Whole-Cell Bioreporter-Based Assay for Detecting Fungal-Derived β-Lactamase Inhibitors
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Bacterial Strains
2.3. Strain Growth Conditions
2.4. Bioluminescence Assay
2.5. Optimization of Bacterial Response to β-Lactam Antibiotics
2.6. Optimization of the Bioassay Using Commercial β-Lactamase Inhibitors
2.7. Collection of Coral-Associated Fungi
2.8. Fungal DNA Extraction and Sequence Analysis
2.9. Coral-Associated Fungal Processing
2.10. Using the Developed Screening Platform to Test Coral-Associated Fungi as BLI Producers
3. Results
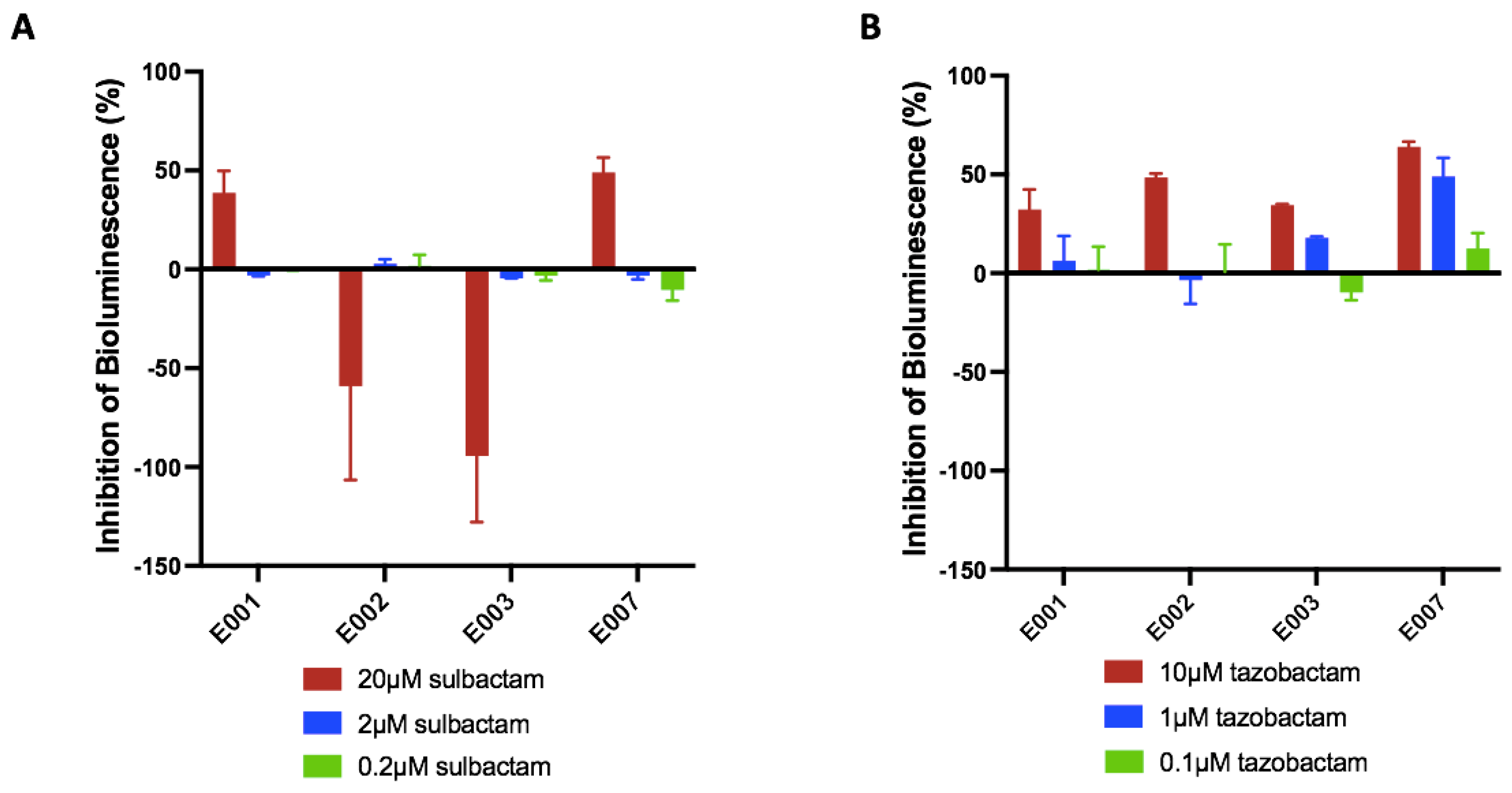
3.1. Bioassay Optimization, Validation, and Proof of Concept
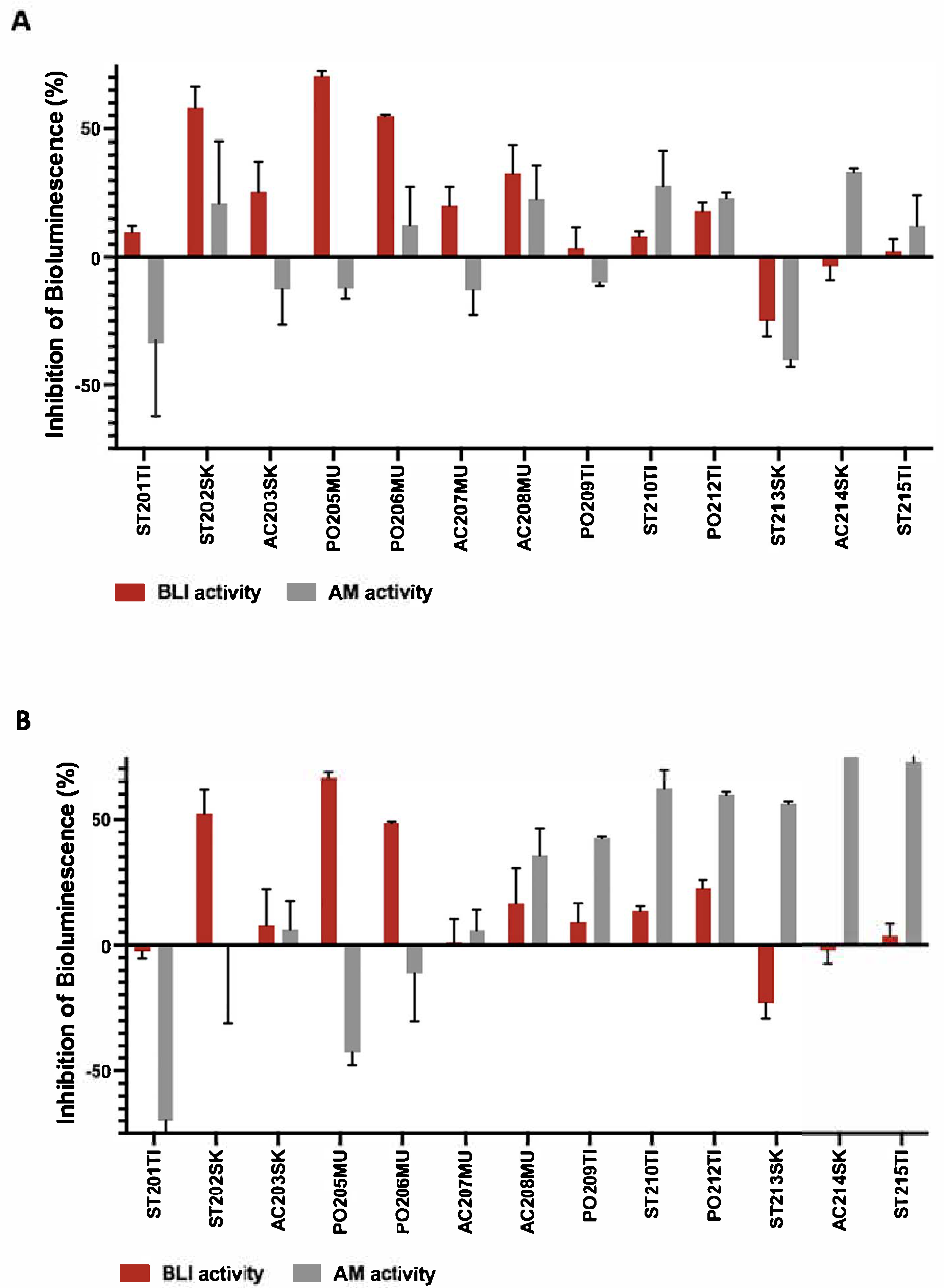
3.2. Detection of Potential β-Lactamase Inhibitor Agents from Fungal Isolates
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| XDR | Extensive drug-resistant |
| MDR | Multidrug-resistant |
| BLAs | β-lactam antibiotics |
| PBPs | Penicillin-binding proteins |
| ESBLs | Extended-spectrum β-lactamases |
| BLI | β-lactamases inhibitor |
| BLA | β-lactam antibiotic |
| MICs | Minimum inhibitory concentrations |
| LB | Luria–Bertani |
| RLU | Relative light units |
| DDW | Double-distilled water |
| IC50 | 50% inhibitory concentrations |
| PDA | Potato Dextrose agar |
| GPY | Glucose Peptone Yeast |
| ASW | Artificial seawater |
| cUTI | Complicated urinary tract |
| cIAI | Complicated intra-abdominal infections |
| HAP | Hospital-acquired pneumonia |
| VAP | Ventilator-associated pneumonia |
| HTS | High-throughput screening |
| NMR | Nuclear magnetic resonance |
| FBDD | Fragment-based drug discovery |
References
- Dabhi, M.; Patel, R.; Shah, V.; Soni, R.; Saraf, M.; Rawal, R.; Goswami, D. Penicillin-binding proteins: The master builders and breakers of bacterial cell walls and its interaction with β-lactam antibiotics. J. Proteins Proteom. 2024, 15, 215–232. [Google Scholar] [CrossRef]
- De Rosa, M.; Verdino, A.; Soriente, A.; Marabotti, A. The odd couple(s): An overview of beta-lactam antibiotics bearing more than one pharmacophoric group. Int. J. Mol. Sci. 2021, 22, 617. [Google Scholar] [CrossRef]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef]
- Husna, A.; Rahman, M.M.; Badruzzaman, A.; Sikder, M.H.; Islam, M.R.; Rahman, M.T.; Alam, J.; Ashour, H.M. Extended-spectrum β-lactamases (ESBL): Challenges and opportunities. Biomedicines 2023, 11, 2937. [Google Scholar] [CrossRef]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, G.; Del Giacomo, P.; Posteraro, B.; Sanguinetti, M.; Tumbarello, M. Molecular mechanisms, epidemiology, and clinical importance of β-lactam resistance in Enterobacteriaceae. Int. J. Mol. Sci. 2020, 21, 5090. [Google Scholar] [CrossRef] [PubMed]
- Perez-Llarena, F.J.; Bou, G. β-Lactamase inhibitors: The story so far. Curr. Med. Chem. 2009, 16, 3740–3765. [Google Scholar] [CrossRef] [PubMed]
- Sang, V.T.; Dat, T.T.H.; Vinh, L.B.; Cuong, L.C.V.; Oanh, P.T.T.; Ha, H.; Kim, Y.H.; Anh, H.L.T.; Yang, S.Y. Coral and coral-associated microorganisms: A prolific source of potential bioactive natural products. Mar. Drugs 2019, 17, 468. [Google Scholar] [CrossRef]
- Connelly, M.T.; Snyder, G.; Palacio-Castro, A.M.; Gillette, P.R.; Baker, A.C.; Traylor-Knowles, N. Antibiotics reduce Pocillopora coral-associated bacteria diversity, decrease holobiont oxygen consumption and activate immune gene expression. Mol. Ecol. 2023, 32, 4677–4694. [Google Scholar] [CrossRef]
- Mascuch, S.J.; Demko, A.; Viulu, S.; Ginigini, J.; Soapi, K.; Jensen, P.; Kubanek, J. Antibiotic activity altered by competitive interactions between two coral reef–associated bacteria. Microb. Ecol. 2023, 85, 1226–1235. [Google Scholar] [CrossRef]
- Sigwart, J.D.; Blasiak, R.; Jaspars, M.; Jouffray, J.-B.; Tasdemir, D. Unlocking the potential of marine biodiscovery. Nat. Prod. Rep. 2021, 38, 1235–1242. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Rahman, T.M.; Tharwat, N.A.; Abo El-Souad, S.M.; El-Beih, A.A.; El-Diwany, A.I. Biological activities and variation of symbiotic fungi isolated from Coral reefs collected from Red Sea in Egypt. Mycology 2020, 11, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Pang, X.; He, Y.; Lin, X.; Zhou, X.; Liu, Y.; Yang, B. Secondary metabolites from coral-associated fungi: Source, chemistry and bioactivities. J. Fungi 2022, 8, 1043. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Chen, J.; Kang, X.; Shen, X.; Lao, X.; Zheng, H. Approaches for the discovery of metallo-β-lactamase inhibitors: A review. Chem. Biol. Drug Des. 2019, 94, 1427–1440. [Google Scholar] [CrossRef] [PubMed]
- Babaoglu, K.; Simeonov, A.; Irwin, J.J.; Nelson, M.E.; Feng, B.; Thomas, C.J.; Cancian, L.; Costi, M.P.; Maltby, D.A.; Jadhav, A. Comprehensive mechanistic analysis of hits from high-throughput and docking screens against β-lactamase. J. Med. Chem. 2008, 51, 2502–2511. [Google Scholar] [CrossRef]
- Brown, E.D.; Wright, G.D. Antibacterial drug discovery in the resistance era. Nature 2016, 529, 336–343. [Google Scholar] [CrossRef]
- Hoess, R.H. Protein design and phage display. Chem. Rev. 2001, 101, 3205–3218. [Google Scholar] [CrossRef]
- Huang, W.; Beharry, Z.; Zhang, Z.; Palzkill, T. A broad-spectrum peptide inhibitor of β-lactamase identified using phage display and peptide arrays. Protein Eng. 2003, 16, 853–860. [Google Scholar] [CrossRef]
- Vella, P.; Hussein, W.M.; Leung, E.W.; Clayton, D.; Ollis, D.L.; Mitić, N.; Schenk, G.; McGeary, R.P. The identification of new metallo-β-lactamase inhibitor leads from fragment-based screening. Bioorganic Med. Chem. Lett. 2011, 21, 3282–3285. [Google Scholar] [CrossRef]
- Mureddu, L.; Vuister, G. Fragment-based drug discovery by NMR. Where are the successes and where can it be improved? Front. Mol. Biosci. 2022, 9, 834453. [Google Scholar] [CrossRef]
- Bian, Y.; Feng, Z.; Yang, P.; Xie, X.-Q. Integrated in silico fragment-based drug design: Case study with allosteric modulators on metabotropic glutamate receptor 5. AAPS J. 2017, 19, 1235–1248. [Google Scholar] [CrossRef]
- Belkin, S. Microbial whole-cell sensing systems of environmental pollutants. Curr. Opin. Microbiol. 2003, 6, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Michelini, E.; Cevenini, L.; Calabretta, M.M.; Spinozzi, S.; Camborata, C.; Roda, A. Field-deployable whole-cell bioluminescent biosensors: So near and yet so far. Anal. Bioanal. Chem. 2013, 405, 6155–6163. [Google Scholar] [CrossRef]
- Sagi, E.; Hever, N.; Rosen, R.; Bartolome, A.J.; Premkumar, J.R.; Ulber, R.; Lev, O.; Scheper, T.; Belkin, S. Fluorescence and bioluminescence reporter functions in genetically modified bacterial sensor strains. Sens. Actuators B Chem. 2003, 90, 2–8. [Google Scholar] [CrossRef]
- Davidov, Y.; Rozen, R.; Smulski, D.R.; Van Dyk, T.K.; Vollmer, A.C.; Elsemore, D.A.; LaRossa, R.A.; Belkin, S. Improved bacterial SOS promoter: Lux fusions for genotoxicity detection. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2000, 466, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Yagi, K. Applications of whole-cell bacterial sensors in biotechnology and environmental science. Appl. Microbiol. Biotechnol. 2007, 73, 1251–1258. [Google Scholar] [CrossRef]
- Chmelnitsky, I.; Carmeli, Y.; Leavitt, A.; Schwaber, M.J.; Navon-Venezia, S. CTX-M-2 and a new CTX-M-39 enzyme are the major extended-spectrum beta-lactamases in multiple Escherichia coli clones isolated in Tel Aviv, Israel. Antimicrob. Agents Chemother. 2005, 49, 4745–4750. [Google Scholar] [CrossRef]
- Schlesinger, J.; Navon-Venezia, S.; Chmelnitsky, I.; Hammer-Münz, O.; Leavitt, A.; Gold, H.S.; Schwaber, M.J.; Carmeli, Y. Extended-spectrum beta-lactamases among Enterobacter isolates obtained in Tel Aviv, Israel. Antimicrob. Agents Chemother. 2005, 49, 1150–1156. [Google Scholar] [CrossRef]
- Eltzov, E.; Pennybaker, S.; Shanit-Orland, M.; Marks, R.S.; Kushmaro, A. Multi-resistance as a tool for detecting novel beta-lactam antibiotics in the environment. Sens. Actuators B Chem. 2012, 174, 342–348. [Google Scholar] [CrossRef]
- Appelbaum, P.; Philippon, A.; Jacobs, M.; Spangler, S.; Gutmann, L. Characterization of beta-lactamases from non-Bacteroides fragilis group Bacteroides spp. belonging to seven species and their role in beta-lactam resistance. Antimicrob. Agents Chemother. 1990, 34, 2169–2176. [Google Scholar] [CrossRef]
- Zhang, S.; Liao, X.; Ding, T.; Ahn, J. Role of β-lactamase inhibitors as potentiators in antimicrobial chemotherapy targeting Gram-negative bacteria. Antibiotics 2024, 13, 260. [Google Scholar] [CrossRef]
- Paz, Z.; Komon-Zelazowska, M.; Druzhinina, I.; Aveskamp, M.; Shnaiderman, A.; Aluma, Y.; Carmeli, S.; Ilan, M.; Yarden, O. Diversity and potential antifungal properties of fungi associated with a Mediterranean sponge. Fungal Divers. 2010, 42, 17–26. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. 1990, 18, 315–322. [Google Scholar]
- Kalp, M.; Bethel, C.R.; Bonomo, R.A.; Carey, P.R. Why the extended-spectrum β-lactamases SHV-2 and SHV-5 are “hypersusceptible” to mechanism-based inhibitors. Biochemistry 2009, 48, 9912–9920. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.; Singh, A.; Sobia, F.; Patel, M.; Khan, H.M.; Malik, A.; Shukla, I. An overview of CTX-M β-lactamases. Rev. Res. Med. Microbiol. 2011, 22, 28–40. [Google Scholar] [CrossRef]
- Altmann, K.-H. Drugs from the oceans: Marine natural products as leads for drug discovery. Chimia 2017, 71, 646–652. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Hasan, S.; Ansari, M.I.; Ahmad, A.; Mishra, M. Major bioactive metabolites from marine fungi: A Review. Bioinformation 2015, 11, 176. [Google Scholar] [CrossRef]
- Torres, M.J.; Blanca, M. The complex clinical picture of β-lactam hypersensitivity: Penicillins, cephalosporins, monobactams, carbapenems, and clavams. Med. Clin. 2010, 94, 805–820. [Google Scholar] [CrossRef]
- Vasudevan, A.; Kesavan, D.K.; Wu, L.; Su, Z.; Wang, S.; Ramasamy, M.K.; Hopper, W.; Xu, H. In silico and in vitro screening of natural compounds as broad-spectrum β-lactamase inhibitors against Acinetobacter baumannii New Delhi metallo-β-lactamase-1 (NDM-1). BioMed Res. Int. 2022, 2022, 4230788. [Google Scholar] [CrossRef]
- Liu, S.; Zhou, Y.; Niu, X.; Wang, T.; Li, J.; Liu, Z.; Wang, J.; Tang, S.; Wang, Y.; Deng, X. Magnolol restores the activity of meropenem against NDM-1-producing Escherichia coli by inhibiting the activity of metallo-beta-lactamase. Cell Death Discov. 2018, 4, 28. [Google Scholar] [CrossRef] [PubMed]
- King, A.M.; Reid-Yu, S.A.; Wang, W.; King, D.T.; De Pascale, G.; Strynadka, N.C.; Walsh, T.R.; Coombes, B.K.; Wright, G.D. Aspergillomarasmine A overcomes metallo-β-lactamase antibiotic resistance. Nature 2014, 510, 503–506. [Google Scholar] [CrossRef] [PubMed]
- Maoluo, G.; Yufeng, L.; Yinlei, B.; Yan, G.; Li, L.; Rongmei, G.; Weiying, H.; Xuefu, Y.; Yuhuan, L.; Liyan, Y. Polyketides with New Delhi Metallo-β-lactamase 1 Inhibitory Activity from Penicillium sp. J. Nat. Prod. 2013, 76, 1535–1540. [Google Scholar]
- Soriano, A.; Carmeli, Y.; Omrani, A.S.; Moore, L.S.; Tawadrous, M.; Irani, P. Ceftazidime-avibactam for the treatment of serious gram-negative infections with limited treatment options: A systematic literature review. Infect. Dis. Ther. 2021, 10, 1989–2034. [Google Scholar] [CrossRef]
- Wu, G.; Cheon, E. Meropenem-vaborbactam for the treatment of complicated urinary tract infections including acute pyelonephritis. Expert Opin. Pharmacother. 2018, 19, 1495–1502. [Google Scholar] [CrossRef]
- Sellarès-Nadal, J.; Eremiev, S.; Burgos, J.; Almirante, B. An overview of cilastatin + imipenem + relebactam as a therapeutic option for hospital-acquired and ventilator-associated bacterial pneumonia: Evidence to date. Expert Opin. Pharmacother. 2021, 22, 1521–1531. [Google Scholar] [CrossRef]
- Carcione, D.; Siracusa, C.; Sulejmani, A.; Leoni, V.; Intra, J. Old and new beta-lactamase inhibitors: Molecular structure, mechanism of action, and clinical use. Antibiotics 2021, 10, 995. [Google Scholar] [CrossRef]
- Jungbluth, S.P.; Del Rio, T.G.; Tringe, S.G.; Stepanauskas, R.; Rappé, M.S. Genomic comparisons of a bacterial lineage that inhabits both marine and terrestrial deep subsurface systems. PeerJ 2017, 5, e3134. [Google Scholar] [CrossRef]
- Béjà, O.; Suzuki, M.T.; Koonin, E.V.; Aravind, L.; Hadd, A.; Nguyen, L.P.; Villacorta, R.; Amjadi, M.; Garrigues, C.; Jovanovich, S.B. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2000, 2, 516–529. [Google Scholar] [CrossRef]
- Canabal, R.; González-Bello, C. Chemical sensors for the early diagnosis of bacterial resistance to β-lactam antibiotics. Bioorganic Chem. 2024, 150, 107528. [Google Scholar] [CrossRef] [PubMed]
- Bebrone, C.; Moali, C.; Mahy, F.; Rival, S.; Docquier, J.D.; Rossolini, G.M.; Fastrez, J.; Pratt, R.F.; Frère, J.-M.; Galleni, M. CENTA as a chromogenic substrate for studying β-lactamases. Antimicrob. Agents Chemother. 2001, 45, 1868–1871. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Vosbeek, A.; Corbella, K.; Severson, J.; Schesser, J.; Sutton, L.D. A chromogenic cephalosporin for β-lactamase inhibitor screening assays. Anal. Biochem. 2012, 428, 96–98. [Google Scholar] [CrossRef] [PubMed]
- Makena, A.; van Berkel, S.S.; Lejeune, C.; Owens, R.J.; Verma, A.; Salimraj, R.; Spencer, J.; Brem, J.; Schofield, C.J. Chromophore-Linked Substrate (CLS405): Probing Metallo-β-Lactamase Activity and Inhibition. ChemMedChem 2013, 8, 1923–1929. [Google Scholar] [CrossRef]
- Klingler, F.-M.; Proschak, E. Approved Drugs containing Thiols as Inhibitors of Metallo-β-Lactamases: A Strategy to Combat Multidrug-Resistant Bacteria: PB-014. Protein Sci. 2015, 24, 10–11. [Google Scholar]
- van Berkel, S.S.; Brem, J.; Rydzik, A.M.; Salimraj, R.; Cain, R.; Verma, A.; Owens, R.J.; Fishwick, C.W.; Spencer, J.; Schofield, C.J. Assay platform for clinically relevant metallo-β-lactamases. J. Med. Chem. 2013, 56, 6945–6953. [Google Scholar] [CrossRef]
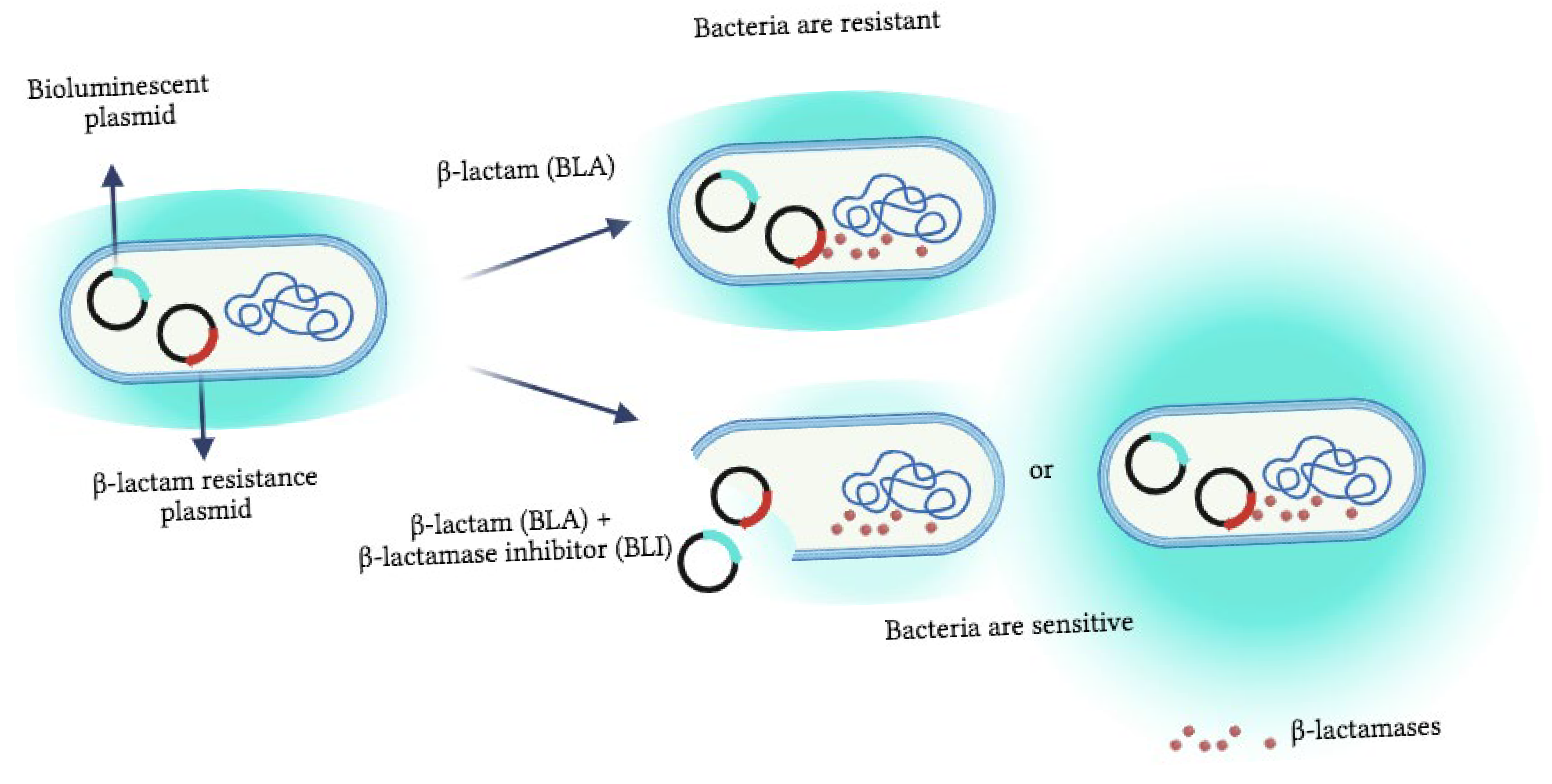
| Strains | Bioluminescent Plasmids | β-Lactam Resistance Gene |
|---|---|---|
| E001 | recA::lux | blaCTX-M-2 |
| E002 | recA::lux | blaSHV-12 |
| E003 | grpE::lux | blaSHV-12 |
| E007 | grpE::lux | blaCTX-M-2 |
| Isolate Name | Source Coral Genus | Medium | Most Related Strains by BLAST + Accession Number + Identity (%) |
|---|---|---|---|
| ST201TI | Stylophora (coral No. 1) | GPY + ASW | Cladosporium sphaerospermum KP701988.1 (100%) |
| ST210TI | Stylophora No. 1 | PDA + ASW | Cladosporium sphaerospermum KP701988.1 (100%) |
| PO209TI | Pocillopora No. 1 | PDA + Bupirimate + ASW | Cladosporium sphaerospermum KP701988.1 (100%) |
| ST202SK | Stylophora No. 2 | GPY + ASW | Aspergillus terreus KC119206.1 (100%) |
| ST213SK | Stylophora No. 2 | Czapex-Dox + ASW | Aspergillus terreus KC119206.1 (100%) |
| AC203SK | Acropora No. 1 | Malt Extract + ASW | Cladosporium cladosporioides AJ300335.1 (100%) |
| PO205TI | Pocillopora No. 1 | GPY + ASW | Alternaria alternata KU182490.1 (100%) |
| PO206MU | Pocillopora No. 2 | GPY + ASW | Penicillium spinulosum KF646101.1 (99%) |
| ST215TI | Stylophora No. 2 | Czapex-Dox + ASW | Penicillium spinulosum KF646101.1 (100%) |
| AC207MU | Acropora No. 2 | Malt Extract + ASW | Cladosporium halotolerans KP701958.1 (100%) |
| AC208MU | Acropora No. 2 | Malt Extract + ASW | Cladosporium perangustum KP701968.1 (99%) |
| PO212TI | Pocillopora No. 1 | PDA+ ASW | Cladosporium cladosporioides KU182497.1 (100%) |
| AC214SK | Acropora No. 1 | Czapex-Dox + ASW | Cladosporium cladosporioides KU182497.1 (100%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benou, R.; Marks, R.S.; Sivan, A.; Kramarsky-Winter, E.; Golberg, K.; Kushmaro, A. Whole-Cell Bioreporter-Based Assay for Detecting Fungal-Derived β-Lactamase Inhibitors. Biosensors 2025, 15, 594. https://doi.org/10.3390/bios15090594
Benou R, Marks RS, Sivan A, Kramarsky-Winter E, Golberg K, Kushmaro A. Whole-Cell Bioreporter-Based Assay for Detecting Fungal-Derived β-Lactamase Inhibitors. Biosensors. 2025; 15(9):594. https://doi.org/10.3390/bios15090594
Chicago/Turabian StyleBenou, Raz, Robert S. Marks, Alex Sivan, Esti Kramarsky-Winter, Karina Golberg, and Ariel Kushmaro. 2025. "Whole-Cell Bioreporter-Based Assay for Detecting Fungal-Derived β-Lactamase Inhibitors" Biosensors 15, no. 9: 594. https://doi.org/10.3390/bios15090594
APA StyleBenou, R., Marks, R. S., Sivan, A., Kramarsky-Winter, E., Golberg, K., & Kushmaro, A. (2025). Whole-Cell Bioreporter-Based Assay for Detecting Fungal-Derived β-Lactamase Inhibitors. Biosensors, 15(9), 594. https://doi.org/10.3390/bios15090594







