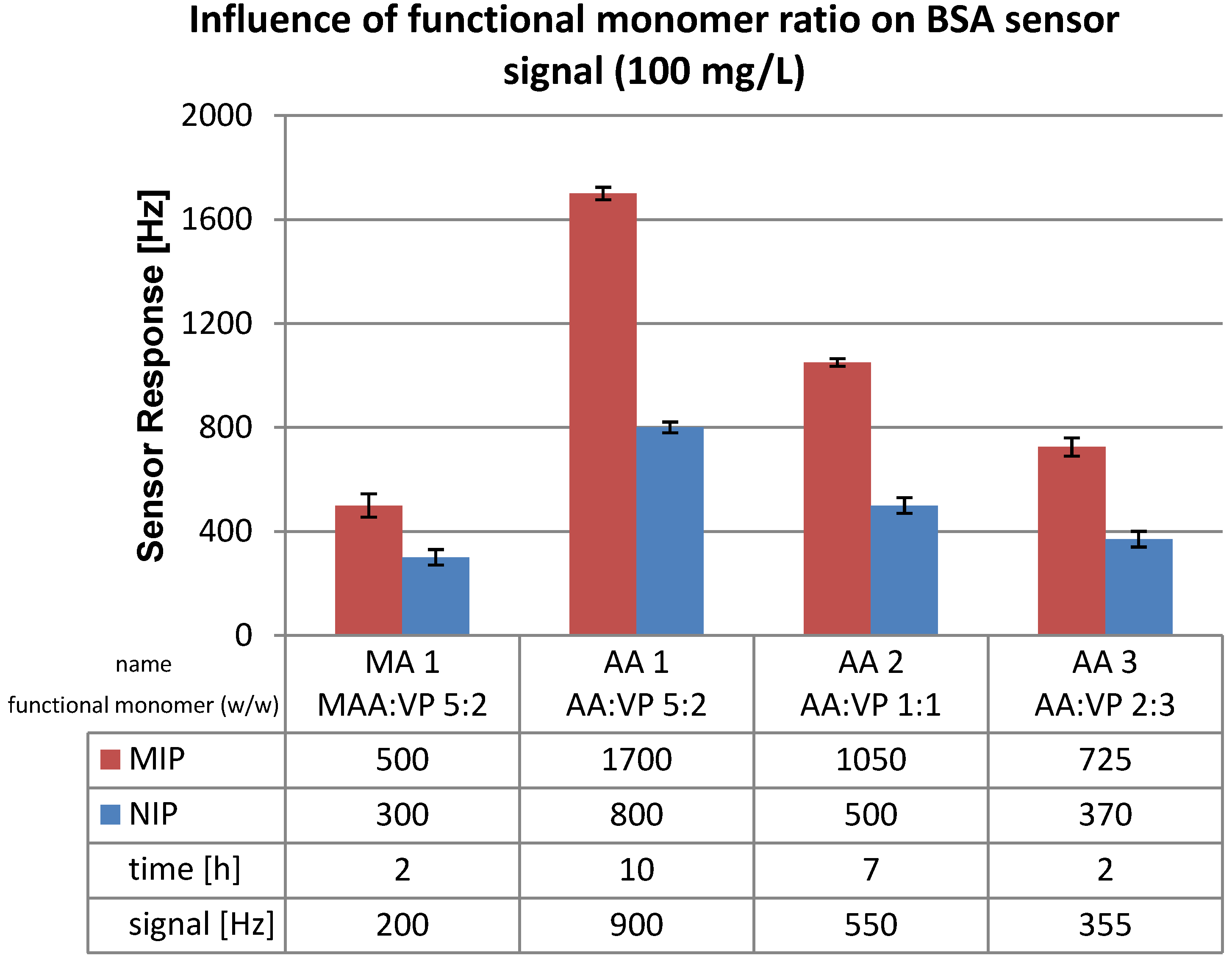
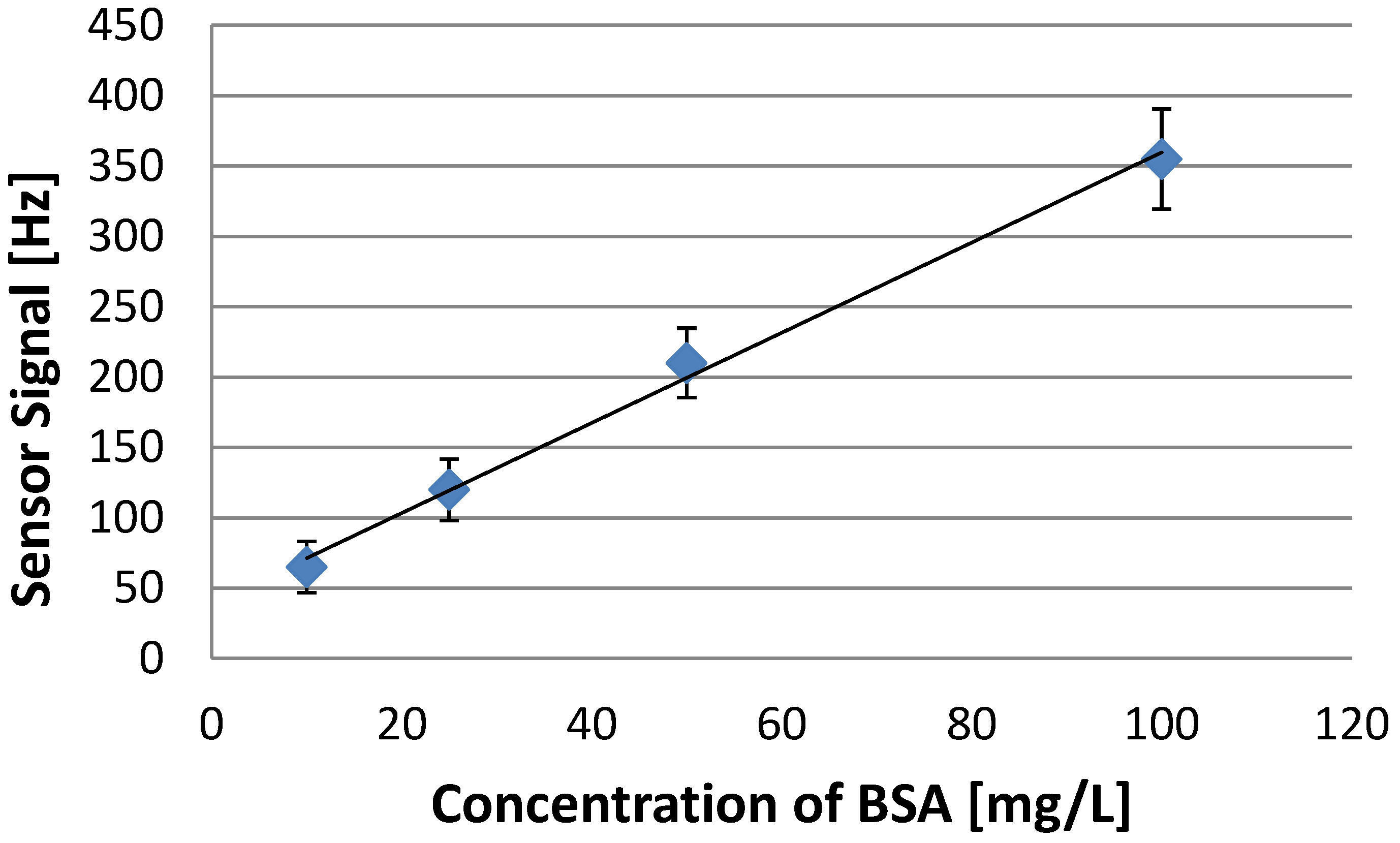
3.3. Polymerization Solvent
3.4. Polymer Morphology
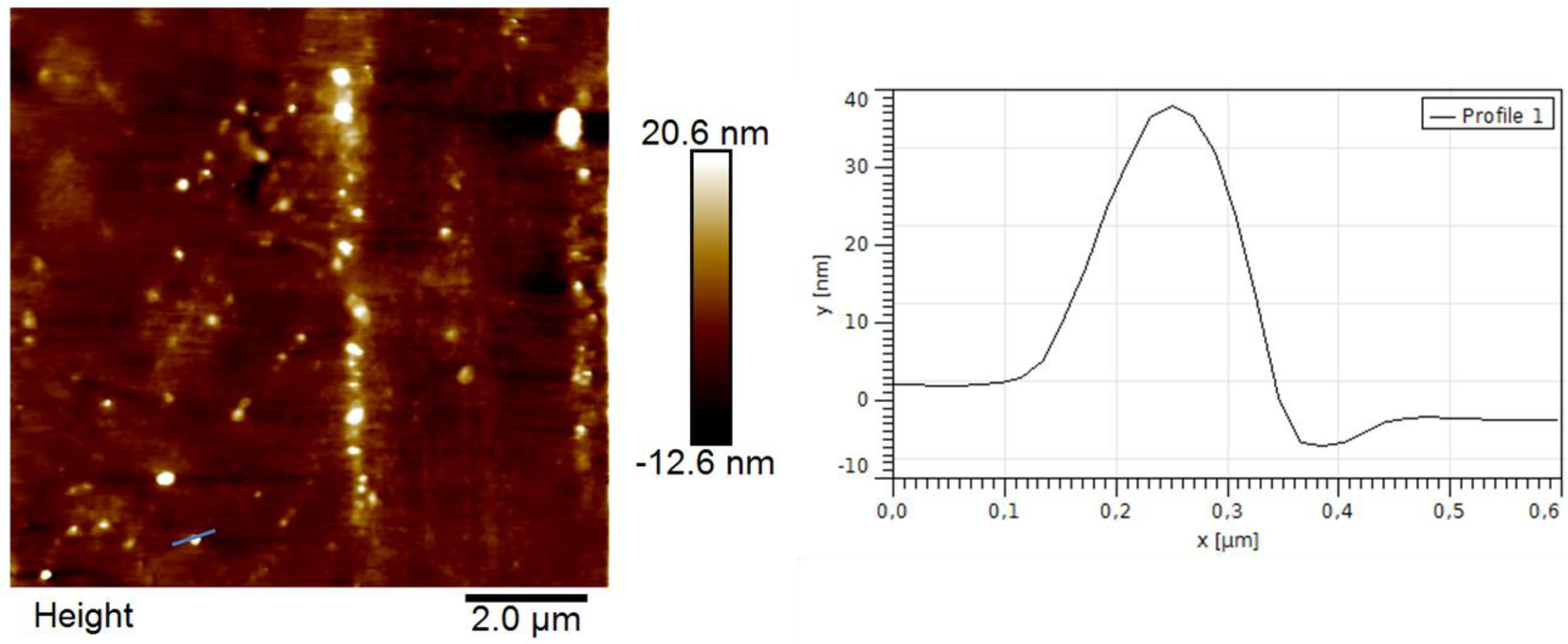
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kotova, K.; Hussain, M.; Mustafa, G.; Lieberzeit, P.A. MIP sensors on the way to biotech applications: Targeting selectivity. Sens. Actuator. B Chem. 2013, 189, 199–202. [Google Scholar] [CrossRef]
- Cakir, P.; Cutivet, A.; Resmini, M.; Bui, B.T.S.; Haupt, K. Protein-size molecularly imprinted polymer nanogels as synthetic antibodies, by localized polymerization with multi-initiators. Adv. Mater. 2013, 25, 1048–1051. [Google Scholar] [CrossRef]
- Sauerbrey, G. Verwendung Von Schwingquarzen zur wägung dünner Schichten und zur mikrowägung. Z Phys. 1959, 155, 206–222. [Google Scholar] [CrossRef]
- Nopper, D.; Lammershop, O.; Wulff, G.; Gauglitz, G. Amidine-based molecularly imprinted polymers—New sensitive elements for chiral chemosensors. Anal. Bioanal. Chem. 2003, 377, 608–613. [Google Scholar] [CrossRef]
- Alenus, J.; Ethirajan, A.; Horemans, F.; Weustenraed, A.; Csipai, P.; Gruber, J.; Peeters, M.; Cleij, T.J.; Wagner, P. Molecularly imprinted polymers as synthetic receptors for the QCM-D-based detection of l-nicotine in diluted saliva and urine samples. Anal. Bioanal. Chem. 2013, 405, 6479–6487. [Google Scholar] [CrossRef]
- Seidler, K.; Lieberzeit, P.A.; Dickert, F.L. Application of yeast imprinting in biotechnology and process control. Analyst 2009, 134, 361–366. [Google Scholar] [CrossRef]
- Lieberzeit, P.A.; Rehman, A.; Najafi, B.; Dickert, F.L. Real-life application of a QCM-based E-nose: Quantitative characterization different plant-degradation processes. Anal. Bioanal. Chem. 2008, 391, 2897–2903. [Google Scholar] [CrossRef]
- Turner, N.W.; Jeans, C.W.; Brain, K.R.; Allender, C.J.; Hlady, V.; Britt, D.W. From 3D to 2D: A review of the molecular imprinting of proteins. Biotechnol. Progr. 2006, 22, 1474–1489. [Google Scholar] [CrossRef]
- Mallik, S.; Plunkett, S.D.; Dhal, P.K.; Johnson, R.D.; Pack, D.; Shnek, D.; Arnold, F.H. Towards materials for the specific recognition and separation of proteins. New J. Chem. 1994, 18, 299–304. [Google Scholar]
- Ramstrom, O.; Ansell, R.J. Molecular imprinting technology: Challenges and prospects for the future. Chirality 1998, 10, 195–209. [Google Scholar] [CrossRef]
- Rothschild, M.A.; Oratz, M.; Schreiber, S.S. Serum-Albumin. Hepatology 1988, 8, 385–401. [Google Scholar] [CrossRef]
- Lieberzeit, P.A.; Rehman, A.; Yaqub, S.; Dickert, F.L. Nanostructured particles and layers for sensing contaminants in air and water. Nano 2008, 3, 205–208. [Google Scholar] [CrossRef]
- Yoshimatsu, K.; Reimhult, K.; Krozer, A.; Mosbach, K.; Sode, K.; Ye, L. Uniform molecularly imprinted microspheres and nanoparticles prepared by precipitation polymerization: The control of particle size suitable for different analytical applications. Anal. Chim. Acta 2007, 584, 112–121. [Google Scholar] [CrossRef]
- Vaidya, A.A.; Lele, B.S.; Kulkarni, M.G.; Mashelkar, R.A. Creating a macromolecular receptor by affinity imprinting. J. Appl. Polym. Sci. 2001, 81, 1075–1083. [Google Scholar] [CrossRef]
- Bossi, A.; Bonini, F.; Turner, A.P.F.; Piletsky, S.A. Molecularly imprinted polymers for the recognition of proteins: The state of the art. Biosens. Bioelectron. 2007, 22, 1131–1137. [Google Scholar]
- Muratsugu, M.; Ohta, F.; Miya, Y.; Hosokawa, T.; Kurosawa, S.; Kamo, N.; Ikeda, H. Quartz-crystal microbalance for the detection of microgram quantities of human serum-albumin—Relationship between the frequency change and the mass of protein adsorbed. Anal Chem 1993, 65, 2933–2937. [Google Scholar] [CrossRef]
- Poma, A.; Turner, A.P.F.; Piletsky, S.A. Advances in the manufacture of MIP nanoparticles. Trends Biotechnol. 2010, 28, 629–637. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Phan, N.V.H.; Sussitz, H.F.; Lieberzeit, P.A. Polymerization Parameters Influencing the QCM Response Characteristics of BSA MIP. Biosensors 2014, 4, 161-171. https://doi.org/10.3390/bios4020161
Phan NVH, Sussitz HF, Lieberzeit PA. Polymerization Parameters Influencing the QCM Response Characteristics of BSA MIP. Biosensors. 2014; 4(2):161-171. https://doi.org/10.3390/bios4020161
Chicago/Turabian StylePhan, Nam V. H., Hermann F. Sussitz, and Peter A. Lieberzeit. 2014. "Polymerization Parameters Influencing the QCM Response Characteristics of BSA MIP" Biosensors 4, no. 2: 161-171. https://doi.org/10.3390/bios4020161
APA StylePhan, N. V. H., Sussitz, H. F., & Lieberzeit, P. A. (2014). Polymerization Parameters Influencing the QCM Response Characteristics of BSA MIP. Biosensors, 4(2), 161-171. https://doi.org/10.3390/bios4020161





