Development of a Fluorescence Resonance Energy Transfer (FRET)-Based DNA Biosensor for Detection of Synthetic Oligonucleotide of Ganoderma boninense
Abstract
:1. Introduction
2. Experimental Section
2.1. Material
- Probe DNA-amine modified single stranded DNA (ssDNA) (20-mer):
- 5′-/NH2C12/TGG GTT GTA GCT GGC CTT CC-3′
- Complementary target DNA (35-mer):
- 5′-GCT AGT CAA GGT AAC GGA AGG CCA GCT ACA ACC CA-3′
- Non-complementary DNA (35-mer):
- 5′-GTA AGG TGC TTG AAT TCG TTA GGC TTG GTT TCG AT-3′
- Reporter probe (15-mer):
- 5′-GTT ACC TTG ACT AGC/Cy5/-3′
2.2. Method
2.2.1. Preparation of CdSe QD
2.2.2. Preparation of Water Soluble CdSe QD
2.2.3. Attachment of CdSe QD with ssDNA (CdSe QD-ssDNA Conjugate)
2.2.4. Hybridization
2.2.5. Fluorescence Emission
2.2.6. Transmission Electron Microscopy (TEM)
3. Results and Discussion
3.1. Water Soluble CdSe QD
3.2. Activation of COOH Functional Group on CdSe QD with EDC and NHS
3.3. Sensing Characterization Based on Fluorescence Emission
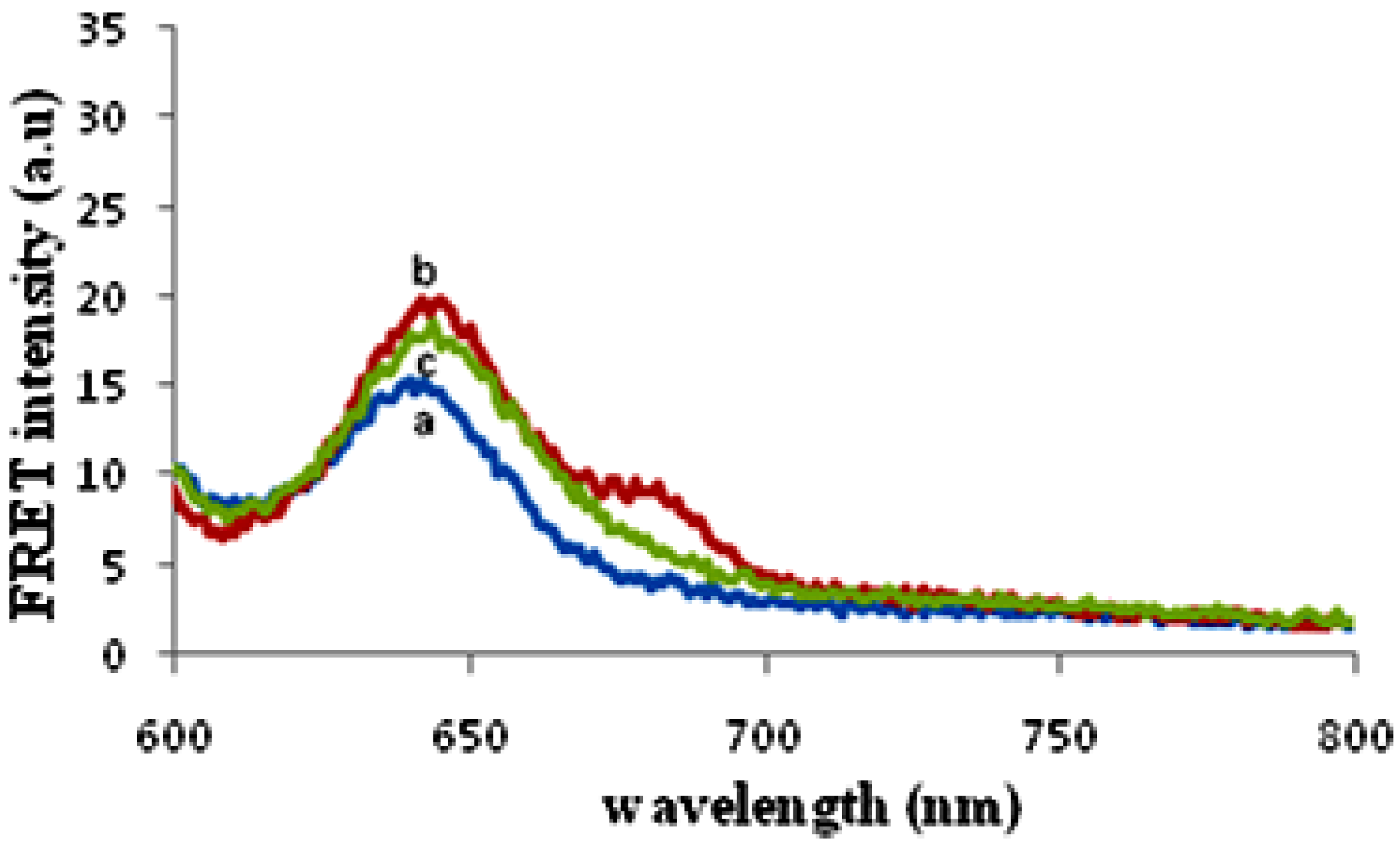
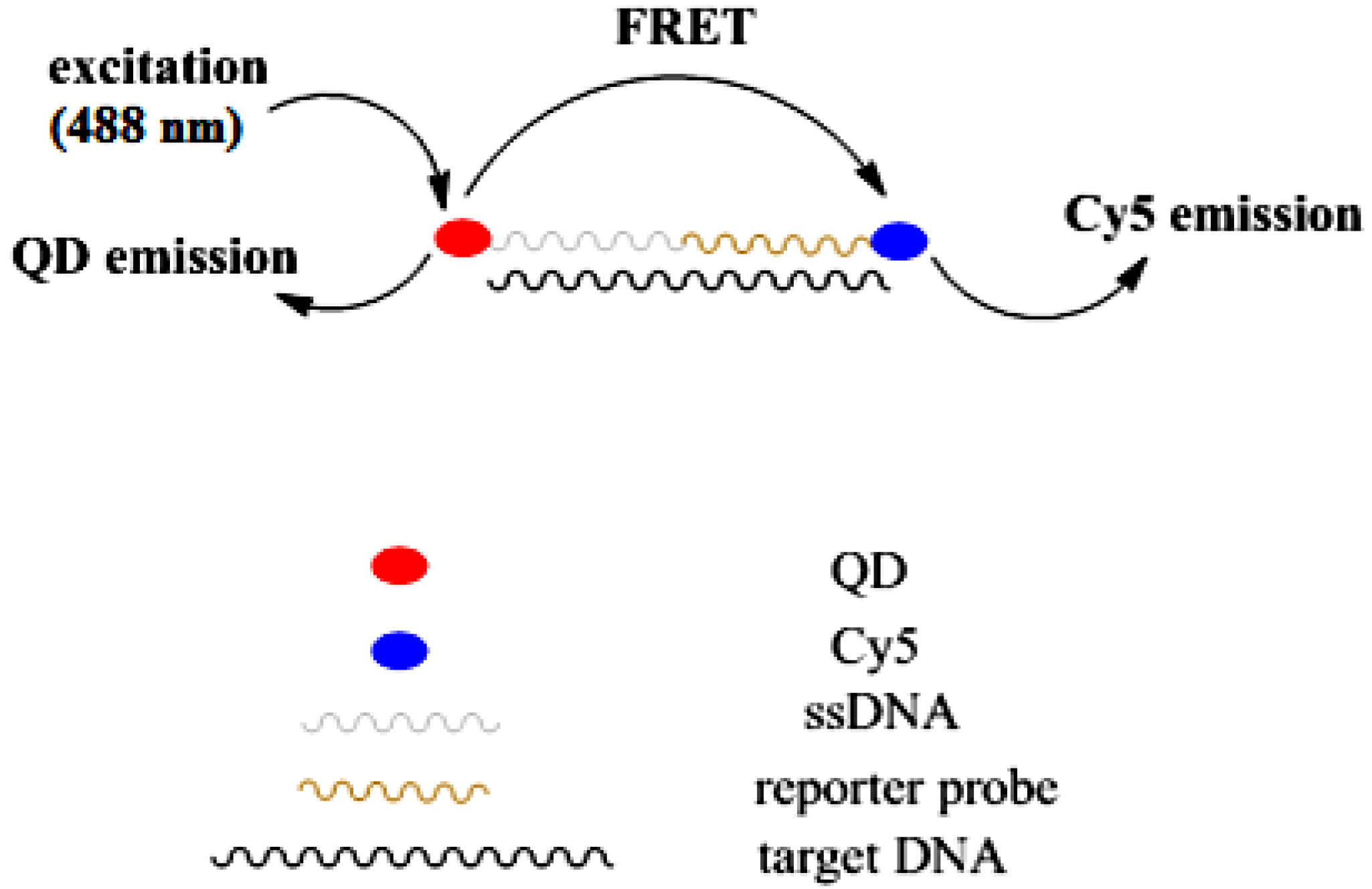
3.4. Factors Affecting the Hybridization Signal Intensity
3.5. TEM Characterization
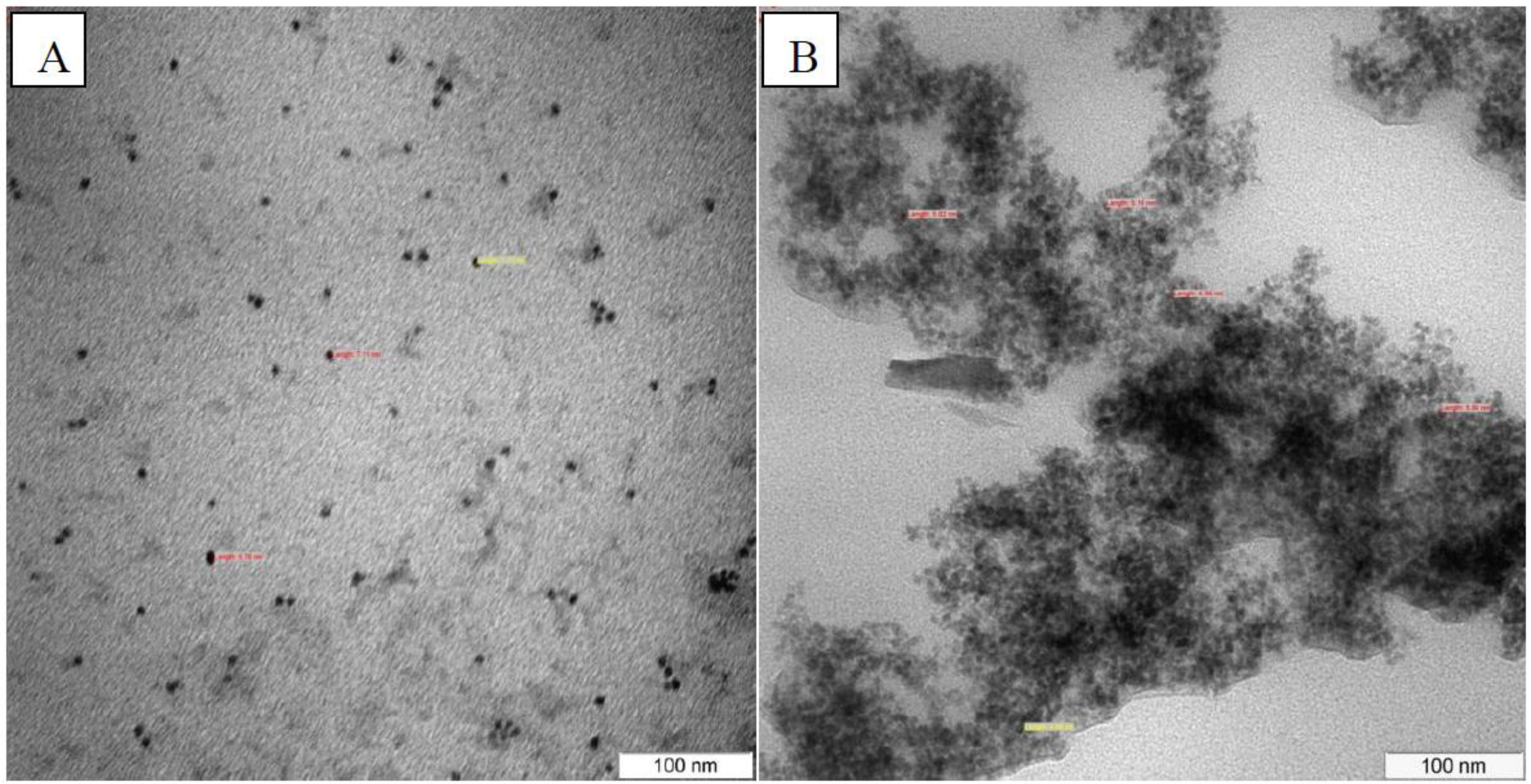
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Panchal, G.; Bridge, P.D. Following basal stem rot in young oil palm plantings. Mycopathologia 2005, 159, 123–127. [Google Scholar] [CrossRef]
- Breton, F.; Hasan, Y.; Hariadi, L.Z.; de Franqueville, H. Characterization of parameters for the development of an early screening test for basal stem rot tolerance in oil palm progenies. J. Oil Palm Res. 2006, 2006, 24–36. [Google Scholar]
- Smith, A.M.; Nie, S. Chemical analysis and cellular imaging with quantum dots. Analyst 2004, 129, 672–677. [Google Scholar] [CrossRef]
- Sutherland, A.J. Quantum dots as luminescence probes in biological systems. Solid State Mater. Sci. 2002, 6, 365–370. [Google Scholar] [CrossRef]
- Sharma, H.; Sharma, S.N.; Kumar, U.; Singh, V.N.; Mehta, B.R.; Singh, G.; Shivaprasad, S.M.; Kakkar, R. Formation of water-soluble and biocompatibe TOPO-capped CdSe quantum dots with efficient photoluminescence. J. Mater. Sci. Mater. Med. 2008, 20, 123–130. [Google Scholar]
- Algar, W.R.; Krull, U.J. Quantum dots as donors in fluorescence resonance energy transfer for the bioanalysis of nucleic acids, proteins and other biological molecules. Anal. Bioanal. Chem. 2008, 391, 1609–1618. [Google Scholar] [CrossRef]
- Aldana, J.; Lavelle, N.; Wang, Y.; Peng, X. Size-dependent dissociation pH of thiolate ligands from cadmium chalcogenide nanocrystals. J. Am. Chem. Soc. 2005, 127, 2496–2504. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Yeh, H.C.; Kuroki, M.T.; Wang, T.H. Single-quantum-dot-based DNA nanosensor. Nat. Mater. 2005, 4, 826–831. [Google Scholar] [CrossRef]
- Guilbault, G.G. Practical Fluorescence; Taylor & Francis Publisher: New York, NY, USA, 1990; pp. 28–29. [Google Scholar]
- Peng, H.; Zhang, L.; Kjallman, T.H.M.; Soeller, C.; Travas-Sejdic, J. DNA hybridization detection with blue luminescent quantum dots and dye-labelled single-stranded DNA. Am. Chem. Soc. 2007, 129, 1–2. [Google Scholar]
- Technical Note: An Introduction to Fluorescence Measurement. Available online: http://www.turnerdesigns.com/t2/doc/appnotes/998-0050.pdf (accessed on 25 September 2012).
- Christian, G.D. Analytical Chemistry; John Wiley and Sons: New York, NY, USA, 2004; p. 113. [Google Scholar]
Appendix
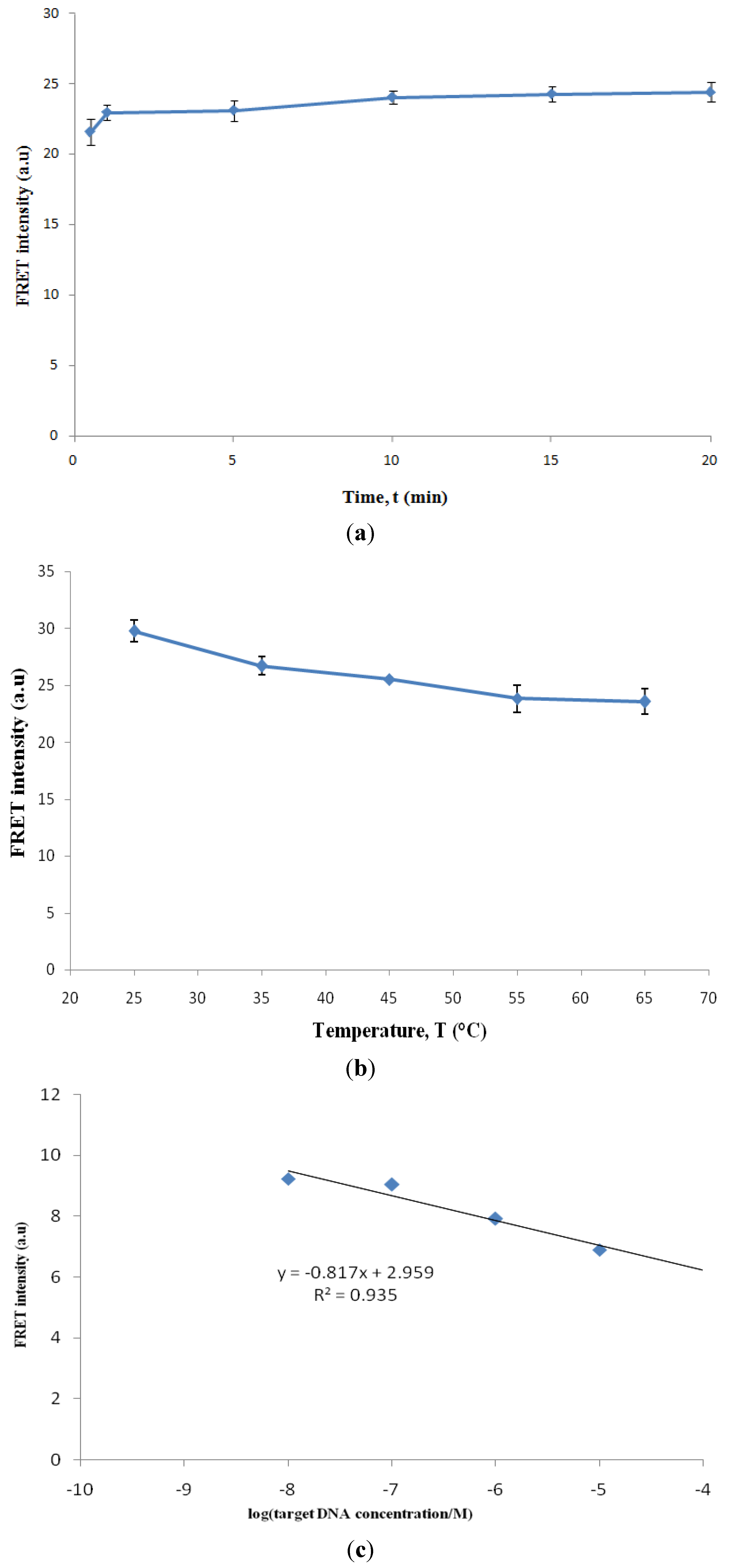
Calculation of Limit of Detection (LOD)
- LOD = 3.3 (s/S) [12]
- s = Standard deviation of response (y-intercept of the regression line)
- S = Slope of calibration curve
- y = −0.817x + 2.959
- s = 2.092, S = −0.817
- LOD = 3.3 (2.092/−0.817)
- = 8.45
- = anti log (−8.45)
- = 3.55 × 10−9 M
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Bakhori, N.M.; Yusof, N.A.; Abdullah, A.H.; Hussein, M.Z. Development of a Fluorescence Resonance Energy Transfer (FRET)-Based DNA Biosensor for Detection of Synthetic Oligonucleotide of Ganoderma boninense. Biosensors 2013, 3, 419-428. https://doi.org/10.3390/bios3040419
Bakhori NM, Yusof NA, Abdullah AH, Hussein MZ. Development of a Fluorescence Resonance Energy Transfer (FRET)-Based DNA Biosensor for Detection of Synthetic Oligonucleotide of Ganoderma boninense. Biosensors. 2013; 3(4):419-428. https://doi.org/10.3390/bios3040419
Chicago/Turabian StyleBakhori, Noremylia Mohd, Nor Azah Yusof, Abdul Halim Abdullah, and Mohd Zobir Hussein. 2013. "Development of a Fluorescence Resonance Energy Transfer (FRET)-Based DNA Biosensor for Detection of Synthetic Oligonucleotide of Ganoderma boninense" Biosensors 3, no. 4: 419-428. https://doi.org/10.3390/bios3040419
APA StyleBakhori, N. M., Yusof, N. A., Abdullah, A. H., & Hussein, M. Z. (2013). Development of a Fluorescence Resonance Energy Transfer (FRET)-Based DNA Biosensor for Detection of Synthetic Oligonucleotide of Ganoderma boninense. Biosensors, 3(4), 419-428. https://doi.org/10.3390/bios3040419



