Kinetic Analysis of SARS-CoV-2 S1–Integrin Binding Using Live-Cell, Label-Free Optical Biosensing
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Sample Preparation
2.2. S1 Coating Measurement
2.3. S1 Protein Inhibition Assay
2.4. Basic Sensing Mechanism of the Epic BT RWG Biosensor System
2.5. Assessment of S1 Surface-Density-Dependent Adhesion
2.6. Statistics and Kinetic Analysis
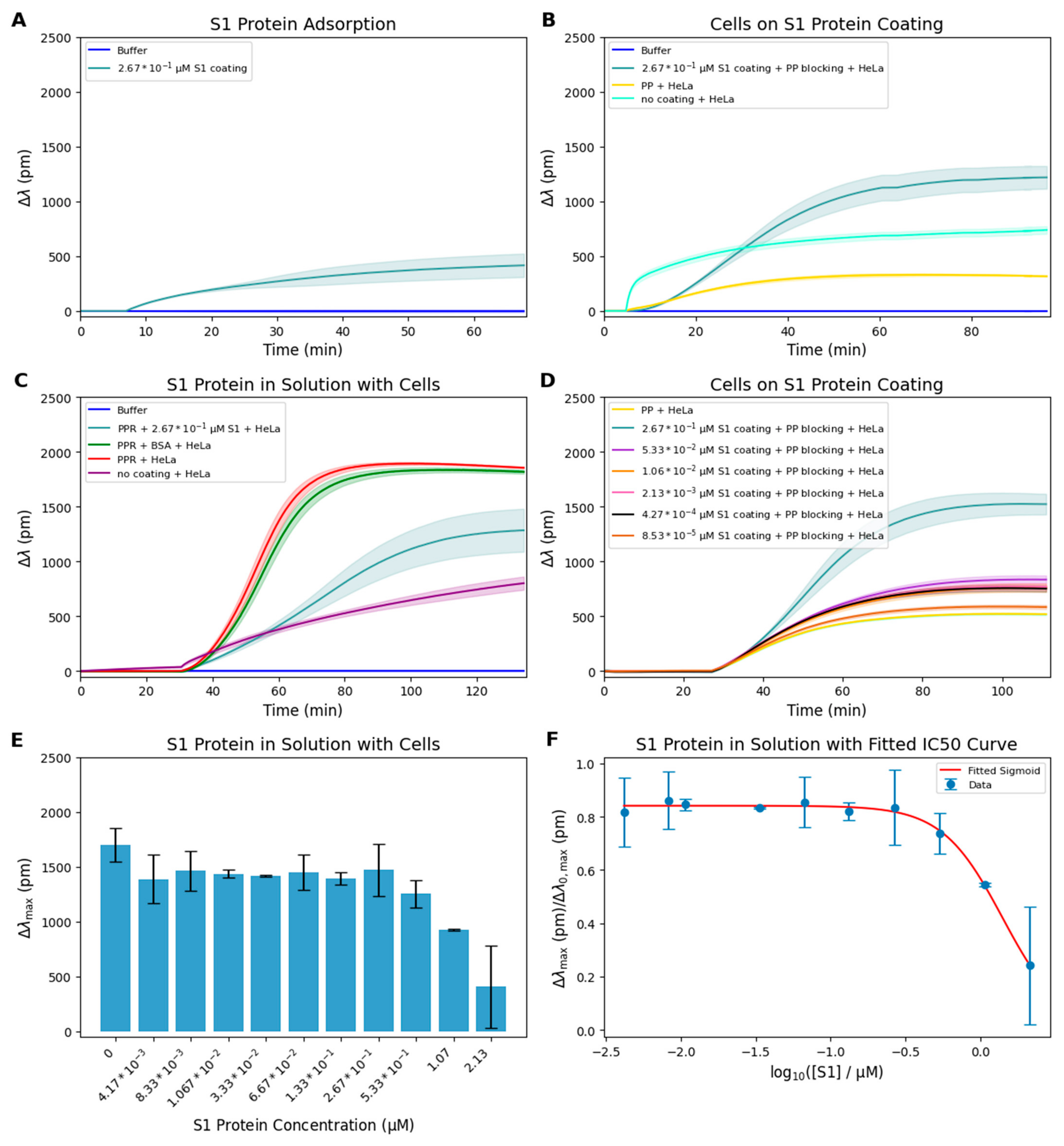
3. Results and Discussion
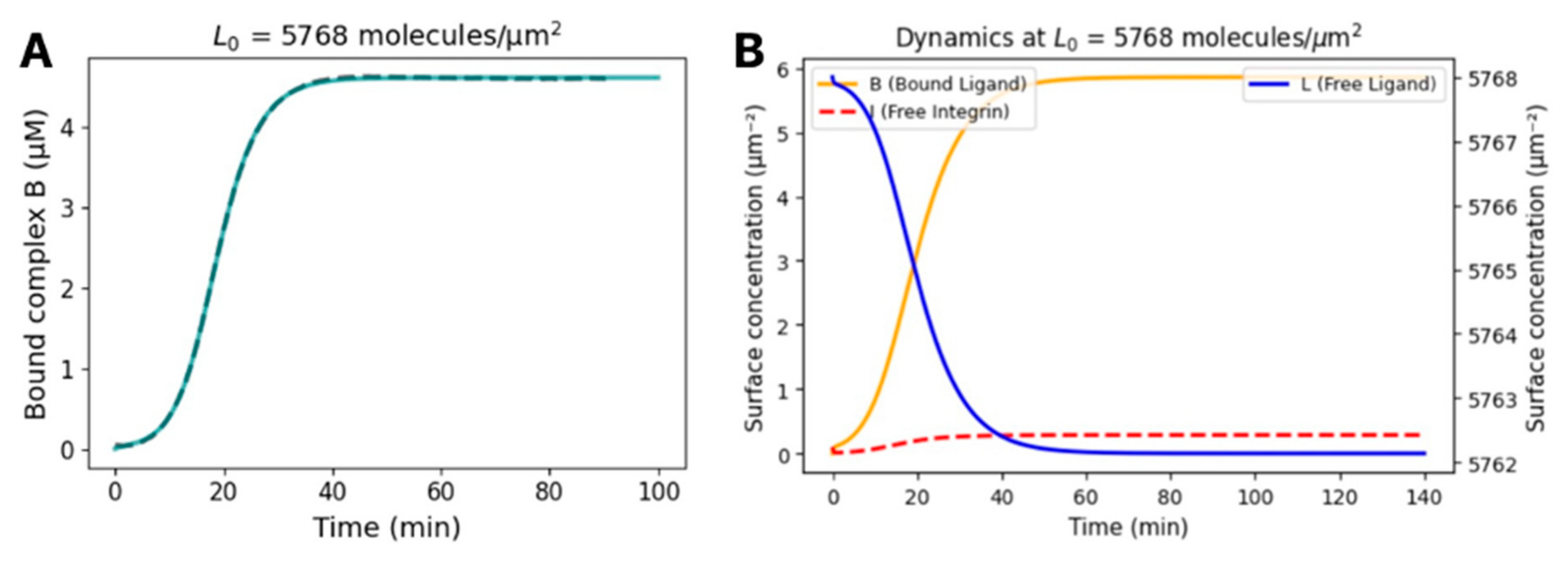
Kinetic Modeling and Prediction of the Dissociation Constant of S1-Integrin Interaction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Paszek, M.J.; Boettiger, D.; Weaver, V.M.; Hammer, D.A. Integrin clustering is driven by mechanical resistance from the glycocalyx and the substrate. PLoS Comput. Biol. 2009, 5, e1000604. [Google Scholar] [CrossRef]
- Orgovan, N.; Peter, B.; Bosze, S.; Ramsden, J.J.; Szabó, B.; Horvath, R. Dependence of cancer cell adhesion kinetics on integrin ligand surface density measured by a high-throughput label-free resonant waveguide grating biosensor. Sci. Rep. 2014, 4, 4034. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Valero-Rello, A.; Baeza-Delgado, C.; Andreu-Moreno, I.; Sanjuán, R. Cellular receptors for mammalian viruses. PLoS Pathog. 2024, 20, e1012021. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, T.D.; Bang-Christensen, S.R.; Jørgensen, A.M.; Løppke, C.; Spliid, C.B.; Sand, N.T.; Clausen, T.M.; Salanti, A.; Agerbæk, M.O. The Role of Proteoglycans in Cancer Metastasis and Circulating Tumor Cell Analysis. Front. Cell Dev. Biol. 2020, 8, 749. [Google Scholar] [CrossRef] [PubMed]
- Bhella, D. The role of cellular adhesion molecules in virus attachment and entry. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140035. [Google Scholar] [CrossRef]
- Sigrist, C.J.; Bridge, A.; Le Mercier, P. A potential role for integrins in host cell entry by SARS-CoV-2. Antivir. Res. 2020, 177, 104759. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Liu, J.; Lu, F.; Chen, Y.; Plow, E.; Qin, J. Integrin mediates cell entry of the SARS-CoV-2 virus independent of cellular receptor ACE2. J. Biol. Chem. 2022, 298, 101710. [Google Scholar] [CrossRef]
- Bugatti, A.; Filippini, F.; Bardelli, M.; Zani, A.; Chiodelli, P.; Messali, S.; Caruso, A.; Caccuri, F. SARS-CoV-2 Infects Human ACE2-Negative Endothelial Cells through an αvβ3 Integrin-Mediated Endocytosis Even in the Presence of Vaccine-Elicited Neutralizing Antibodies. Viruses 2022, 14, 705. [Google Scholar] [CrossRef]
- Norris, E.G.; Pan, X.S.; Hocking, D.C. Receptor-binding domain of SARS-CoV-2 is a functional αv-integrin agonist. J. Biol. Chem. 2023, 299, 102922. [Google Scholar] [CrossRef]
- Jiang, B.; Yang, Y.; Zhao, R.; Chen, D.; Wang, Y.; Liu, J.; Long, F.; Chen, R.; Hao, R. A multifunctional evanescent wave biosensor for the universal assay of SARS-CoV-2 variants and affinity analysis of coronavirus spike protein-hACE2 interactions. Biosens. Bioelectron. 2024, 260, 116426. [Google Scholar] [CrossRef]
- Sivagnanam, S.; Das, K.; Pan, I.; Stewart, A.; Barik, A.; Maity, B.; Das, P. Engineered triphenylphosphonium-based, mitochondrial-targeted liposomal drug delivery system facilitates cancer cell killing actions of chemotherapeutics. RSC Chem. Biol. 2023, 5, 236–248. [Google Scholar] [CrossRef]
- Tekin, Y.S.; Kul, S.M.; Sagdic, O.; Rodthongkum, N.; Geiss, B.; Ozer, T. Optical biosensors for diagnosis of COVID-19: Nanomaterial-enabled particle strategies for post pandemic era. Microchim. Acta 2024, 191, 320. [Google Scholar] [CrossRef]
- Kanyo, N.; Kovacs, K.D.; Saftics, A.; Szekacs, I.; Peter, B.; Santa-Maria, A.R.; Walter, F.R.; Dér, A.; Deli, M.A.; Horvath, R. Glycocalyx regulates the strength and kinetics of cancer cell adhesion revealed by biophysical models based on high resolution label-free optical data. Sci. Rep. 2020, 10, 22422. [Google Scholar] [CrossRef]
- Szekacs, I.; Farkas, E.; Gemes, B.L.; Takacs, E.; Szekacs, A.; Horvath, R. Integrin targeting of glyphosate and its cell adhesion modulation effects on osteoblastic MC3T3-E1 cells revealed by label-free optical biosensing. Sci. Rep. 2018, 8, 17401. [Google Scholar] [CrossRef]
- Maginnis, M.S.; Forrest, J.C.; Kopecky-Bromberg, S.A.; Dickeson, S.K.; Santoro, S.A.; Zutter, M.M.; Nemerow, G.R.; Bergelson, J.M.; Dermody, T.S. β1 Integrin Mediates Internalization of Mammalian Reovirus. J. Virol. 2006, 80, 2760–2770. [Google Scholar] [CrossRef] [PubMed]
- Santos, P.A.; Rocha, C.S.; Baptista, M.S. Adhesion and proliferation of HeLa and fibroblast cells on chemically-modified gold surfaces. Colloids Surf. B Biointerfaces 2014, 123, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Kanyo, N.; Kovács, K.D.; Kovács, S.V.; Béres, B.; Peter, B.; Székács, I.; Horvath, R. Single-cell adhesivity distribution of glycocalyx digested cancer cells from high spatial resolution label-free biosensor measurements. Matrix Biol. Plus 2022, 14, 100103. [Google Scholar] [CrossRef]
- Peter, B.; Lagzi, I.; Teraji, S.; Nakanishi, H.; Cervenak, L.; Zámbó, D.; Deák, A.; Molnár, K.; Truszka, M.; Szekacs, I.; et al. Interaction of Positively Charged Gold Nanoparticles with Cancer Cells Monitored by an in Situ Label-Free Optical Biosensor and Transmission Electron Microscopy. ACS Appl. Mater. Interfaces 2018, 10, 26841–26850. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C. Kinetics and mechanics of cell adhesion. J. Biomech. 2000, 33, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Zarnitsyna, V.I.; Zhu, C. Adhesion frequency assay for in situ kinetics analysis of cross-junctional molecular interactions at the cell-cell interface. J. Vis. Exp. 2011, 45, e3519. [Google Scholar] [CrossRef]
- Orgovan, N.; Kovacs, B.; Farkas, E.; Szabó, B.; Zaytseva, N.; Fang, Y.; Horvath, R. Bulk and surface sensitivity of a resonant waveguide grating imager. Appl. Phys. Lett. 2014, 104, 083506. [Google Scholar] [CrossRef]
- Stuible, M.; Gervais, C.; Lord-Dufour, S.; Perret, S.; L’Abbé, D.; Schrag, J.; St-Laurent, G.; Durocher, Y. Rapid, high-yield production of full-length SARS-CoV-2 spike ectodomain by transient gene expression in CHO cells. J. Biotechnol. 2021, 326, 21–27. [Google Scholar] [CrossRef]
- Hook, F.F.; Vörös, J.; Rodahl, M.; Kurrat, R.; Böni, P.; Ramsden, J.J.; Textor, M.; Spencer, N.D.; Tengvall, P.; Gold, J.; et al. A comparative study of protein adsorption on titanium oxide surfaces using in situ ellipsometry, optical waveguide lightmode spectroscopy, and quartz crystal microbalance/dissipation. Colloids Surf. B Biointerfaces 2002, 24, 155–170. [Google Scholar] [CrossRef]
- Available online: https://www.abcam.com/en-us/products/proteins-peptides/recombinant-human-coronavirus-sars-cov-2-spike-glycoprotein-s1-active-ab273068?srsltid=AfmBOooG7_0JBah7-mQWdYlPUvurzAuGdJ1h07UgpSb9Ky6kn1Dux2G0 (accessed on 12 May 2021).
- Deschout, H.; Platzman, I.; Sage, D.; Feletti, L.; Spatz, J.P.; Radenovic, A. Investigating Focal Adhesion Substructures by Localization Microscopy. Biophys. J. 2017, 113, 2508–2518. [Google Scholar] [CrossRef]
- Zaidel-Bar, R.; Milo, R.; Kam, Z.; Geiger, B. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J. Cell Sci. 2007, 120, 137–148. [Google Scholar] [CrossRef]
- Zaidel-Bar, R.; Itzkovitz, S.; Ma’ayan, A.; Iyengar, R.; Geiger, B. Functional atlas of the integrin adhesome. Nat. Cell Biol. 2007, 9, 858–867. [Google Scholar] [CrossRef] [PubMed]
- Othman, H.; Messaoud, H.B.; Khamessi, O.; Ben-Mabrouk, H.; Ghedira, K.; Bharuthram, A.; Treurnicht, F.; Achilonu, I.; Sayed, Y.; Srairi-Abid, N. SARS-CoV-2 Spike Protein Unlikely to Bind to Integrins via the Arg-Gly-Asp (RGD) Motif of the Receptor Binding Domain: Evidence From Structural Analysis and Microscale Accelerated Molecular Dynamics. Front. Mol. Biosci. 2022, 9, 834857. [Google Scholar] [CrossRef] [PubMed]

| Integrin Target | Viral Ligand | Reported (nM) | Method | Reference Study |
|---|---|---|---|---|
| αVβ6 | recombinant SARS-CoV-2 S1-RBD (RGD-containing) | 230 ± 180 | SPR | Norris, 2022 [11] |
| α5β1 | recombinant full S1 domain (not just the RBD, but the larger S1 region of the spike protein) (biotin-S1) | 31 ± n.d. | SPR (1:1 kinetic fit) | Liu, 2022 [9] |
| αVβ3 | recombinant SARS-CoV-2 S1-RBD peptide (RGD-containing) | >500 | SPR | Norris, 2022 [11] |
| αVβ1 | recombinant RGD-peptide from S-protein | 50–100 | In vitro integrin-binding assay | Bugatti, 2022 [10] |
| α5β1, αvβ5, αvβ3 | recombinant human coronavirus SARS-CoV-2 Spike Glycoprotein S1 (Active) | 4616 ± 252 and 1116 ± 0.040 | RWG biosensor-based whole cell interaction assay (from adhesion kinetics and competitive adhesion assay) | present study |
| Concentration (µM) | Δλ_corr (pm) | Γ (ng/cm2) | (nmol/m2) | dRGD-RGD (nm) | νRGD (μm−2) |
|---|---|---|---|---|---|
| 2.67 × 10−1 | 1003.10 | 116 | 9.28 × 10−4 | 14.40 | 5768 |
| 5.33 × 10−2 | 315.64 | 23.2 | 1.85 × 10−4 | 32.1 | 1113 |
| 1.06 × 10−2 | 240.55 | 4.64 | 3.71 × 10−5 | 71.9 | 223 |
| 2.13 × 10−3; | 249.78 | 0.928 | 7.42 × 10−6 | 160.7 | 44.5 |
| 4.27 × 10−4 | 233.64 | 0.19 | 1.48 × 10−6 | 359.4 | 8.9 |
| 8.53 × 10−5 | 64.98 | 0.03712 | 2.97 × 10−7 | 803.6 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanyo, N.; Borbely, K.; Peter, B.; Kovacs, K.D.; Balogh, A.; Magyaródi, B.; Kurunczi, S.; Szekacs, I.; Horvath, R. Kinetic Analysis of SARS-CoV-2 S1–Integrin Binding Using Live-Cell, Label-Free Optical Biosensing. Biosensors 2025, 15, 534. https://doi.org/10.3390/bios15080534
Kanyo N, Borbely K, Peter B, Kovacs KD, Balogh A, Magyaródi B, Kurunczi S, Szekacs I, Horvath R. Kinetic Analysis of SARS-CoV-2 S1–Integrin Binding Using Live-Cell, Label-Free Optical Biosensing. Biosensors. 2025; 15(8):534. https://doi.org/10.3390/bios15080534
Chicago/Turabian StyleKanyo, Nicolett, Krisztina Borbely, Beatrix Peter, Kinga Dora Kovacs, Anna Balogh, Beatrix Magyaródi, Sandor Kurunczi, Inna Szekacs, and Robert Horvath. 2025. "Kinetic Analysis of SARS-CoV-2 S1–Integrin Binding Using Live-Cell, Label-Free Optical Biosensing" Biosensors 15, no. 8: 534. https://doi.org/10.3390/bios15080534
APA StyleKanyo, N., Borbely, K., Peter, B., Kovacs, K. D., Balogh, A., Magyaródi, B., Kurunczi, S., Szekacs, I., & Horvath, R. (2025). Kinetic Analysis of SARS-CoV-2 S1–Integrin Binding Using Live-Cell, Label-Free Optical Biosensing. Biosensors, 15(8), 534. https://doi.org/10.3390/bios15080534





