DNA/RNA Detection Using DNA-Templated Few-Atom Silver Nanoclusters
Abstract
:1. Introduction
2. DNA-Templated Silver Nanoclusters
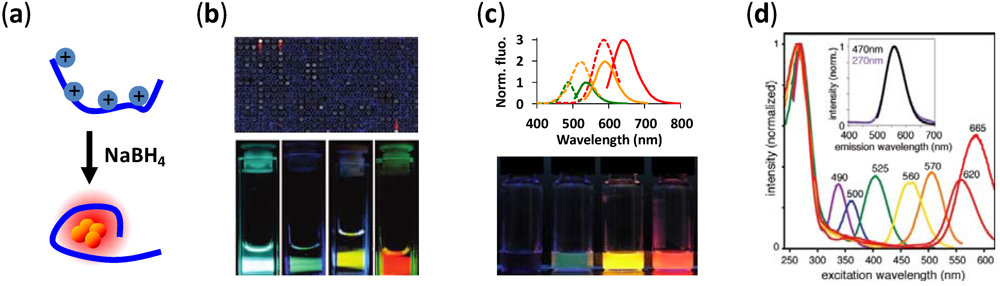
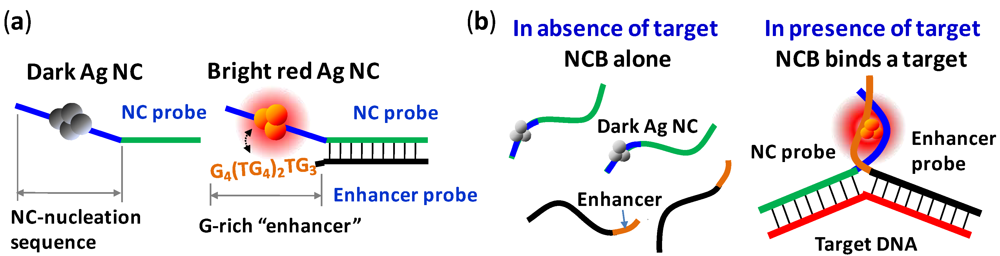
3. NanoCluster Beacons

4. Other DNA/Ag NC-Based DNA/RNA Detection Methods
4.1. Detection of DNA Targets

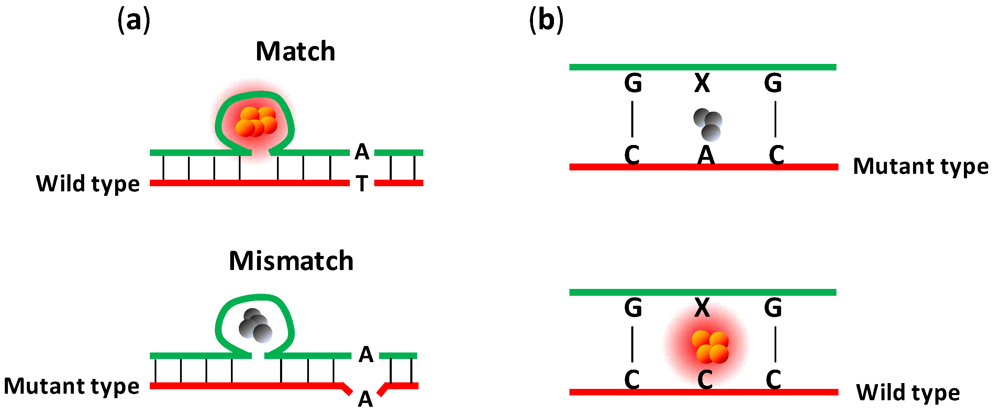
4.2. Detection of Single-Nucleotide Polymorphisms
4.3. Detection of RNA Targets
5. Future Prospects
Acknowledgments
References
- Cheng, X.; Chen, G.; Rodriguez, W.R. Micro-and nanotechnology for viral detection. Anal. Bioanal. Chem. 2009, 393, 487–501. [Google Scholar] [CrossRef]
- Frazer, K.A.; Ballinger, D.G.; Cox, D.R.; Hinds, D.A.; Stuve, L.L.; Gibbs, R.A.; Belmont, J.W.; Boudreau, A.; Hardenbol, P.; Leal, S.M. A second generation human haplotype map of over 3.1 million SNPs. Nature 2007, 449, 851–861. [Google Scholar] [CrossRef]
- Budowle, B.; Allard, M.W.; Wilson, M.R.; Chakraborty, R. Forensics and mitochondrial DNA: Applications, debates, and foundations. Ann. Rev. Genom. Hum. Genet. 2003, 4, 119–141. [Google Scholar] [CrossRef]
- Pinkel, D.; Segraves, R.; Sudar, D.; Clark, S.; Poole, I.; Kowbel, D.; Collins, C.; Kuo, W.-L.; Chen, C.; Zhai, Y. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 1998, 20, 207–211. [Google Scholar] [CrossRef]
- Sozzi, G.; Conte, D.; Mariani, L.; Vullo, S.L.; Roz, L.; Lombardo, C.; Pierotti, M.A.; Tavecchio, L. Analysis of circulating tumor DNA in plasma at diagnosis and during follow-up of lung cancer patients. Canc. Res. 2001, 61, 4675–4678. [Google Scholar]
- Fan, H.C.; Gu, W.; Wang, J.; Blumenfeld, Y.J.; El-Sayed, Y.Y.; Quake, S.R. Non-invasive prenatal measurement of the fetal genome. Nature 2012, 487, 320–324. [Google Scholar]
- Raj, A.; van den Bogaard, P.; Rifkin, S.A.; van Oudenaarden, A.; Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Meth. 2008, 5, 877–879. [Google Scholar]
- Munsky, B.; Neuert, G.; van Oudenaarden, A. Using gene expression noise to understand gene regulation. Science 2012, 336, 183–187. [Google Scholar] [CrossRef]
- Taniguchi, Y.; Choi, P.J.; Li, G.-W.; Chen, H.; Babu, M.; Hearn, J.; Emili, A.; Xie, X.S. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 2010, 329, 533–538. [Google Scholar]
- Heid, C.A.; Stevens, J.; Livak, K.J.; Williams, P.M. Real time quantitative PCR. Genome Res. 1996, 6, 986–994. [Google Scholar] [CrossRef]
- Hunkapiller, T.; Kaiser, R.; Koop, B.; Hood, L. Large-scale and automated DNA sequence determination. Science 1991, 254, 59–67. [Google Scholar]
- Brown, O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270, 467–470. [Google Scholar]
- Tyagi, S.; Kramer, F.R. Molecular beacons: Probes that fluoresce upon hybridization. Nat. Biotechnol. 1996, 14, 303–308. [Google Scholar]
- Kolpashchikov, D.M. Binary malachite green aptamer for fluorescent detection of nucleic acids. J. Am. Chem. Soc. 2005, 127, 12442–12443. [Google Scholar] [CrossRef]
- Storhoff, J.J.; Lucas, A.D.; Garimella, V.; Bao, Y.P.; Müller, U.R. Homogeneous detection of unamplified genomic DNA sequences based on colorimetric scatter of gold nanoparticle probes. Nat. Biotechnol. 2004, 22, 883–887. [Google Scholar]
- Holland, P.M.; Abramson, R.D.; Watson, R.; Gelfand, D.H. Detection of specific polymerase chain reaction product by utilizing the 5'----3' exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 1991, 88, 7276–7280. [Google Scholar] [CrossRef]
- Förster, T. Intermolecular Energy Transfer and Fluorescence; National Research Council of Canada: Ottawa, Canada, 1955. [Google Scholar]
- Stryer, L.; Haugland, R.P. Energy transfer: A spectroscopic ruler. Proc. Natl. Acad. Sci. USA 1967, 58, 719–726. [Google Scholar] [CrossRef]
- Elghanian, R.; Storhoff, J.J.; Mucic, R.C.; Letsinger, R.L.; Mirkin, C.A. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science 1997, 277, 1078–1081. [Google Scholar] [CrossRef]
- Sato, K.; Hosokawa, K.; Maeda, M. Rapid aggregation of gold nanoparticles induced by non-cross-linking DNA hybridization. J. Am. Chem. Soc. 2003, 125, 8102–8103. [Google Scholar]
- Li, H.; Rothberg, L. Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc. Natl. Acad. Sci. USA 2004, 101, 14036–14039. [Google Scholar] [CrossRef]
- Heller, D.A.; Jeng, E.S.; Yeung, T.K.; Martinez, B.M.; Moll, A.E.; Gastala, J.B.; Strano, M.S. Optical detection of DNA conformational polymorphism on single-walled carbon nanotubes. Science 2006, 311, 508–511. [Google Scholar] [CrossRef]
- Heller, D.A.; Jin, H.; Martinez, B.M.; Patel, D.; Miller, B.M.; Yeung, T.-K.; Jena, P.V.; Hobartner, C.; Ha, T.; Silverman, S.K.; Strano, M.S. Multimodal optical sensing and analyte specificity using single-walled carbon nanotubes. Nat. Nanotechnol. 2009, 4, 114–120. [Google Scholar]
- Petty, J.T.; Zheng, J.; Hud, N.V.; Dickson, R.M. DNA-templated Ag nanocluster formation. J. Am. Chem. Soc. 2004, 126, 5207–5212. [Google Scholar] [CrossRef]
- Vosch, T.; Antoku, Y.; Hsiang, J.-C.; Richards, C.I.; Gonzalez, J.I.; Dickson, R.M. Strongly emissive individual DNA-encapsulated Ag nanoclusters as single-molecule fluorophores. Proc. Natl. Acad. Sci. USA 2007, 104, 12616–12621. [Google Scholar]
- Gwinn, E.G.; O’Neill, P.; Guerrero, A.J.; Bouwmeester, D.; Fygenson, D.K. Sequence-dependent fluorescence of DNA-hosted silver nanoclusters. Adv. Mater. 2008, 20, 279–283. [Google Scholar] [CrossRef]
- Richards, C.I.; Choi, S.; Hsiang, J.-C.; Antoku, Y.; Vosch, T.; Bongiorno, A.; Tzeng, Y.-L.; Dickson, R.M. Oligonucleotide-stabilized Ag nanocluster fluorophores. J. Am. Chem. Soc. 2008, 130, 5038–5039. [Google Scholar] [CrossRef]
- Sharma, J.; Yeh, H.-C.; Yoo, H.; Werner, J.H.; Martinez, J.S. A complementary palette of fluorescent silver nanoclusters. Chem. Commun. 2010, 46, 3280–3282. [Google Scholar]
- Yeh, H.-C.; Sharma, J.; Han, J.J.; Martinez, J.S.; Werner, J.H. A DNA-silver nanocluster probe that fluoresces upon hybridization. Nano Lett. 2010, 10, 3106–3110. [Google Scholar] [CrossRef]
- Yeh, H.-C.; Sharma, J.; Han, J.J.; Martinez, J.S.; Werner, J.H. A beacon of light—A new molecular probe for homogeneous detection of nucleic acid targets. IEEE Nanotechnol. Mag. 2011, 5, 28–33. [Google Scholar]
- Yeh, H.-C.; Sharma, J.; Shih, I.-M.; Vu, D.M.; Martinez, J.S.; Werner, J.H. A fluorescence light-up Ag nanocluster probe that discriminates single-nucleotide variants by emission color. J. Am. Chem. Soc. 2012, 134, 11550–11558. [Google Scholar]
- Guo, W.; Yuan, J.; Dong, Q.; Wang, E. Highly sequence-dependent formation of fluorescent silver nanoclusters in hybridized DNA duplexes for single nucleotide mutation identification. J. Am. Chem. Soc. 2010, 132, 932–934. [Google Scholar] [CrossRef]
- Petty, J.T.; Fan, C.; Story, S.P.; Sengupta, B.; Sartin, M.; Hsiang, J.-C.; Perry, J.W.; Dickson, R.M. Optically enhanced, near-IR, silver cluster emission altered by single base changes in the DNA template. J. Phys. Chem. B 2011, 115, 7996–8003. [Google Scholar] [CrossRef]
- Zheng, J.; Nicovich, P.R.; Dickson, R.M. Highly fluorescent noble-metal quantum dots. Ann. Rev. Phys. Chem. 2007, 58, 409–431. [Google Scholar] [CrossRef]
- Schmid, G. Large clusters and colloids. Metals in the embryonic state. Chem. Rev. 1992, 92, 1709–1727. [Google Scholar]
- Wilcoxon, J.; Martin, J.; Parsapour, F.; Wiedenman, B.; Kelley, D. Photoluminescence from nanosize gold clusters. J. Chem. Phys. 1998, 108, 9137–9143. [Google Scholar] [CrossRef]
- Huang, T.; Murray, R.W. Visible luminescence of water-soluble monolayer-protected gold clusters. J. Phys. Chem. B 2001, 105, 12498–12502. [Google Scholar] [CrossRef]
- Link, S.; Beeby, A.; FitzGerald, S.; El-Sayed, M.A.; Schaaff, T.G.; Whetten, R.L. Visible to infrared luminescence from a 28-atom gold cluster. J. Phys. Chem. B 2002, 106, 3410–3415. [Google Scholar]
- Zheng, J.; Zhang, C.; Dickson, R.M. Highly fluorescent, water-soluble, size-tunable gold quantum dots. Phys. Rev. Lett. 2004, 93, 077402. [Google Scholar] [CrossRef]
- Zheng, J.; Dickson, R.M. Individual water-soluble dendrimer-encapsulated silver nanodot fluorescence. J. Am. Chem. Soc. 2002, 124, 13982–13983. [Google Scholar]
- Sun, T.; Seff, K. Silver clusters and chemistry in zeolites. Chem. Rev. 1994, 94, 857–870. [Google Scholar]
- De Cremer, G.; Sels, B.F.; Hotta, J.i.; Roeffaers, M.B.; Bartholomeeusen, E.; Coutiño‐Gonzalez, E.; Valtchev, V.; de Vos, D.E.; Vosch, T.; Hofkens, J. Optical encoding of silver zeolite microcarriers. Adv. Mater. 2010, 22, 957–960. [Google Scholar]
- Lesniak, W.; Bielinska, A.U.; Sun, K.; Janczak, K.W.; Shi, X.; Baker, J.R., Jr.; Balogh, L.P. Silver/dendrimer nanocomposites as biomarkers: Fabrication, characterization, in vitro toxicity, and intracellular detection. Nano Lett. 2005, 5, 2123–2130. [Google Scholar]
- Shang, L.; Dong, S.J. Facile preparation of water-soluble fluorescent silver nanoclusters using a polyelectrolyte template. Chem. Commun. 2008, 9, 1088–1090. [Google Scholar] [CrossRef]
- Ershov, B.G.; Henglein, A. Reduction of Ag+ on polyacrylate chains in aqueous solution. J. Phys. Chem. B 1998, 102, 10663–10666. [Google Scholar]
- Shen, Z.; Duan, H.W.; Frey, H. Water-soluble fluorescent Ag nanoclusters obtained from multiarm star poly(acrylic acid) as “molecular hydrogel” templates. Adv. Mater. 2007, 19, 349–352. [Google Scholar] [CrossRef]
- Zhang, J.G.; Xu, S.Q.; Kumacheva, E. Photogeneration of fluorescent silver nanoclusters in polymer microgels. Adv. Mater. 2005, 17, 2336–2340. [Google Scholar]
- Pal, S.; Varghese, R.; Deng, Z.T.; Zhao, Z.; Kumar, A.; Yan, H.; Liu, Y. Site-specific synthesis and in situ immobilization of fluorescent silver nanoclusters on DNA nanoscaffolds by use of the tollens reaction. Angew. Chem. Int. Ed. 2011, 50, 4176–4179. [Google Scholar]
- Rao, T.U.B.; Pradeep, T. Luminescent Ag7 and Ag8 clusters by interfacial synthesis. Angew. Chem. Int. Ed. 2010, 49, 3925–3929. [Google Scholar] [CrossRef]
- Adhikari, B.; Banerjee, A. Facile synthesis of water-soluble fluorescent silver nanoclusters and HgII sensing. Chem. Mater. 2010, 22, 4364–4371. [Google Scholar] [CrossRef]
- Cathcart, N.; Mistry, P.; Makra, C.; Pietrobon, B.; Coombs, N.; Jelokhani-Niaraki, M.; Kitaev, V. Chiral thiol-stabilized silver nanoclusters with well-resolved optical transitions synthesized by a facile etching procedure in aqueous solutions. Langmuir 2009, 25, 5840–5846. [Google Scholar] [CrossRef]
- Yu, J.; Patel, S.A.; Dickson, R.M. In vitro and intracellular production of peptide-encapsulated fluorescent silver nanoclusters. Angew. Chem. Int. Ed. 2007, 46, 2028–2030. [Google Scholar] [CrossRef]
- O’Neill, P.R.; Gwinn, E.G.; Fygenson, D.K. UV excitation of DNA stabilized Ag cluster fluorescence via the DNA bases. J. Phys. Chem. C 2011, 115, 24061–24066. [Google Scholar]
- Yeh, H.C.; Sharma, J.; Yoo, H.; Martinez, J.S.; Werner, J.H. Photophysical characterization of fluorescent metal nanoclusters synthesized using oligonucleotides, proteins and small molecule ligands. Proc. SPIE 2010, 7576, 75760N1–75760N9. [Google Scholar]
- Sharma, J.; Rocha, R.C.; Phipps, M.L.; Yeh, H.-C.; Balatsky, K.A.; Vu, D.M.; Shreve, A.P.; Werner, J.H.; Martinez, J.S. A DNA-templated fluorescent silver nanocluster with enhanced stability. Nanoscale 2012, 4, 4107–4110. [Google Scholar]
- Antoku, Y.; Hotta, J.; Mizuno, H.; Dickson, R.M.; Hofkens, J.; Vosch, T. Transfection of living HeLa cells with fluorescent poly-cytosine encapsulated Ag nanoclusters. Photochem. Photobiol. Sci. 2010, 9, 716–721. [Google Scholar]
- Le Guevel, X.; Spies, C.; Daum, N.; Jung, G.; Schneider, M. Highly fluorescent silver nanoclusters stabilized by glutathione: A promising fluorescent label for bioimaging. Nano Res. 2012, 5, 379–387. [Google Scholar]
- Richards, C.I.; Hsiang, J.-C.; Senapati, D.; Patel, S.A.; Yu, J.; Vosch, T.; Dickson, R.M. Optically modulated fluorophores for selective fluorescence signal recovery. J. Am. Chem. Soc. 2009, 131, 4619–4621. [Google Scholar]
- Choi, S.; Yu, J.; Patel, S.A.; Tzeng, Y.-L.; Dickson, R.M. Tailoring silver nanodots for intracellular staining. Photochem. Photobiol. Sci. 2011, 10, 109–115. [Google Scholar] [CrossRef]
- Ritchie, C.M.; Johnsen, K.R.; Kiser, J.R.; Antoku, Y.; Dickson, R.M.; Petty, J.T. Ag nanocluster formation using a cytosine oligonucleotide template. J. Phys. Chem. C 2007, 111, 175–181. [Google Scholar]
- O’Neill, P.R.; Velazquez, L.R.; Dunn, D.G.; Gwinn, E.G.; Fygenson, D.K. Hairpins with Poly-C loops stabilize four types of fluorescent Agn:DNA. J. Phys. Chem. C 2009, 113, 4229–4233. [Google Scholar]
- Schultz, D.; Gwinn, E.G. Silver atom and strand numbers in fluorescent and dark Ag:DNAs. Chem. Commun. 2012, 48, 5748–5750. [Google Scholar] [CrossRef]
- Petty, J.T.; Fan, C.; Story, S.P.; Sengupta, B.; St. John, I.A.; Prudowsky, Z.; Dickson, R.M. DNA encapsulation of 10 silver atoms producing a bright, modulatable, near-infrared-emitting cluster. The J. Phys. Chem. Lett. 2010, 1, 2524–2529. [Google Scholar]
- Petty, J.T.; Giri, B.; Miller, I.C.; Nicholson, D.A.; Sergev, O.O.; Banks, T.M.; Story, S.P. Silver clusters as both chromophoric reporters and DNA ligands. Anal. Chem. 2013, 85, 2183–2190. [Google Scholar] [CrossRef]
- Yu, J.H.; Choi, S.M.; Richards, C.I.; Antoku, Y.; Dickson, R.M. Live cell surface labeling with fluorescent Ag nanocluster conjugates. Photochem. Photobiol. 2008, 84, 1435–1439. [Google Scholar] [CrossRef]
- Yu, J.H.; Choi, S.; Dickson, R.M. Shuttle-based fluorogenic silver-cluster biolabels. Angew. Chem. Int. Ed. 2009, 48, 318–320. [Google Scholar]
- Li, T.; Zhang, L.B.; Ai, J.; Dong, S.J.; Wang, E.K. Ion-tuned DNA/Ag fluorescent nanoclusters as versatile logic device. ACS Nano 2011, 5, 6334–6338. [Google Scholar] [CrossRef]
- Guo, W.; Yuan, J.; Wang, E. Oligonucleotide-stabilized Ag nanoclusters as novel fluorescence probes for the highly selective and sensitive detection of the Hg2+ ion. Chem. Commun. 2009, 3395–3397. [Google Scholar]
- Lan, G.Y.; Huang, C.C.; Chang, H.T. Silver nanoclusters as fluorescent probes for selective and sensitive detection of copper ions. Chem. Commun. 2010, 46, 1257–1259. [Google Scholar] [CrossRef]
- Sharma, J.; Yeh, H.C.; Yoo, H.; Werner, J.H.; Martinez, J.S. Silver nanocluster aptamers: In situ generation of intrinsically fluorescent recognition ligands for protein detection. Chem. Commun. 2011, 47, 2294–2296. [Google Scholar]
- Li, J.; Zhong, X.; Zhang, H.; Le, X.C.; Zhu, J.J. Binding-induced fluorescence turn-on assay using aptamer-functionalized silver nanocluster DNA probes. Anal. Chem. 2012, 84, 5170–5174. [Google Scholar]
- Liu, J.J.; Song, X.R.; Wang, Y.W.; Zheng, A.X.; Chen, G.N.; Yang, H.H. Label-free and fluorescence turn-on aptasensor for protein detection via target-induced silver nanoclusters formation. Anal. Chim. Acta 2012, 749, 70–74. [Google Scholar] [CrossRef]
- Seidel, C.A.M.; Schulz, A.; Sauer, M.H.M. Nucleobase-specific quenching of fluorescent dyes. 1. nucleobase one-electron redox potentials and their correlation with static and dynamic quenching efficiencies. J. Phys. Chem. 1996, 100, 5541–5553. [Google Scholar] [CrossRef]
- Knemeyer, J.P.; Marme, N.; Sauer, M. Probes for detection of specific DNA sequences at the single-molecule level. Anal. Chem. 2000, 72, 3717–3724. [Google Scholar] [CrossRef]
- Heinlein, T.; Knemeyer, J.-P.; Piestert, O.; Sauer, M.H.M. Photoinduced electron transfer between fluorescent dyes and guanosine residues in DNA-hairpins. J. Phys. Chem. B 2003, 107, 7957–7964. [Google Scholar]
- Zhang, M.; Guo, S.M.; Li, Y.R.; Zuo, P.; Ye, B.C. A label-free fluorescent molecular beacon based on DNA-templated silver nanoclusters for detection of adenosine and adenosine deaminase. Chem. Commun. 2012, 48, 5488–5490. [Google Scholar] [CrossRef]
- Kostrikis, L.G.; Tyagi, S.; Mhlanga, M.M.; Ho, D.D.; Kramer, F.R. Molecular beacons-spectral genotyping of human alleles. Science 1998, 279, 1228–1229. [Google Scholar]
- Park, S.J.; Taton, T.A.; Mirkin, C.A. Array-based electrical detection of DNA with nanoparticle probes. Science 2002, 295, 1503–1506. [Google Scholar]
- Subramanian, H.K.K.; Chakraborty, B.; Sha, R.; Seeman, N.C. The label-free unambiguous detection and symbolic display of single nucleotide polymorphisms on DNA origami. Nano Lett. 2011, 11, 910–913. [Google Scholar] [CrossRef]
- Petty, J.T.; Sengupta, B.; Story, S.P.; Degtyareva, N.N. DNA sensing by amplifying the number of near-infrared emitting, oligonucleotide-encapsulated silver clusters. Anal. Chem. 2011, 83, 5957–5964. [Google Scholar]
- Lan, G.Y.; Chen, W.Y.; Chang, H.T. One-pot synthesis of fluorescent oligonucleotide Ag nanoclusters for specific and sensitive detection of DNA. Biosens. Bioelectron. 2011, 26, 2431–2435. [Google Scholar]
- Shah, P.; Rørvig-Lund, A.; Chaabane, S.B.; Thulstrup, P.W.; Kjaergaard, H.G.; Fron, E.; Hofkens, J.; Yang, S.W.; Vosch, T. Design aspects of bright red emissive silver nanoclusters/DNA probes for microRNA detection. ACS Nano 2012, 6, 8803–8814. [Google Scholar]
- He, S.; Song, B.; Li, D.; Zhu, C.; Qi, W.; Wen, Y.; Wang, L.; Song, S.; Fang, H.; Fan, C. A graphene nanoprobe for rapid, sensitive, and multicolor fluorescent DNA analysis. Adv. Func. Mater. 2010, 20, 453–459. [Google Scholar] [CrossRef]
- Loh, K.P.; Bao, Q.; Eda, G.; Chhowalla, M. Graphene oxide as a chemically tunable platform for optical applications. Nat. Chem. 2010, 2, 1015–1024. [Google Scholar] [CrossRef]
- Tao, Y.; Lin, Y.; Huang, Z.; Ren, J.; Qu, X. DNA-templated silver nanoclusters-graphene oxide nanohybrid materials: A platform for label-free and sensitive fluorescence turn-on detection of multiple nucleic acid targets. Analyst 2012, 137, 2588–2592. [Google Scholar]
- Ma, K.; Cui, Q.; Liu, G.; Wu, F.; Xu, S.; Shao, Y. DNA abasic site-directed formation of fluorescent silver nanoclusters for selective nucleobase recognition. Nanotechnology 2011, 22, 305502. [Google Scholar] [CrossRef]
- Liu, Y.Q.; Zhang, M.; Yin, B.C.; Ye, B.C. Attomolar ultrasensitive microRNA detection by DNA-scaffolded silver-nanocluster probe based on isothermal amplification. Anal. Chem. 2012, 84, 5165–5169. [Google Scholar]
- Yang, S.W.; Vosch, T. Rapid detection of microRNA by a silver nanocluster DNA probe. Anal. Chem. 2011, 83, 6935–6939. [Google Scholar] [CrossRef]
- Dong, H.; Jin, S.; Ju, H.; Hao, K.; Xu, L.P.; Lu, H.; Zhang, X. Trace and label-free microRNA detection using oligonucleotide encapsulated silver nanoclusters as probes. Anal. Chem. 2012, 84, 8670–8674. [Google Scholar]
- Bertrand, E.; Chartrand, P.; Schaefer, M.; Shenoy, S.M.; Singer, R.H.; Long, R.M. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 1998, 2, 437–445. [Google Scholar] [CrossRef]
- Lessard, G.A.; Goodwin, P.M.; Werner, J.H. Three-dimensional tracking of individual quantum dots. Appl. Phys. Lett. 2007, 91. [Google Scholar] [CrossRef]
- Wells, N.P.; Lessard, G.A.; Werner, J.H. Confocal, three-dimensional tracking of individual quantum dots in high-background environments. Anal. Chem. 2008, 80, 9830–9834. [Google Scholar] [CrossRef]
- Wells, N.P.; Lessard, G.A.; Goodwin, P.M.; Phipps, M.E.; Cutler, P.J.; Lidke, D.S.; Wilson, B.S.; Werner, J.H. Time-resolved three-dimensional molecular tracking in live cells. Nano Lett. 2010, 10, 4732–4737. [Google Scholar] [CrossRef]
- Han, J.J.; Kiss, C.; Bradbury, A.R.M.; Werner, J.H. Time-resolved, confocal single-molecule tracking of individual organic dyes and fluorescent proteins in three dimensions. ACS Nano 2012, 6, 8922–8932. [Google Scholar]
- Han, M.; Gao, X.; Su, J.Z.; Nie, S. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat. Biotechnol. 2001, 19, 631–635. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Obliosca, J.M.; Liu, C.; Batson, R.A.; Babin, M.C.; Werner, J.H.; Yeh, H.-C. DNA/RNA Detection Using DNA-Templated Few-Atom Silver Nanoclusters. Biosensors 2013, 3, 185-200. https://doi.org/10.3390/bios3020185
Obliosca JM, Liu C, Batson RA, Babin MC, Werner JH, Yeh H-C. DNA/RNA Detection Using DNA-Templated Few-Atom Silver Nanoclusters. Biosensors. 2013; 3(2):185-200. https://doi.org/10.3390/bios3020185
Chicago/Turabian StyleObliosca, Judy M., Cong Liu, Robert Austin Batson, Mark C. Babin, James H. Werner, and Hsin-Chih Yeh. 2013. "DNA/RNA Detection Using DNA-Templated Few-Atom Silver Nanoclusters" Biosensors 3, no. 2: 185-200. https://doi.org/10.3390/bios3020185
APA StyleObliosca, J. M., Liu, C., Batson, R. A., Babin, M. C., Werner, J. H., & Yeh, H.-C. (2013). DNA/RNA Detection Using DNA-Templated Few-Atom Silver Nanoclusters. Biosensors, 3(2), 185-200. https://doi.org/10.3390/bios3020185



