Comparative Evaluation of Combined Denoising and Resolution Enhancement Algorithms for Intravital Two-Photon Imaging of Organs
Abstract
1. Introduction
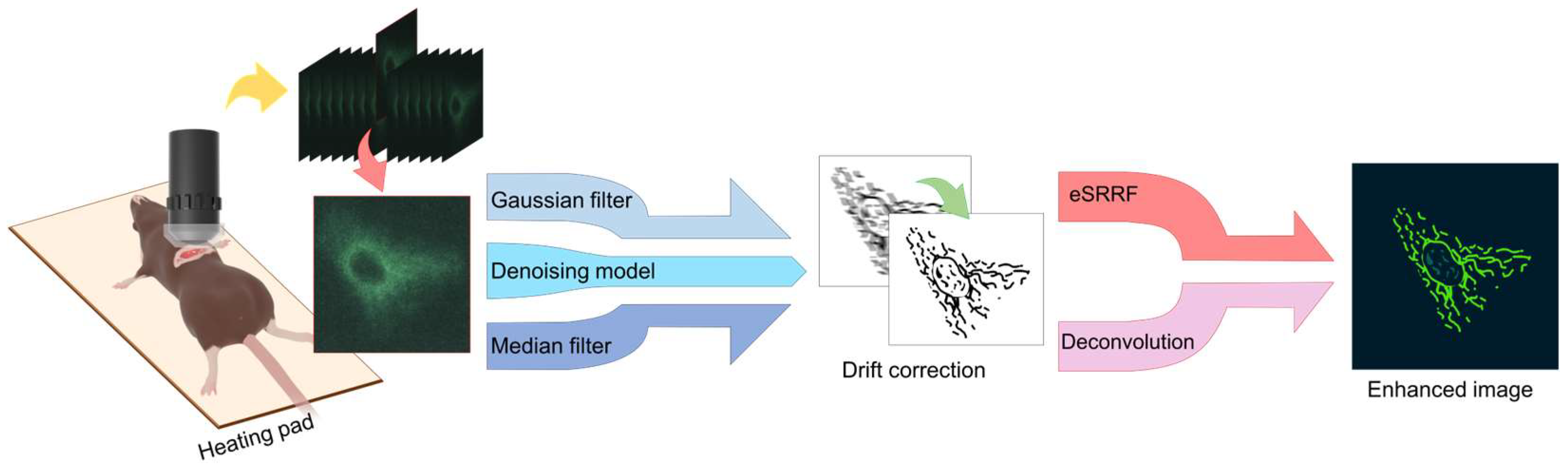
2. Materials and Methods
2.1. Animal Preparation
2.2. Intravital Imaging Procedure
2.3. Image Analysis
2.4. Alcoholic Liver Disease Model
3. Results
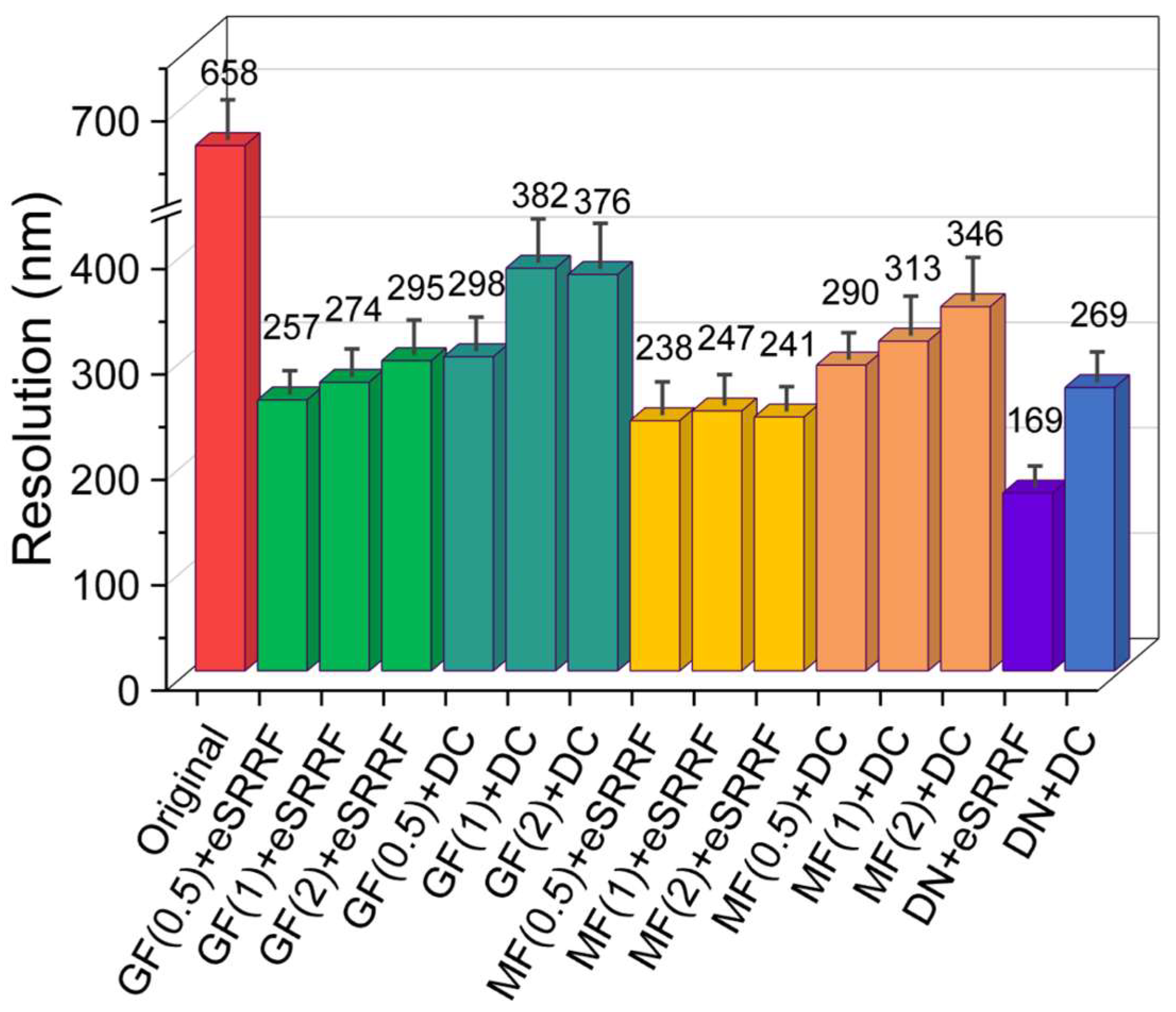
3.1. Quantitative Evaluation of Resolution Enhancement Techniques
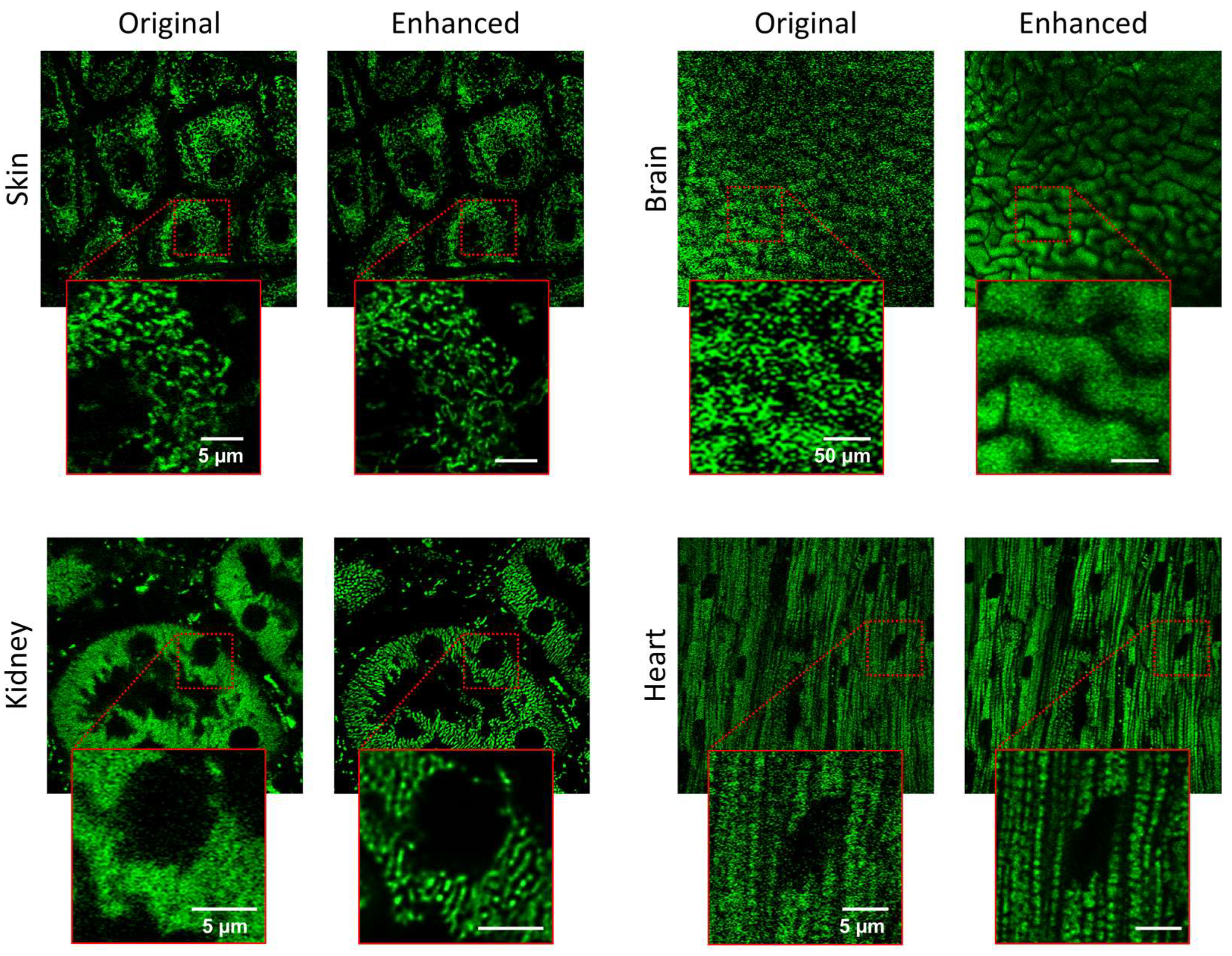
3.2. Imaging Various Organs
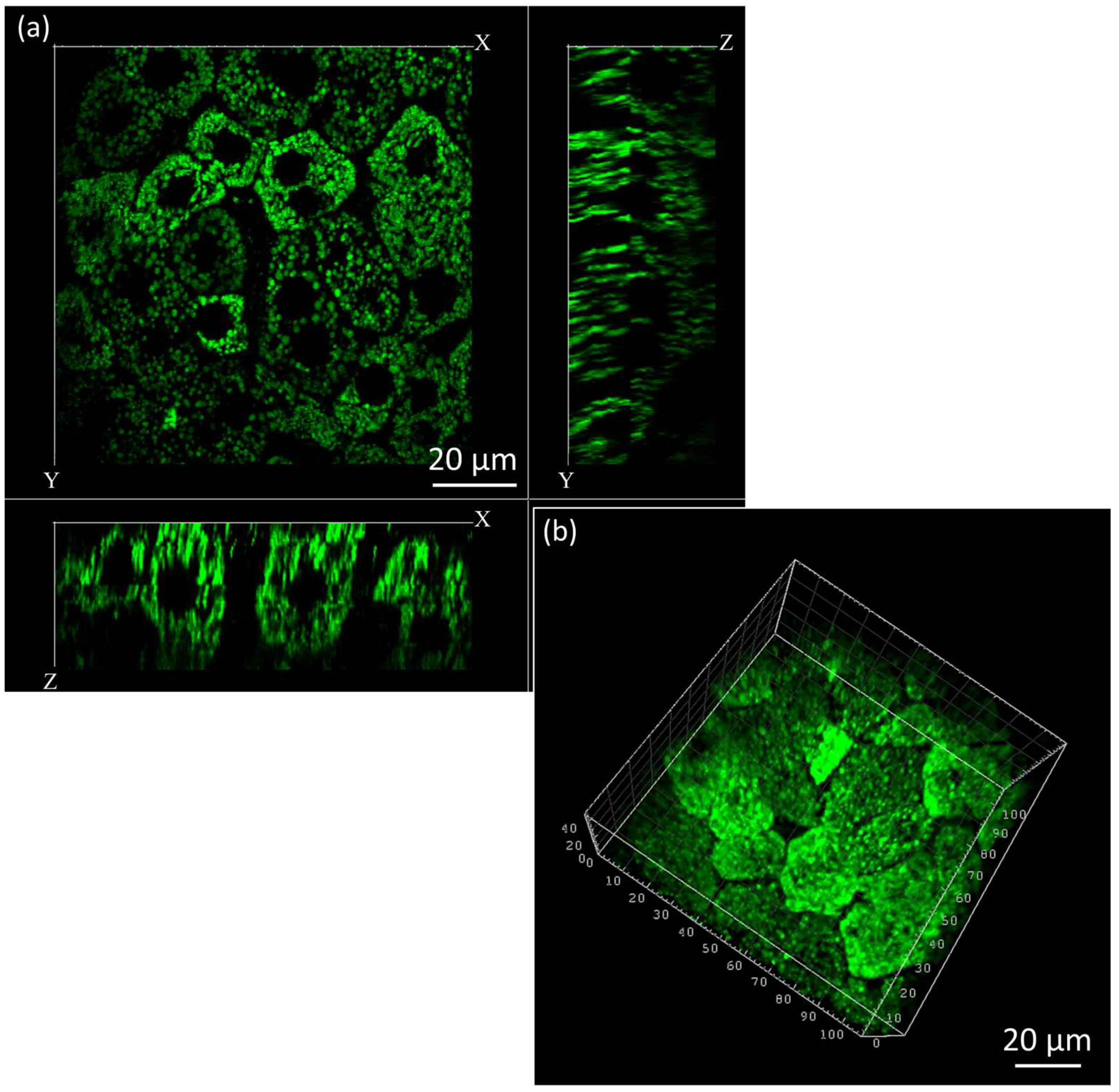
3.3. Three-Dimensional Mitochondrial Mapping of Hepatocytes
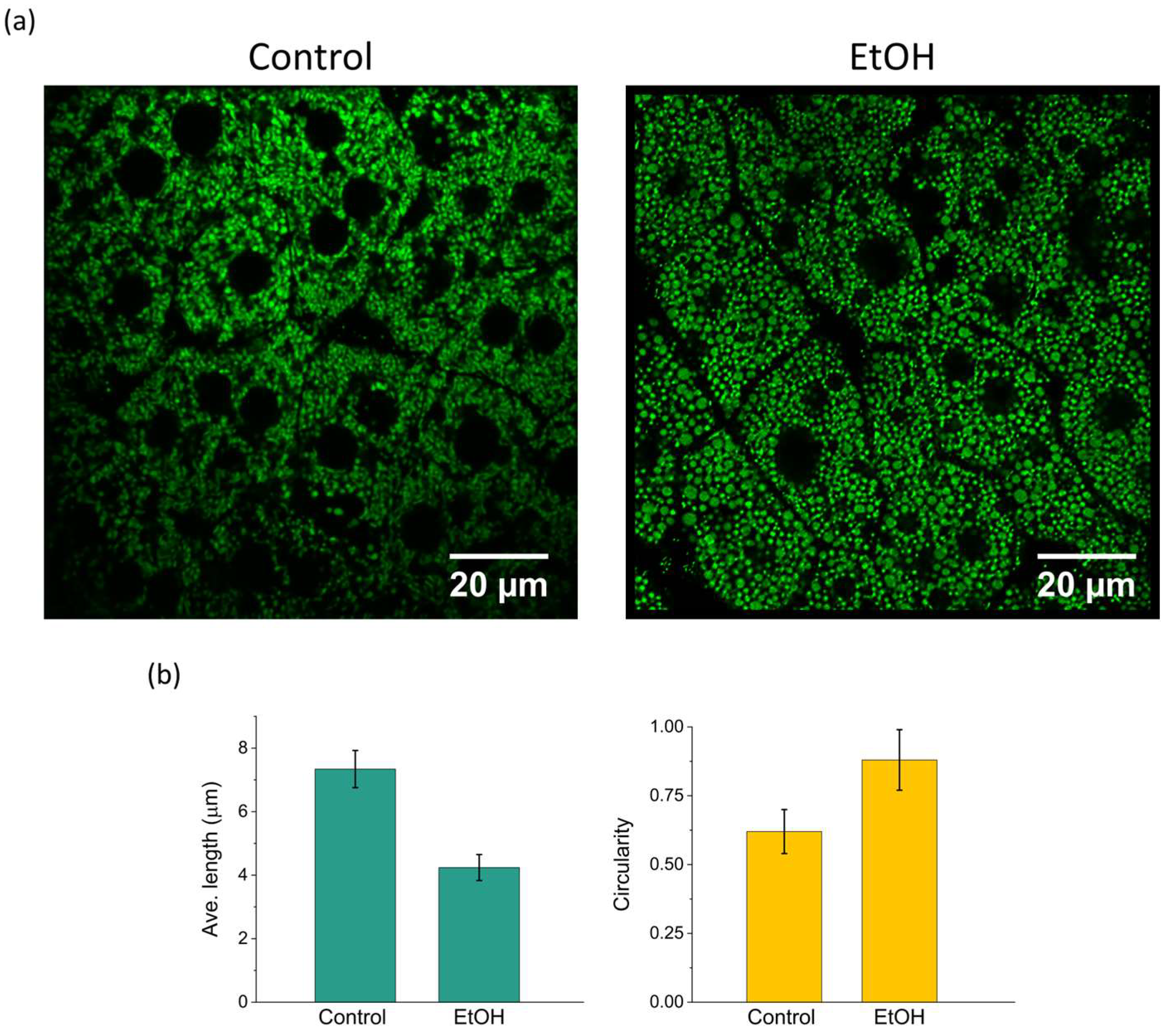
3.4. Alcohol-Induced Mitochondrial Fragmentation Detection
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| eSRRF | Enhanced super-resolution radial fluctuations |
| EtOH | Ethanol |
| N2N | noise2noise |
| PSF | Point Spread Function |
| QnR | quality and resolution |
| rFRC | rolling Fourier ring correlation |
References
- Pittet, M.J.; Weissleder, R. Intravital imaging. Cell 2011, 147, 983–991. [Google Scholar] [CrossRef]
- Masedunskas, A.; Milberg, O.; Porat-Shliom, N.; Sramkova, M.; Wigand, T.; Amornphimoltham, P.; Weigert, R. Intravital microscopy: A practical guide on imaging intracellular structures in live animals. Bioarchitecture 2012, 2, 143–157. [Google Scholar] [CrossRef]
- Müller, J.; Wunder, A.; Licha, K. Optical imaging. In Molecular Imaging in Oncology; Springer: Berlin/Heidelberg, Germany, 2012; pp. 221–246. [Google Scholar]
- Dhawan, A.P.; D’Alessandro, B.; Fu, X. Optical imaging modalities for biomedical applications. IEEE Rev. Biomed. Eng. 2010, 3, 69–92. [Google Scholar] [CrossRef]
- Pirovano, G.; Roberts, S.; Kossatz, S.; Reiner, T. Optical imaging modalities: Principles and applications in preclinical research and clinical settings. J. Nucl. Med. 2020, 61, 1419–1427. [Google Scholar] [CrossRef]
- Luker, G.D.; Luker, K.E. Optical imaging: Current applications and future directions. J. Nucl. Med. 2008, 49, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, M.; Oh, J.; Jeong, S.; Lee, M.; Bohlooli Darian, S.; Oh, K.; Kim, J.K. Fabry–Perot Interferometric Fiber-Optic Sensor for Rapid and Accurate Thrombus Detection. Biosensors 2023, 13, 817. [Google Scholar] [CrossRef]
- Sanderson, J. Multi-photon microscopy. Curr. Protoc. 2023, 3, e634. [Google Scholar] [CrossRef] [PubMed]
- König, K. Multiphoton microscopy in life sciences. J. Microsc. 2000, 200, 83–104. [Google Scholar] [CrossRef]
- Kim, J.K.; Lee, W.M.; Kim, P.; Choi, M.; Jung, K.; Kim, S.; Yun, S.H. Fabrication and operation of GRIN probes for in vivo fluorescence cellular imaging of internal organs in small animals. Nat. Protoc. 2012, 7, 1456–1469. [Google Scholar] [CrossRef]
- Paulson, B.; Darian, S.B.; Kim, Y.; Oh, J.; Ghasemi, M.; Lee, K.; Kim, J.K. Spectral Multiplexing of Fluorescent Endoscopy for Simultaneous Imaging with Multiple Fluorophores and Multiple Fields of View. Biosensors 2022, 13, 33. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.L. Diffraction and resolving power. J. Opt. Soc. Am. 1964, 54, 931–936. [Google Scholar] [CrossRef]
- Presson Jr, R.G.; Brown, M.B.; Fisher, A.J.; Sandoval, R.M.; Dunn, K.W.; Lorenz, K.S.; Delp, E.J.; Salama, P.; Molitoris, B.A.; Petrache, I. Two-photon imaging within the murine thorax without respiratory and cardiac motion artifact. Am. J. Pathol. 2011, 179, 75–82. [Google Scholar] [CrossRef]
- Surre, J.; Saint-Ruf, C.; Collin, V.; Orenga, S.; Ramjeet, M.; Matic, I. Strong increase in the autofluorescence of cells signals struggle for survival. Sci. Rep. 2018, 8, 12088. [Google Scholar] [CrossRef]
- Stanciu, S.G.; Hristu, R.; Stanciu, G.A.; Tranca, D.E.; Eftimie, L.; Dumitru, A.; Costache, M.; Stenmark, H.A.; Manders, H.; Cherian, A.; et al. Super-resolution re-scan second harmonic generation microscopy. Proc. Natl. Acad. Sci. USA 2022, 119, e2214662119. [Google Scholar] [CrossRef]
- Gregor, I.; Spiecker, M.; Petrovsky, R.; Großhans, J.; Ros, R.; Enderlein, J. Rapid nonlinear image scanning microscopy. Nat. Methods 2017, 14, 1087–1089. [Google Scholar] [CrossRef]
- Annesley, S.J.; Fisher, P.R. Mitochondria in health and disease. Cells 2019, 8, 680. [Google Scholar] [CrossRef] [PubMed]
- McBride, H.M.; Neuspiel, M.; Wasiak, S. Mitochondria: More than just a powerhouse. Curr. Biol. 2006, 16, R551–R560. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, J.B.; Haigis, M.C. The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 2018, 20, 745–754. [Google Scholar] [CrossRef]
- Lee, Y.-H.; Zhang, S.; Mitchell, C.K.; Lin, Y.-P.; O’Brien, J. Calcium Imaging with Super-Resolution Radial Fluctuations. Biosci. Bioeng. 2018, 4, 78. [Google Scholar]
- Nagata, S. Apoptosis and clearance of apoptotic cells. Annu. Rev. Immunol. 2018, 36, 489–517. [Google Scholar] [CrossRef] [PubMed]
- Youle, R.J.; Van Der Bliek, A.M. Mitochondrial fission, fusion, and stress. Science 2012, 337, 1062–1065. [Google Scholar] [CrossRef]
- Duraisamy, A.J.; Mohammad, G.; Kowluru, R.A. Mitochondrial fusion and maintenance of mitochondrial homeostasis in diabetic retinopathy. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2019, 1865, 1617–1626. [Google Scholar] [CrossRef]
- Chinnery, P.F.; Schon, E.A. Mitochondria. J. Neurol. Neurosurg. Psychiatry 2003, 74, 1188–1199. [Google Scholar] [CrossRef]
- Bohlooli, D.S.; Jeongmin, O.; Bjorn, P.; Minju, C.; Globinna, K.; Eunyoung, T.; Inki, K.; Chan-Gi, P.; Jung-Man, N.; In-Jeoung, B.; et al. Multiphoton intravital microscopy in small animals of long-term mitochondrial dynamics based on super-resolution radial fluctuations. Opto-Electron. Adv. 2025, 8, 240311. [Google Scholar] [CrossRef]
- Galbraith, C.G.; Galbraith, J.A. Super-resolution microscopy at a glance. J. Cell Sci. 2011, 124, 1607–1611. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Hoess, P.; Ries, J. Super-resolution microscopy for structural cell biology. Annu. Rev. Biophys. 2022, 51, 301–326. [Google Scholar] [CrossRef]
- Prakash, K.; Diederich, B.; Heintzmann, R.; Schermelleh, L. Super-resolution microscopy: A brief history and new avenues. In Advances in Medical Imaging, Detection, and Diagnosis; Routledge: London, UK, 2023; pp. 1195–1211. [Google Scholar]
- Laine, R.F.; Heil, H.S.; Coelho, S.; Nixon-Abell, J.; Jimenez, A.; Wiesner, T.; Martínez, D.; Galgani, T.; Régnier, L.; Stubb, A.; et al. High-fidelity 3D live-cell nanoscopy through data-driven enhanced super-resolution radial fluctuation. Nat. Methods 2023, 20, 1949–1956. [Google Scholar] [CrossRef] [PubMed]
- Culley, S.; Tosheva, K.L.; Matos Pereira, P.; Henriques, R. SRRF: Universal live-cell super-resolution microscopy. Int. J. Biochem. Cell Biol. 2018, 101, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Sibarita, J.-B. Deconvolution Microscopy. In Microscopy Techniques; Rietdorf, J., Ed.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 201–243. [Google Scholar]
- Lehtinen, J.; Munkberg, J.; Hasselgren, J.; Laine, S.; Karras, T.; Aittala, M.; Aila, T. Noise2Noise: Learning image restoration without clean data. arXiv 2018, arXiv:1803.04189. [Google Scholar] [CrossRef]
- Zhao, W.; Huang, X.; Yang, J.; Qu, L.; Qiu, G.; Zhao, Y.; Wang, X.; Su, D.; Ding, X.; Mao, H.; et al. Quantitatively mapping local quality of super-resolution microscopy by rolling Fourier ring correlation. Light Sci. Appl. 2023, 12, 298. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Laine, R.F.; Tosheva, K.L.; Gustafsson, N.; Gray, R.D.M.; Almada, P.; Albrecht, D.; Risa, G.T.; Hurtig, F.; Lindås, A.-C.; Baum, B.; et al. NanoJ: A high-performance open-source super-resolution microscopy toolbox. J. Phys. D Appl. Phys. 2019, 52, 163001. [Google Scholar] [CrossRef]
- Ikoma, H.; Broxton, M.; Kudo, T.; Wetzstein, G. A convex 3D deconvolution algorithm for low photon count fluorescence imaging. Sci. Rep. 2018, 8, 11489. [Google Scholar] [CrossRef]
- Culley, S.; Albrecht, D.; Jacobs, C.; Pereira, P.M.; Leterrier, C.; Mercer, J.; Henriques, R. Quantitative mapping and minimization of super-resolution optical imaging artifacts. Nat. Methods 2018, 15, 263–266. [Google Scholar] [CrossRef]
- Valente, A.J.; Maddalena, L.A.; Robb, E.L.; Moradi, F.; Stuart, J.A. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 2017, 119, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Edenberg, H.J.; McClintick, J.N. Alcohol dehydrogenases, aldehyde dehydrogenases, and alcohol use disorders: A critical review. Alcohol. Clin. Exp. Res. 2018, 42, 2281–2297. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.; Carr, R. Alcohol effects on hepatic lipid metabolism. J. Lipid Res. 2020, 61, 470–479. [Google Scholar] [CrossRef]
- Ziman, A.P.; Ward, C.W.; Rodney, G.G.; Lederer, W.J.; Bloch, R.J. Quantitative Measurement of Ca2+ in the Sarcoplasmic Reticulum Lumen of Mammalian Skeletal Muscle. Biophys. J. 2010, 99, 2705–2714. [Google Scholar] [CrossRef]
- Tammineni, E.R.; Kraeva, N.; Figueroa, L.; Manno, C.; Ibarra, C.A.; Klip, A.; Riazi, S.; Rios, E. Intracellular calcium leak lowers glucose storage in human muscle, promoting hyperglycemia and diabetes. eLife 2020, 9, e53999. [Google Scholar] [CrossRef]
- Zorova, L.D.; Popkov, V.A.; Plotnikov, E.Y.; Silachev, D.N.; Pevzner, I.B.; Jankauskas, S.S.; Babenko, V.A.; Zorov, S.D.; Balakireva, A.V.; Juhaszova, M. Mitochondrial membrane potential. Anal. Biochem. 2018, 552, 50–59. [Google Scholar] [CrossRef]
- Besseling, T.H.; Jose, J.; Blaaderen, A.V. Methods to calibrate and scale axial distances in confocal microscopy as a function of refractive index. J. Microsc. 2015, 257, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Vendelin, M.; Béraud, N.; Guerrero, K.; Andrienko, T.; Kuznetsov, A.V.; Olivares, J.; Kay, L.; Saks, V.A. Mitochondrial regular arrangement in muscle cells: A “crystal-like” pattern. Am. J. Physiol.-Cell Physiol. 2005, 288, C757–C767. [Google Scholar] [CrossRef] [PubMed]
- BenÍTez-Bribiesca, L.; GÓMez-Camarillo, M.; Castellanos-JuÁRez, E.; Mravko, E.; SÁNchez-SuÁRez, P. Morphologic, Biochemical and Molecular Mitochondrial Changes during Reperfusion Phase following Brief Renal Ischemia. Ann. N. Y. Acad. Sci. 2006, 926, 165–179. [Google Scholar] [CrossRef]
- Georgakoudi, I.; Mellem, D.; Sattler, M.; Pagel-Wolff, S.; Jaspers, S.; Wenck, H.; Rübhausen, M.A.; Fischer, F. Fragmentation of the mitochondrial network in skin in vivo. PLoS ONE 2017, 12, e0174469. [Google Scholar] [CrossRef]
- Rust, M.J.; Bates, M.; Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 2006, 3, 793–796. [Google Scholar] [CrossRef]
- Nehme, E.; Weiss, L.E.; Michaeli, T.; Shechtman, Y. Deep-STORM: Super-resolution single-molecule microscopy by deep learning. Optica 2018, 5, 458–464. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bohlooli Darian, S.; Choi, W.J.; Oh, J.; Kim, J.K. Comparative Evaluation of Combined Denoising and Resolution Enhancement Algorithms for Intravital Two-Photon Imaging of Organs. Biosensors 2025, 15, 616. https://doi.org/10.3390/bios15090616
Bohlooli Darian S, Choi WJ, Oh J, Kim JK. Comparative Evaluation of Combined Denoising and Resolution Enhancement Algorithms for Intravital Two-Photon Imaging of Organs. Biosensors. 2025; 15(9):616. https://doi.org/10.3390/bios15090616
Chicago/Turabian StyleBohlooli Darian, Saeed, Woo June Choi, Jeongmin Oh, and Jun Ki Kim. 2025. "Comparative Evaluation of Combined Denoising and Resolution Enhancement Algorithms for Intravital Two-Photon Imaging of Organs" Biosensors 15, no. 9: 616. https://doi.org/10.3390/bios15090616
APA StyleBohlooli Darian, S., Choi, W. J., Oh, J., & Kim, J. K. (2025). Comparative Evaluation of Combined Denoising and Resolution Enhancement Algorithms for Intravital Two-Photon Imaging of Organs. Biosensors, 15(9), 616. https://doi.org/10.3390/bios15090616






