Micropatterning of 3D Microenvironments for Living Biosensor Applications
Abstract
:1. Introduction
2. Experimental Section
2.1. Fabrication of the Surface Patterning Tool (SPT)
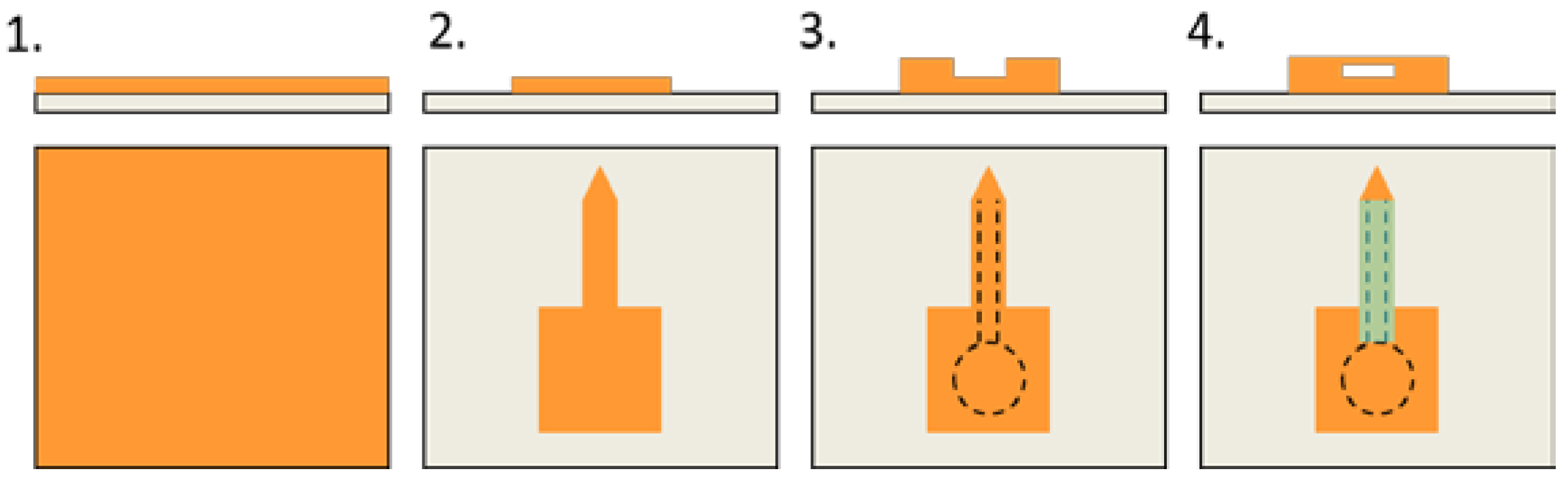

2.2. Micro-Printer Modification
2.3. Hydrogel Matrix Formulation and Printing Matrix Preparation
2.4. Cell Culture
2.5. Cell Printing
2.6. Examination of 3D Growth in Cell Deposits
2.7. Development of a Living ROS Sensor
3. Results and Discussion
3.1. Hydrogel Matrix Formulation and Optimization
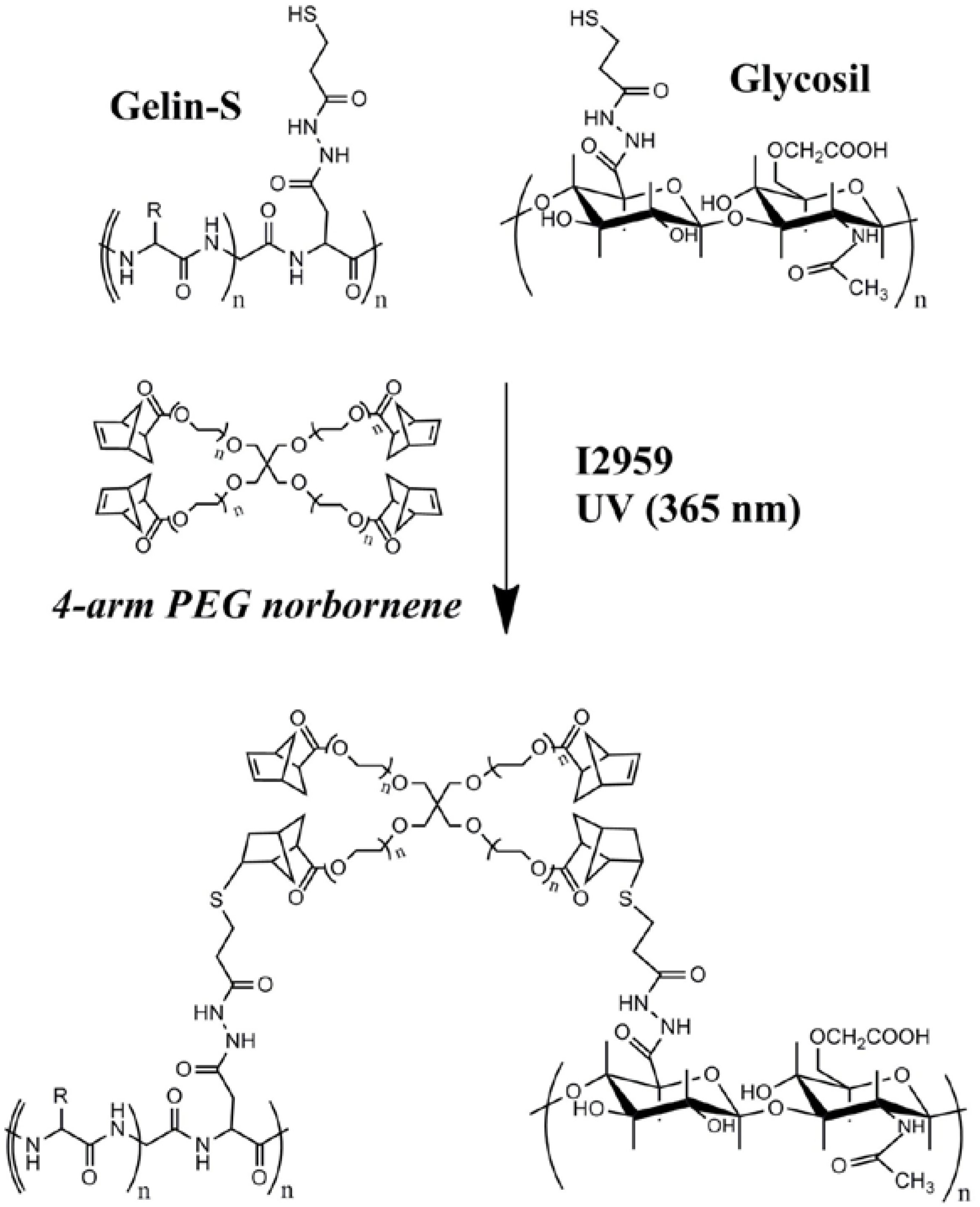
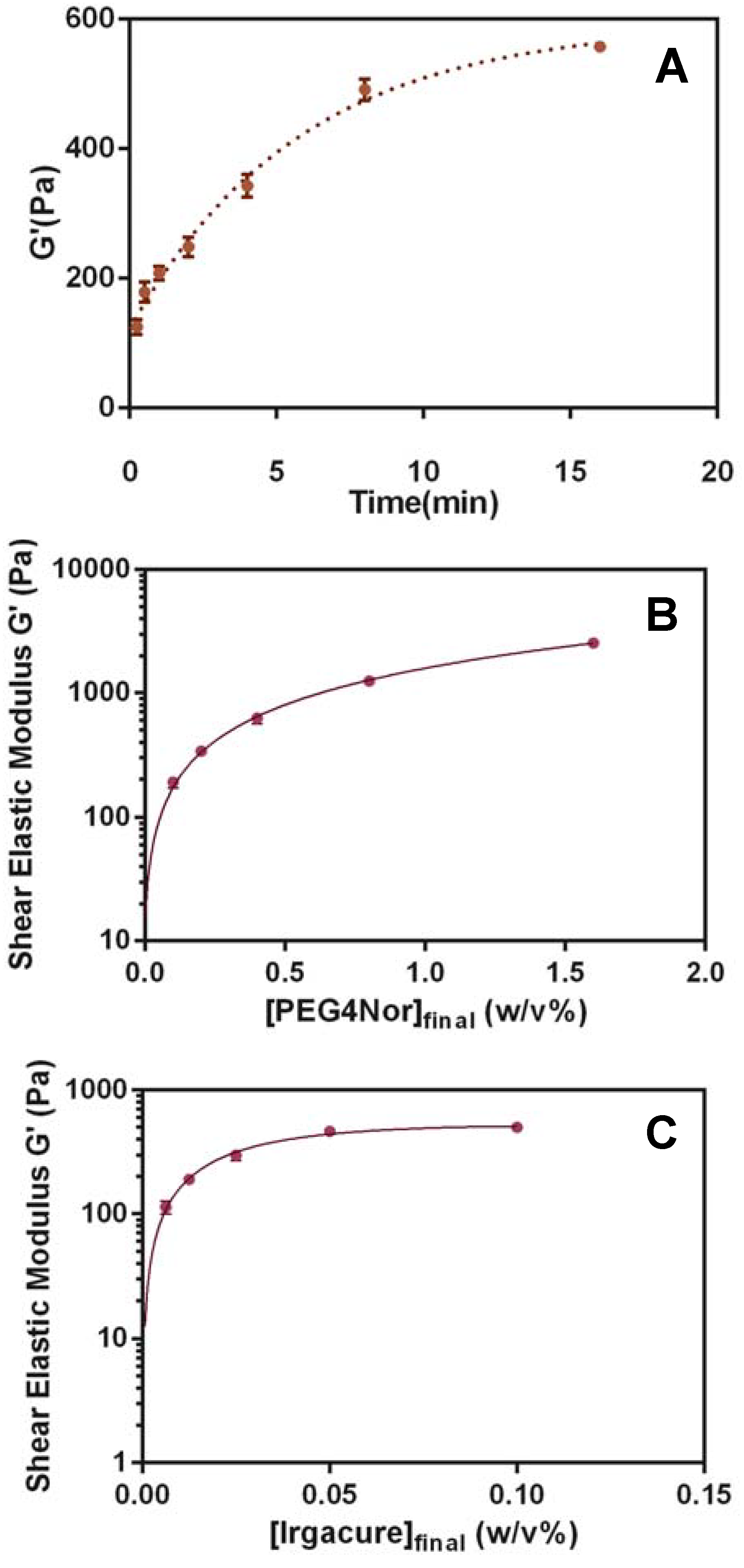
3.2. Biocompatibility of the HyStem-C/PEG Norbornene Hydrogels
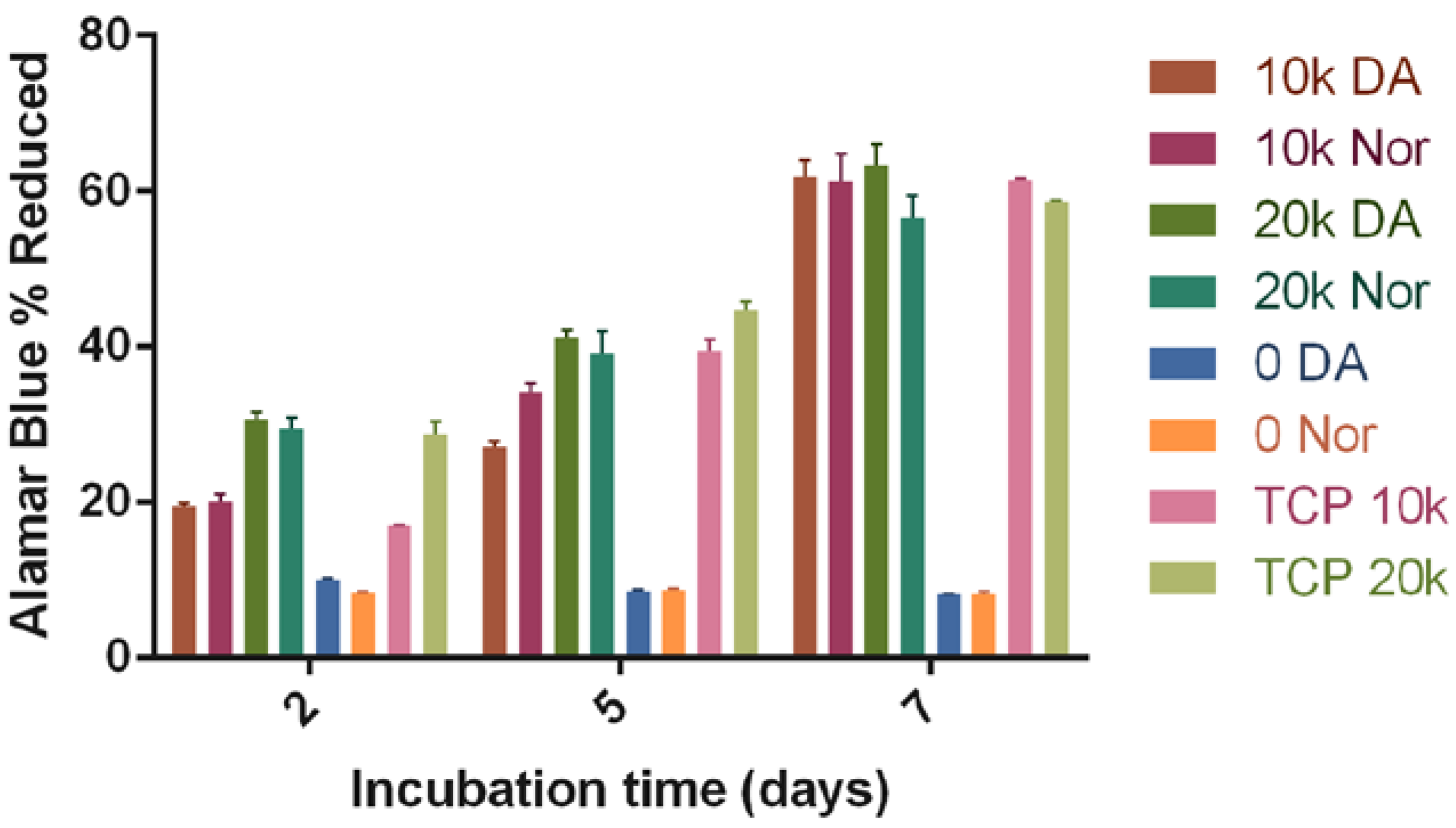
3.3. Cell Proliferation after Printing
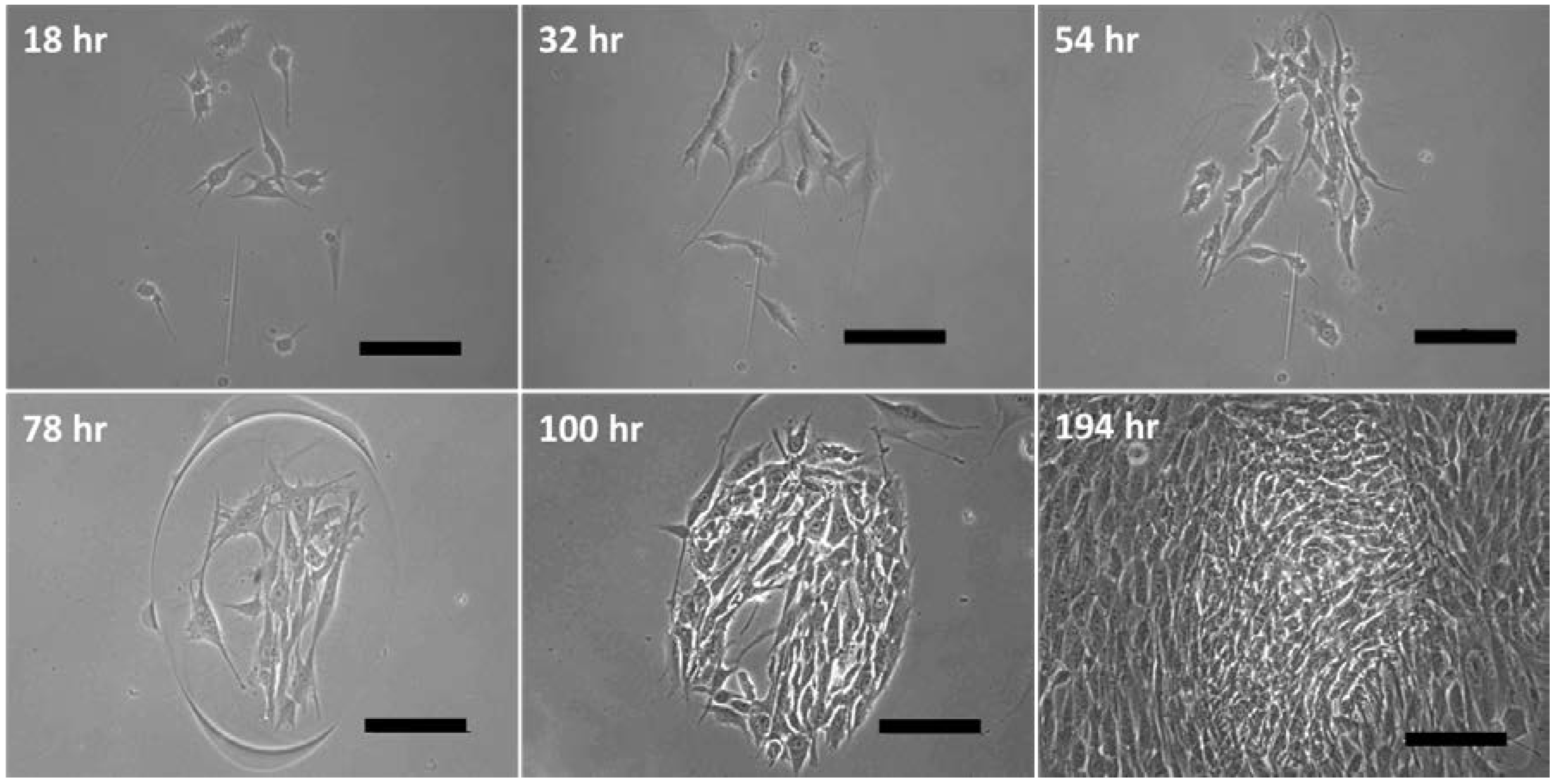
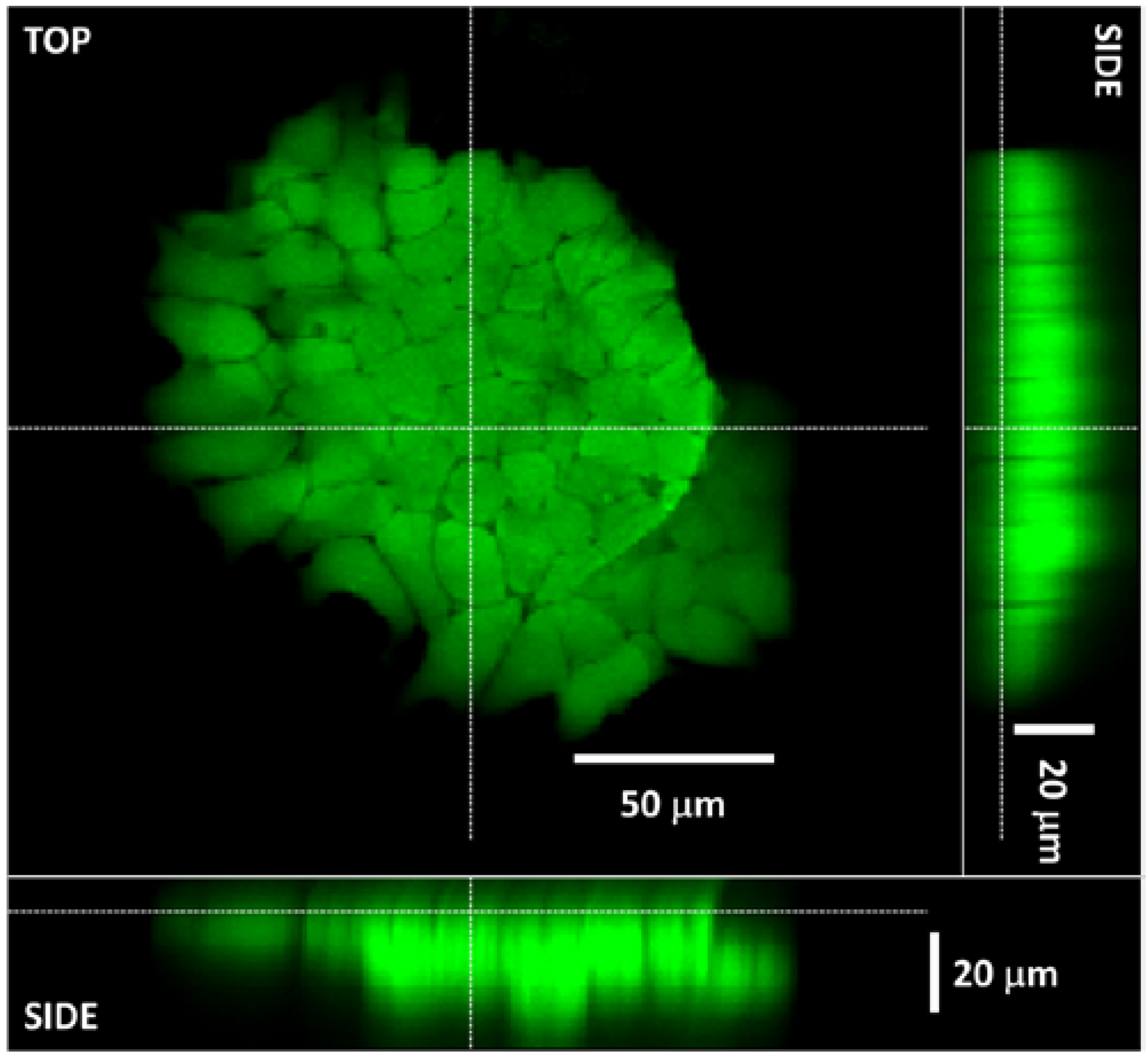
3.4. The 3D Nature of the Printed Cell Matrix
3.5. Verification of Living ROS Sensor Function

4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mironov, V.; Reis, N.; Derby, B. Review: Bioprinting: A beginning. Tissue Eng. 2006, 12, 631–634. [Google Scholar] [CrossRef]
- Jakab, K.; Norotte, C.; Marga, F.; Murphy, K.; Vunjak-Novakovic, G.; Forgacs, G. Tissue engineering by self-assembly and bio-printing of living cells. Biofabrication 2010, 2. [Google Scholar] [CrossRef]
- Xu, F.; Celli, J.; Rizvi, I.; Moon, S.; Hasan, T.; Demirci, U. A three-dimensional in vitro ovarian cancer coculture model using a high-throughput cell patterning platform. Biotechnol. J. 2011, 6, 204–212. [Google Scholar] [CrossRef]
- Rodriguez-Devora, J.I.; Zhang, B.; Reyna, D.; Shi, Z.D.; Xu, T. High throughput miniature drug-screening platform using bioprinting technology. Biofabrication 2012, 4. [Google Scholar] [CrossRef]
- Derby, B. Bioprinting: Inkjet printing proteins and hybrid cell-containing materials and structures. J. Mater. Chem. 2008, 18, 5717–5721. [Google Scholar] [CrossRef]
- Kubisch, R.; Bohrn, U.; Fleischer, M.; Stutz, E. Cell-based sensor system using l6 cells for broad band continuous pollutant monitoring in aquatic environments. Sensors 2012, 12, 3370–3393. [Google Scholar] [CrossRef]
- Arip, M.N.; Heng, L.Y.; Ahmad, M.; Ujang, S. A cell-based potentiometric biosensor using the fungus Lentinus sajor-caju for permethrin determination in treated wood. Talanta 2013, 116, 776–781. [Google Scholar] [CrossRef]
- Jiang, D.; Ji, J.; An, L.; Sun, X.; Zhang, Y.; Zhang, G.; Tang, L. Mast cell-based electrochemical biosensor for quantification of the major shrimp allergen Pen a 1 (tropomyosin). Biosens. Bioelectron. 2013, 50, 150–156. [Google Scholar] [CrossRef]
- Shing, W.L.; Heng, L.Y.; Surif, S. Performance of a cyanobacteria whole cell-based fluorescence biosensor for heavy metal and pesticide detection. Sensors 2013, 13, 6394–6404. [Google Scholar] [CrossRef]
- Cortes-Salazar, F.; Beggah, S.; van der Meer, J.R.; Girault, H.H. Electrochemical As(III) whole-cell based biochip sensor. Biosens. Bioelectron. 2013, 47, 237–242. [Google Scholar] [CrossRef]
- Belkin, S. Microbial whole-cell sensing systems of environmental pollutants. Curr. Opin. Microbiol. 2003, 6, 206–212. [Google Scholar] [CrossRef]
- Hensley, K.; Robinson, K.A.; Gabbita, S.P.; Salsman, S.; Floyd, R.A. Reactive oxygen species, cell signaling, and cell injury. Free Radic. Biol. Med. 2000, 28, 1456–1462. [Google Scholar] [CrossRef]
- Gutteridge, J.M.; Mitchell, J. Redox imbalance in the critically ill. Br. Med. Bull. 1999, 55, 49–75. [Google Scholar] [CrossRef]
- Arias-Barreiro, C.R.; Okazaki, K.; Koutsaftis, A.; Inayat-Hussain, S.H.; Tani, A.; Katsuhara, M.; Kimbara, K.; Mori, I.C. A bacterial biosensor for oxidative stress using the constitutively expressed redox-sensitive protein rogfp2. Sensors 2010, 10, 6290–6306. [Google Scholar] [CrossRef]
- Stadtman, E.R.; Berlett, B.S. Reactive oxygen-mediated protein oxidation in aging and disease. Drug Metab. Rev. 1998, 30, 225–243. [Google Scholar] [CrossRef]
- Cui, X.; Boland, T.; D’Lima, D.D.; Lotz, M.K. Thermal inkjet printing in tissue engineering and regenerative medicine. Recent Pat. Drug Deliv. Formul. 2012, 6, 149–155. [Google Scholar] [CrossRef]
- Xu, T.; Zhao, W.; Zhu, J.M.; Albanna, M.Z.; Yoo, J.J.; Atala, A. Complex heterogeneous tissue constructs containing multiple cell types prepared by inkjet printing technology. Biomaterials 2013, 34, 130–139. [Google Scholar] [CrossRef]
- Arai, K.; Iwanaga, S.; Toda, H.; Genci, C.; Nishiyama, Y.; Nakamura, M. Three-dimensional inkjet biofabrication based on designed images. Biofabrication 2011, 3. [Google Scholar] [CrossRef]
- Merrin, J.; Leibler, S.; Chuang, J.S. Printing multistrain bacterial patterns with a piezoelectric inkjet printer. PLoS One 2007, 2. [Google Scholar] [CrossRef]
- Saunders, R.E.; Gough, J.E.; Derby, B. Delivery of human fibroblast cells by piezoelectric drop-on-demand inkjet printing. Biomaterials 2008, 29, 193–203. [Google Scholar] [CrossRef]
- Barron, J.A.; Wu, P.; Ladouceur, H.D.; Ringeisen, B.R. Biological laser printing: A novel technique for creating heterogeneous 3-dimensional cell patterns. Biomed. Microdevices 2004, 6, 139–147. [Google Scholar] [CrossRef]
- Guillotin, B.; Souquet, A.; Catros, S.; Duocastella, M.; Pippenger, B.; Bellance, S.; Bareille, R.; Remy, M.; Bordenave, L.; Amedee, J.; et al. Laser assisted bioprinting of engineered tissue with high cell density and microscale organization. Biomaterials 2010, 31, 7250–7256. [Google Scholar] [CrossRef]
- Hopp, B.; Smausz, T.; Kresz, N.; Barna, N.; Bor, Z.; Kolozsvari, L.; Chrisey, D.B.; Szabo, A.; Nogradi, A. Survival and proliferative ability of various living cell types after laser-induced forward transfer. Tissue Eng. 2005, 11, 1817–1823. [Google Scholar] [CrossRef]
- Schiele, N.R.; Corr, D.T.; Huang, Y.; Raof, N.A.; Xie, Y.; Chrisey, D.B. Laser-based direct-write techniques for cell printing. Biofabrication 2010, 2. [Google Scholar] [CrossRef]
- Chang, R.; Nam, J.; Sun, W. Direct cell writing of 3D microorgan for in vitro pharmacokinetic model. Tissue Eng. C Meth. 2008, 14, 157–166. [Google Scholar] [CrossRef]
- Norotte, C.; Marga, F.S.; Niklason, L.E.; Forgacs, G. Scaffold-free vascular tissue engineering using bioprinting. Biomaterials 2009, 30, 5910–5917. [Google Scholar] [CrossRef]
- Hynes, W.F.; Gracias, A.; Fahrenkopf, N.M.; Raof, N.A.; Raja, W.K.; Lee, K.; Xie, Y.; Bergkvist, M.; Cady, N.C. Direct cell printing with microfabricated quill-pen cantilevers. MRS Proceedings 2009, 1235. [Google Scholar] [CrossRef]
- Shu, X.Z.; Ghosh, K.; Liu, Y.; Palumbo, F.S.; Luo, Y.; Clark, R.A.; Prestwich, G.D. Attachment and spreading of fibroblasts on an RGD peptide-modified injectable hyaluronan hydrogel. J. Biomed. Mater. Res. A 2004, 68, 365–375. [Google Scholar]
- Vanderhooft, J.L.; Alcoutlabi, M.; Magda, J.J.; Prestwich, G.D. Rheological properties of cross-linked hyaluronan-gelatin hydrogels for tissue engineering. Macromol. Biosci. 2009, 9, 20–28. [Google Scholar] [CrossRef]
- Chen, X.; Thibeault, S.L. Biocompatibility of a synthetic extracellular matrix on immortalized vocal fold fibroblasts in 3-D culture. Acta Biomater. 2010, 6, 2940–2948. [Google Scholar] [CrossRef]
- Hanjaya-Putra, D.; Yee, J.; Ceci, D.; Truitt, R.; Yee, D.; Gerecht, S. Vascular endothelial growth factor and substrate mechanics regulate in vitro tubulogenesis of endothelial progenitor cells. J. Cell. Mol. Med. 2010, 14, 2436–2447. [Google Scholar] [CrossRef]
- Mohand-Kaci, F.; Assoul, N.; Martelly, I.; Allaire, E.; Zidi, M. Optimized hyaluronic acid-hydrogel design and culture conditions for preservation of mesenchymal stem cell properties. Tissue Eng. C Meth. 2013, 19, 288–298. [Google Scholar] [CrossRef]
- Zarembinski, T.I.; Doty, N.J.; Erickson, I.E.; Srinivas, R.; Wirostko, B.M.; Tew, W.P. Thiolated hyaluronan-based hydrogels crosslinked using oxidized glutathione: An injectable matrix designed for ophthalmic applications. Acta Biomater. 2014, 10, 94–103. [Google Scholar] [CrossRef]
- Gerecht, S.; Burdick, J.A.; Ferreira, L.S.; Townsend, S.A.; Langer, R.; Vunjak-Novakovic, G. Hyaluronic acid hydrogel for controlled self-renewal and differentiation of human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 11298–11303. [Google Scholar]
- Fairbanks, B.D.; Schwartz, M.P.; Halevi, A.E.; Nuttelman, C.R.; Bowman, C.N.; Anseth, K.S. A versatile synthetic extracellular matrix mimic via thiol-norbornene photopolymerization. Adv. Mater. 2009, 21, 5005–5010. [Google Scholar] [CrossRef]
- Gramlich, W.M.; Kim, I.L.; Burdick, J.A. Synthesis and orthogonal photopatterning of hyaluronic acid hydrogels with thiol-norbornene chemistry. Biomaterials 2013, 34, 9803–9811. [Google Scholar] [CrossRef]
- Zhang, J.; Tan, K.L.; Hong, G.D.; Yang, L.J.; Gong, H.Q. Polymerization optimization of SU-8 photoresist and its applications in microfluidic systems and MEMS. J. Micromech. Microeng. 2001, 11. [Google Scholar] [CrossRef]
- Zuk, P.A.; Zhu, M.; Mizuno, H.; Huang, J.; Futrell, J.W.; Katz, A.J.; Benhaim, P.; Lorenz, H.P.; Hedrick, M.H. Multilineage cells from human adipose tissue: Implications for cell-based therapies. Tissue Eng. 2001, 7, 211–228. [Google Scholar] [CrossRef]
- Robertson, E.; Bradley, A.; Kuehn, M.; Evans, M. Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vector. Nature 1986, 323, 445–448. [Google Scholar] [CrossRef]
- Keller, G.; Kennedy, M.; Papayannopoulou, T.; Wiles, M.V. Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol. Cell. Biol. 1993, 13, 473–486. [Google Scholar]
- Zonca, M.R., Jr.; Yune, P.S.; Heldt, C.L.; Belfort, G.; Xie, Y. High-throughput screening of substrate chemistry for embryonic stem cell attachment, expansion, and maintaining pluripotency. Macromol. Biosci. 2013, 13, 177–190. [Google Scholar] [CrossRef]
- Melillo, A.A.; Bakshi, C.S.; Melendez, J.A. Francisella tularensis antioxidants harness reactive oxygen species to restrict macrophage signaling and cytokine production. J. Biol. Chem. 2010, 285, 27553–27560. [Google Scholar] [CrossRef]
- Hoyle, C.E.; Bowman, C.N. Thiol-ene click chemistry. Angew. Chem. Int. Ed. Engl. 2010, 49, 1540–1573. [Google Scholar] [CrossRef]
- Schaffler, A.; Buchler, C. Concise review: Adipose tissue-derived stromal cells—Basic and clinical implications for novel cell-based therapies. Stem Cells 2007, 25, 818–827. [Google Scholar] [CrossRef]
- Caplan, A.I. Adult mesenchymal stem cells for tissue engineering versus regenerative medicine. J. Cell. Physiol. 2007, 213, 341–347. [Google Scholar] [CrossRef]
- McKee, C.T.; Last, J.A.; Russell, P.; Murphy, C.J. Indentation versus tensile measurements of young’s modulus for soft biological tissues. Tissue Eng. B Rev. 2011, 17, 155–164. [Google Scholar] [CrossRef]
- Takahashi, A.; Ohtani, N.; Yamakoshi, K.; Iida, S.; Tahara, H.; Nakayama, K.; Nakayama, K.I.; Ide, T.; Saya, H.; Hara, E. Mitogenic signalling and the p16iNK4a-Rb pathway cooperate to enforce irreversible cellular senescence. Nat. Cell. Biol. 2006, 8, 1291–1297. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hynes, W.F.; Doty, N.J.; Zarembinski, T.I.; Schwartz, M.P.; Toepke, M.W.; Murphy, W.L.; Atzet, S.K.; Clark, R.; Melendez, J.A.; Cady, N.C. Micropatterning of 3D Microenvironments for Living Biosensor Applications. Biosensors 2014, 4, 28-44. https://doi.org/10.3390/bios4010028
Hynes WF, Doty NJ, Zarembinski TI, Schwartz MP, Toepke MW, Murphy WL, Atzet SK, Clark R, Melendez JA, Cady NC. Micropatterning of 3D Microenvironments for Living Biosensor Applications. Biosensors. 2014; 4(1):28-44. https://doi.org/10.3390/bios4010028
Chicago/Turabian StyleHynes, William F., Nate J. Doty, Thomas I. Zarembinski, Michael P. Schwartz, Michael W. Toepke, William L. Murphy, Sarah K. Atzet, Ryan Clark, J. Andres Melendez, and Nathaniel C. Cady. 2014. "Micropatterning of 3D Microenvironments for Living Biosensor Applications" Biosensors 4, no. 1: 28-44. https://doi.org/10.3390/bios4010028
APA StyleHynes, W. F., Doty, N. J., Zarembinski, T. I., Schwartz, M. P., Toepke, M. W., Murphy, W. L., Atzet, S. K., Clark, R., Melendez, J. A., & Cady, N. C. (2014). Micropatterning of 3D Microenvironments for Living Biosensor Applications. Biosensors, 4(1), 28-44. https://doi.org/10.3390/bios4010028



