An Isothermal Deoxyribozyme Sensor for Rapid Detection of Enteroviral RNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Bioinformatics Analysis
2.3. Positive Control Construction
2.4. Deoxyribozyme-Based Biosensor Assay
2.5. Reaction Buffers and Cofactor Optimization
2.6. Determination of Detection and Quantification Limits
2.7. Selectivity Assessment
2.8. Viral Isolation and Nucleic Acid Sequence-Based Amplification (NASBA) Integration
3. Results
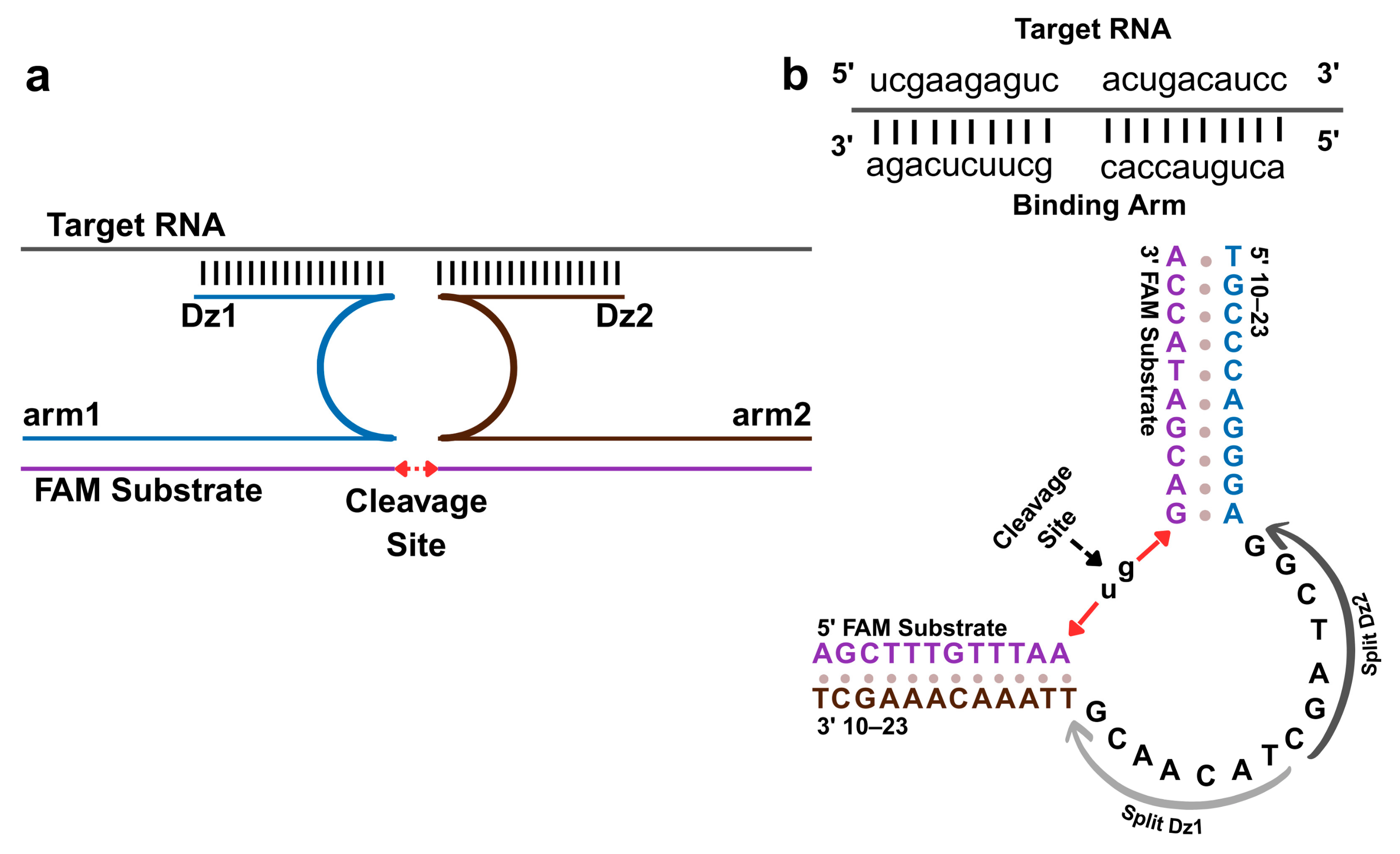
3.1. Target-Induced Reassembly of Split DNAzyme Biosensor
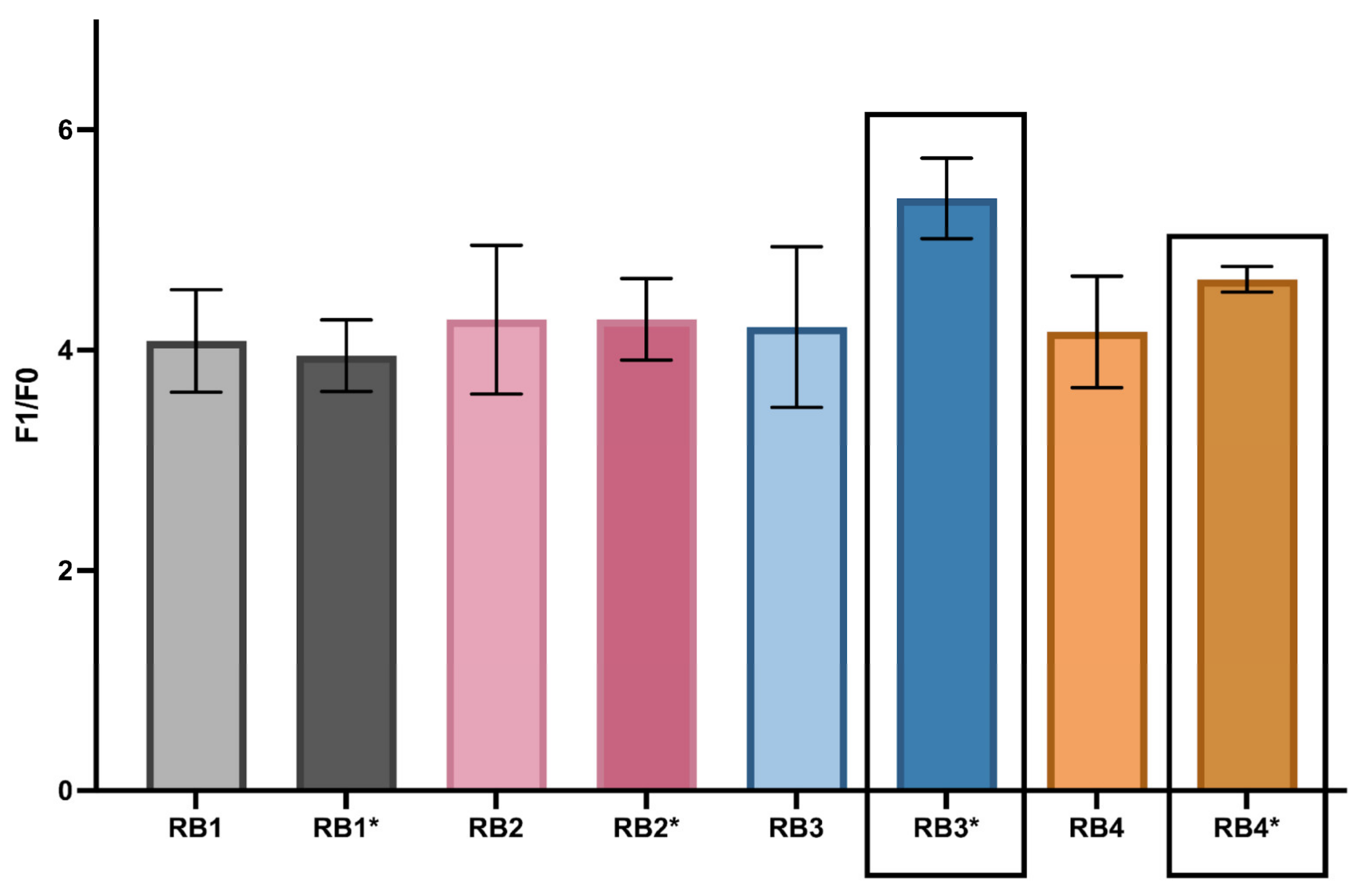
3.2. Buffer Optimization Enhances DNAzyme Catalytic Efficiency
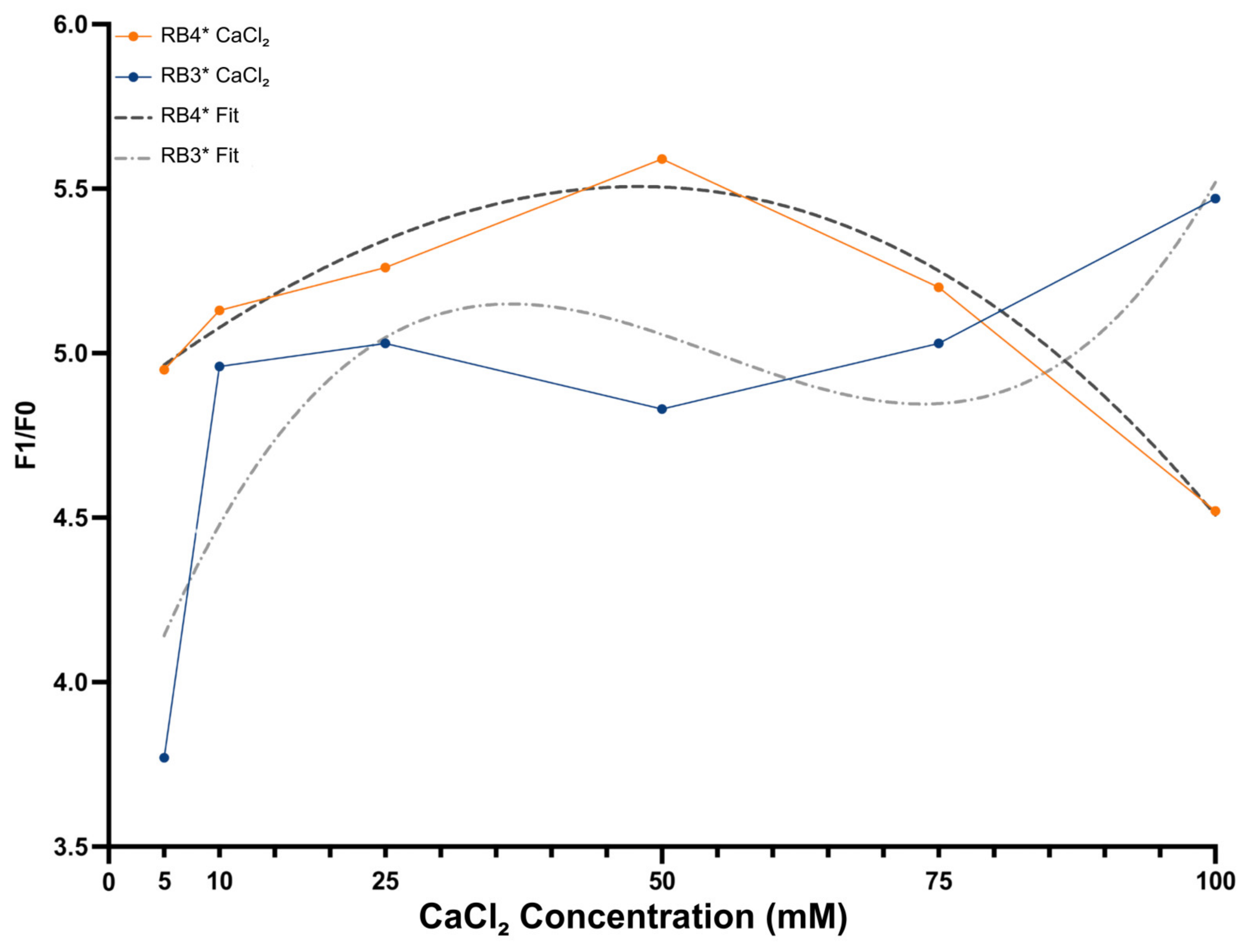
3.3. Calcium-Dependent Catalytic Enhancement
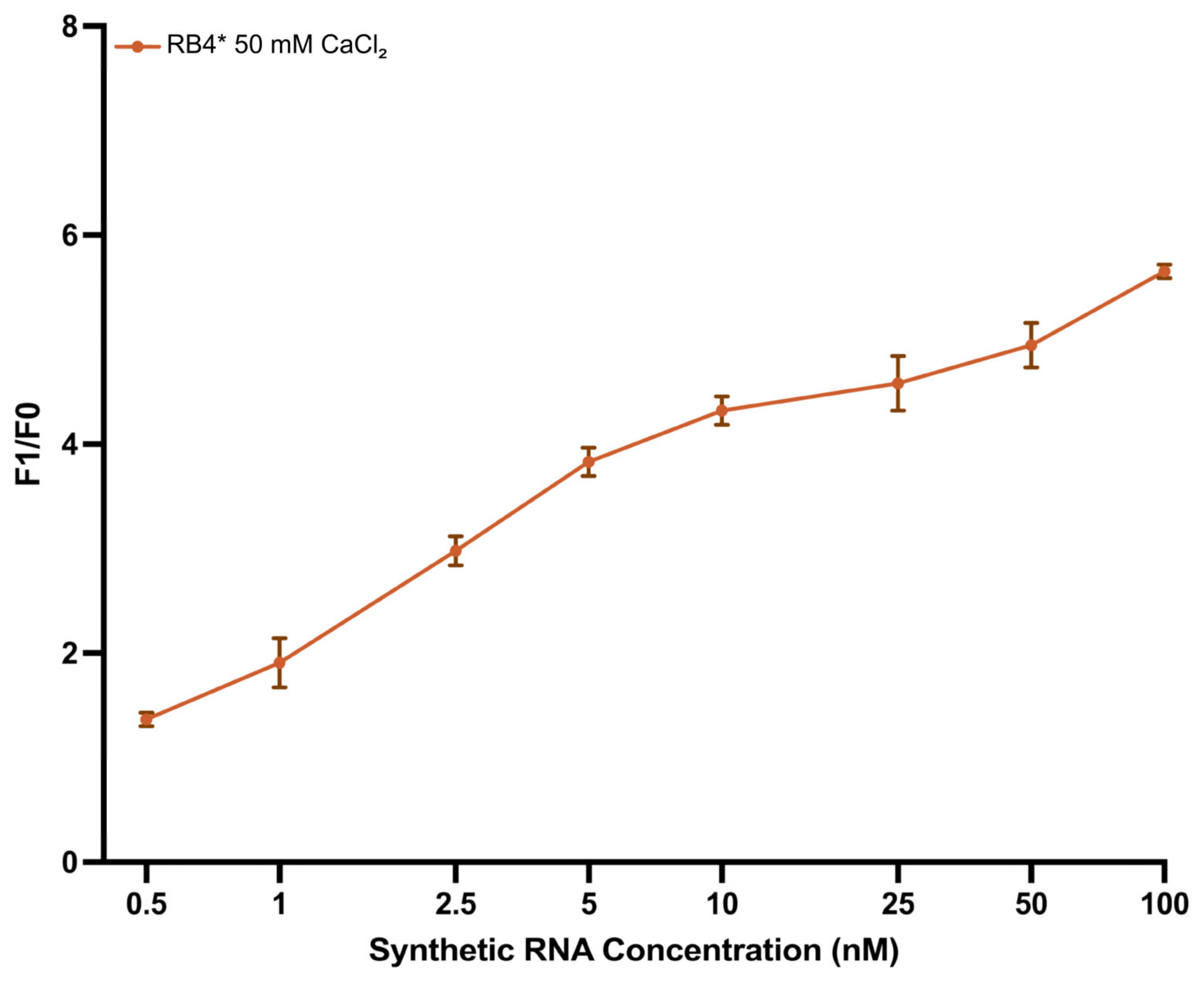
3.4. Quantitative Performance of the DNAzyme Biosensor
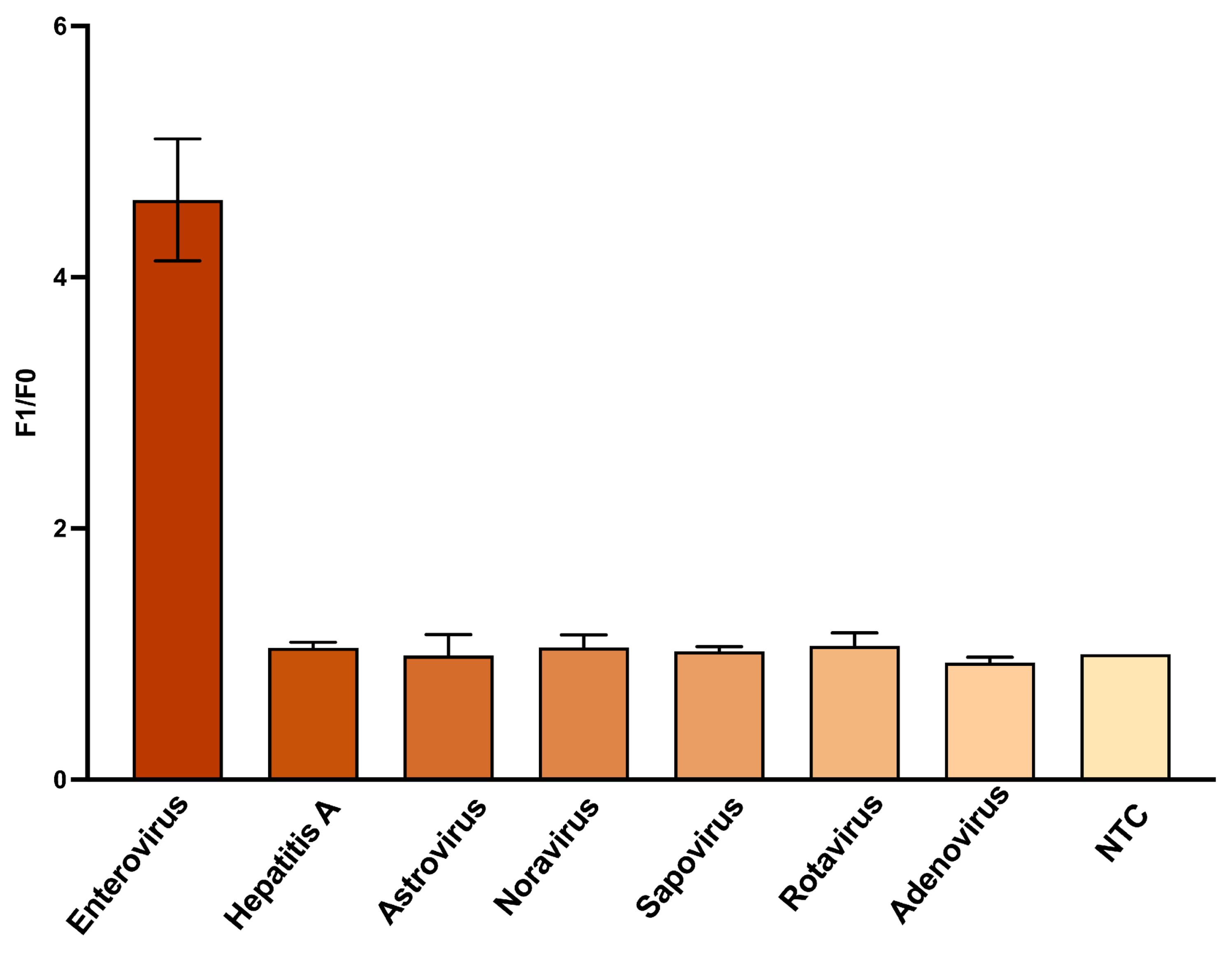
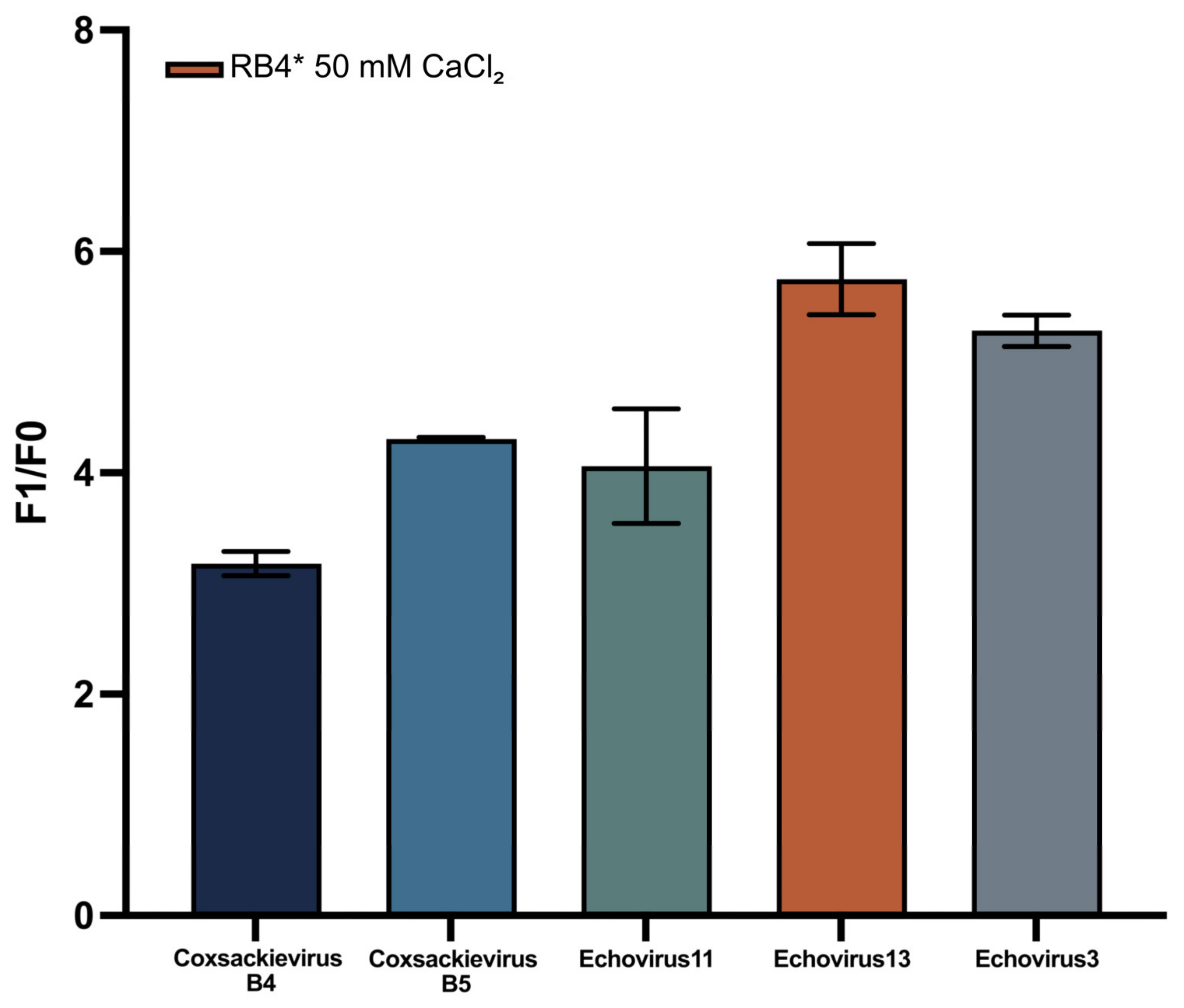
3.5. Sequence Specificity of the DNAzyme Biosensor
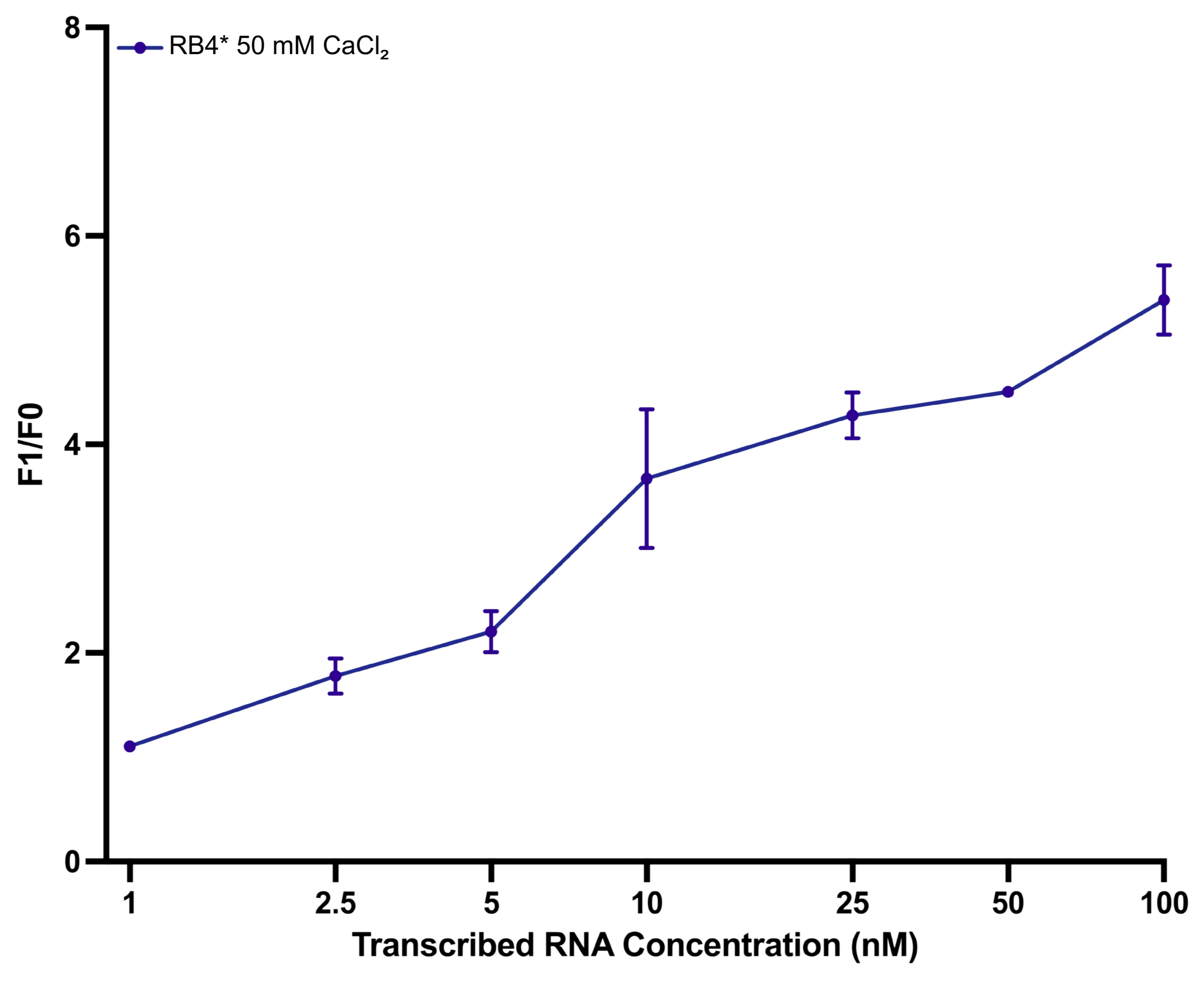
3.6. Sensitivity Analysis Using In Vitro Transcribed Enterovirus RNA
3.7. Adapted NASBA Strategy Enhances DNAzyme-Based Enterovirus Detection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Olaimat, A.N.; Taybeh, A.O.; Al-Nabulsi, A.; Al-Holy, M.; Hatmal, M.M.; Alzyoud, J.; Aolymat, I.; Abughoush, M.H.; Shahbaz, H.; Alzyoud, A.; et al. Common and potential emerging foodborne viruses: A comprehensive review. Life 2024, 14, 190. [Google Scholar] [CrossRef] [PubMed]
- Upfold, N.S.; Luke, G.A.; Knox, C. Occurrence of human enteric viruses in water sources and shellfish: A focus on Africa. Food Environ. Virol. 2021, 13, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Lanrewaju, A.A.; Enitan-Folami, A.M.; Sabiu, S.; Edokpayi, J.N.; Swalaha, F.M. Global public health implications of human exposure to viral contaminated water. Front. Microbiol. 2022, 13, 981896. [Google Scholar] [CrossRef] [PubMed]
- Chandran, S.; Gibson, K.E. Improving the detection and understanding of infectious human norovirus in food and water matrices: A review of methods and emerging models. Viruses 2024, 16, 776. [Google Scholar] [CrossRef]
- Kristanti, R.A.; Hadibarata, T.; Syafrudin, M.; Yılmaz, M.; Abdullah, S. Microbiological contaminants in drinking water: Current status and challenges. Water Air Soil Pollut. 2022, 233, 17. [Google Scholar] [CrossRef]
- Chen, L.; Deng, Y.; Dong, S.; Wang, H.; Li, P.; Zhang, H.; Chu, W. The occurrence and control of waterborne viruses in drinking water treatment: A review. Chemosphere 2021, 281, 130728. [Google Scholar] [CrossRef]
- Burnett, E.; Parashar, U.D.; Winn, A.; Tate, J.E. Trends in rotavirus laboratory detections and internet search volume before and after rotavirus vaccine introduction and in the context of the coronavirus disease 2019 pandemic—United States, 2000–2021. J. Infect. Dis. 2022, 226, 967–974. [Google Scholar] [CrossRef]
- Mehetre, G.T.; Leo, V.V.; Singh, G.; Sorokan, A.; Maksimov, I.; Yadav, M.K.; Upadhyaya, K.; Hashem, A.; Alsaleh, A.N.; Dawoud, T.M.; et al. Current developments and challenges in plant viral diagnostics: A systematic review. Viruses 2021, 13, 412. [Google Scholar] [CrossRef]
- Pilevar, M.; Kim, K.T.; Lee, W.H. Recent advances in biosensors for detecting viruses in water and wastewater. J. Hazard. Mater. 2021, 410, 124656. [Google Scholar] [CrossRef]
- Song, X.; Fredj, Z.; Zheng, Y.; Zhang, H.; Rong, G.; Bian, S.; Sawan, M. Biosensors for waterborne virus detection: Challenges and strategies. J. Pharm. Anal. 2023, 13, 1252–1268. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, H.; Song, G.; Huang, K.; Luo, Y.; Liu, Q.; He, X.; Cheng, N. Intelligent biosensing strategies for rapid detection in food safety: A review. Biosens. Bioelectron. 2022, 202, 114003. [Google Scholar] [CrossRef]
- Curulli, A. Electrochemical biosensors in food safety: Challenges and perspectives. Molecules 2021, 26, 2940. [Google Scholar] [CrossRef] [PubMed]
- Kabay, G.; DeCastro, J.; Altay, A.; Smith, K.; Lu, H.W.; Capossela, A.M.D.; Moarefian, M.; Aran, K.; Dincer, C. Emerging biosensing technologies for the diagnostics of viral infectious diseases. Adv. Mater. 2022, 34, 2201085. [Google Scholar] [CrossRef] [PubMed]
- Walsh, A.T.; Triant, D.A.; Le Tourneau, J.J.; Shamimuzzaman, M.; Elsik, C.G. Hymenoptera genome database: New genomes and annotation datasets for improved GO enrichment and orthologue analyses. Nucleic Acids Res. 2022, 50, D1032–D1039. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Wayment-Steele, H.K.; Kladwang, W.; Strom, A.I.; Lee, J.; Treuille, A.; Becka, A.; Das, R. RNA secondary structure packages evaluated and improved by high-throughput experiments. Nat. Methods 2022, 19, 1234–1242. [Google Scholar] [CrossRef]
- Li, Y.; Liu, K.; Wang, B.; Liu, Z.; Yang, C.; Wang, J.; Ma, X.; Li, H.; Sun, C. Engineering DNAzyme strategies for fluorescent detection of lead ions based on RNA cleavage-propelled signal amplification. J. Hazard. Mater. 2022, 440, 129712. [Google Scholar] [CrossRef]
- Khan, S.; Burciu, B.; Filipe, C.D.M.; Li, Y.; Dellinger, K.; Didar, T.F. DNAzyme-based biosensors: Immobilization strategies, applications, and future prospective. ACS Nano 2021, 15, 13943–13969. [Google Scholar] [CrossRef]
- Kirichenko, A.; Bryushkova, E.; Dedkov, V.; Dolgova, A. A novel DNAzyme-based fluorescent biosensor for detection of RNA-containing Nipah henipavirus. Biosensors 2023, 13, 252. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, H.; Qian, S.; Yu, X.; Chen, H.; Wu, J. Applying CRISPR/Cas system as a signal enhancer for DNAzyme-based lead ion detection. Anal. Chim. Acta 2022, 1192, 339356. [Google Scholar] [CrossRef]
- Wang, H.; Shen, M.; Shen, X.; Liu, J.; Huang, W.; Jiang, X.; Liu, H.; Zeng, S.; Nan, K.; Cai, S. An enzyme-free sensing platform for miRNA detection and in situ imaging in clinical samples based on DNAzyme cleavage-triggered catalytic hairpin assembly. Biosens. Bioelectron. 2024, 256, 116279. [Google Scholar] [CrossRef]
- Li, F.; Song, N.; Dong, Y.; Li, S.; Li, L.; Liu, Y.; Li, Z.; Yang, D. A proton-activatable DNA-based nanosystem enables co-delivery of CRISPR/Cas9 and DNAzyme for combined gene therapy. Angew. Chem. Int. Ed. 2022, 61, e202116569. [Google Scholar] [CrossRef]
- Wang, D.; Chen, G.; Lyu, Y.; Feng, E.; Zhu, L.; Pan, C.; Zhang, W.; Liu, X.; Wang, H. A CRISPR/Cas12a-based DNAzyme visualization system for rapid, non-electrically dependent detection of Bacillus anthracis. Emerg. Microbes Infect. 2022, 11, 428–437. [Google Scholar] [CrossRef]
- Su, J.; Sun, C.; Du, J.; Xing, X.; Wang, F.; Dong, H. RNA-cleaving DNAzyme-based amplification strategies for biosensing and therapy. Adv. Healthc. Mater. 2023, 12, 2300367. [Google Scholar] [CrossRef]
- Jia, G.; Xu, X.; Guo, Z.; Hu, X.; Pan, Q.; Zhu, H.; Wang, Y. A rapid and high sensitivity RNA detection based on NASBA and G4-ThT fluorescent biosensor. Sci. Rep. 2022, 12, 14107. [Google Scholar] [CrossRef]
- Fu, J.; Zhang, Q.; Liu, S.; Puyat, D.; Shah, A.; Ebrahimimojarad, A.; Oh, S.W. Rapid amplification and detection of single-stranded nucleic acids for point-of-care diagnosis. Small Methods 2025, 9, 2401733. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Polio Laboratory Manual; WHO: Geneva, Switzerland, 2004. [Google Scholar]
- Nix, W.A.; Oberste, M.S.; Pallansch, M.A. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J. Clin. Microbiol. 2006, 44, 2698–2704. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Li, J.; Salena, B.J.; Li, Y. Aptamer- and DNAzyme-based colorimetric biosensors for pathogen detection. Angew. Chem. Int. Ed. 2025, 64, e202418725. [Google Scholar] [CrossRef]
- Kolpashchikov, D.M.; Gerasimova, Y.V. Cleavage of structured RNAs is accelerated by high affinity DNAzyme agents. ChemBioChem 2025, 26, e202400950. [Google Scholar] [CrossRef]
- Zhang, W.; He, Y.; Feng, Z.; Zhang, J. Recent advances of functional nucleic acid-based sensors for point-of-care detection of SARS-CoV-2. Microchim. Acta 2022, 189, 18. [Google Scholar] [CrossRef]
- Wang, F.; Duan, Y.; Zhang, Y.; Du, Y.; Zheng, Y.; Shi, J.; Tong, X.; Peng, W.; Zhou, C. Ultra-sensitive detection of femtomolar lead ions using DNAzyme-mediated biosensing on tilted fiber Bragg grating plasmonic sensor. Microchem. J. 2024, 204, 111131. [Google Scholar] [CrossRef]
- Ahsan, M.; Pindi, C.; Palermo, G. Emerging mechanisms of metal-catalyzed RNA and DNA modifications. Annu. Rev. Phys. Chem. 2025, 76, 497–518. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Ma, C.; Tang, Z.; Chen, M.; Zhao, H. A novel fluorescent assay based on DNAzyme-assisted detection of prostate specific antigen for signal amplification. Anal. Chim. Acta 2020, 1104, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Yang, D.; Ma, X.; Pan, L.; Shao, Y.; Arya, G.; Ke, Y.; Zhang, C.; Wang, F.; Zuo, X.; et al. A programmable DNAzyme for the sensitive detection of nucleic acids. Angew. Chem. Int. Ed. 2024, 63, e202320179. [Google Scholar] [CrossRef]
- Bubba, L.; Benschop, K.S.M.; Blomqvist, S.; Duizer, E.; Martin, J.; Shaw, A.G.; Bailly, J.L.; Rasmussen, L.D.; Baicus, A.; Fischer, T.K.; et al. Wastewater Surveillance in Europe for Non-Polio Enteroviruses and Beyond. Microorganisms 2023, 11, 2496. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Şaş, B.; Kirichenko, A.D.; Kapitonova, M.A.; Shabalina, A.V.; Kanaeva, O.I.; El-Messery, T.M.; Dedkov, V.G.; Dolgova, A.S. An Isothermal Deoxyribozyme Sensor for Rapid Detection of Enteroviral RNA. Biosensors 2025, 15, 562. https://doi.org/10.3390/bios15090562
Şaş B, Kirichenko AD, Kapitonova MA, Shabalina AV, Kanaeva OI, El-Messery TM, Dedkov VG, Dolgova AS. An Isothermal Deoxyribozyme Sensor for Rapid Detection of Enteroviral RNA. Biosensors. 2025; 15(9):562. https://doi.org/10.3390/bios15090562
Chicago/Turabian StyleŞaş, Begüm, Anastasiia Dmitrievna Kirichenko, Marina Anatolyevna Kapitonova, Anna Vyacheslavovna Shabalina, Olga Ilyinichna Kanaeva, Tamer Mohammed El-Messery, Vladimir Georgievich Dedkov, and Anna Sergeevna Dolgova. 2025. "An Isothermal Deoxyribozyme Sensor for Rapid Detection of Enteroviral RNA" Biosensors 15, no. 9: 562. https://doi.org/10.3390/bios15090562
APA StyleŞaş, B., Kirichenko, A. D., Kapitonova, M. A., Shabalina, A. V., Kanaeva, O. I., El-Messery, T. M., Dedkov, V. G., & Dolgova, A. S. (2025). An Isothermal Deoxyribozyme Sensor for Rapid Detection of Enteroviral RNA. Biosensors, 15(9), 562. https://doi.org/10.3390/bios15090562







