Sensing Cellular Damages Induced by Food Safety Hazards Using Bacterial Stress-Responsive Biosensors
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Constructions
2.2. Fluorescence Measurement with a Microplate Reader
2.3. Mathematical Model and Data Fitting
2.4. Calculation of LOD
2.5. Milk Sample Pretreatment and Hazard Detection
2.6. HPLC Analysis of Norfloxacin in Samples
3. Results and Discussion
3.1. Construction and Optimization of a RecA-LexA-Based DNA Damage Biosensor
3.2. Construction of Biosensors Targeting Oxidative, Proteotoxic, and Membrane Stress
3.3. Screening and Quantitative Detection of Food Safety Hazard-Induced Cellular Damage Using Constructed Biosensors
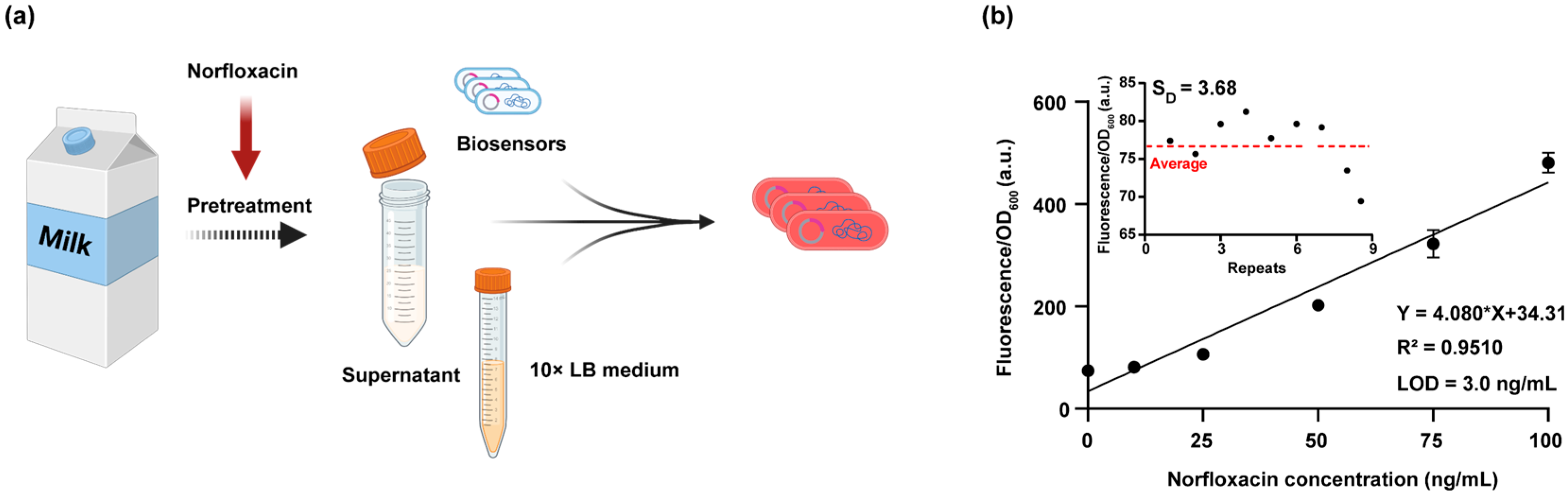
3.4. Application of the DNA Damage Biosensor for Detecting Norfloxacin-Induced Genotoxicity in Milk Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thakali, A.; MacRae, J.D. A Review of Chemical and Microbial Contamination in Food: What Are the Threats to a Circular Food System? Environ. Res. 2021, 194, 110635. [Google Scholar] [CrossRef]
- Koszucka, A.; Nowak, A. Thermal Processing Food-Related Toxicants: A Review. Crit. Rev. Food Sci. Nutr. 2019, 59, 3579–3596. [Google Scholar] [CrossRef] [PubMed]
- Rushing, B.R.; Selim, M.I. Aflatoxin B1: A Review on Metabolism, Toxicity, Occurrence in Food, Occupational Exposure, and Detoxification Methods. Food Chem. Toxicol. 2019, 124, 81–100. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Song, Y.; He, F.; Jing, M.; Tang, J.; Liu, R. A Review of Human and Animals Exposure to Polycyclic Aromatic Hydrocarbons: Health Risk and Adverse Effects, Photo-Induced Toxicity and Regulating Effect of Microplastics. Sci. Total Environ. 2021, 773, 145403. [Google Scholar] [CrossRef] [PubMed]
- Jaishankar, M.; Tseten, T.; Anbalagan, N.; Mathew, B.B.; Beeregowda, K.N. Toxicity, Mechanism and Health Effects of Some Heavy Metals. Interdiscip. Toxicol. 2014, 7, 60–72. [Google Scholar] [CrossRef]
- Chen, J.; Ying, G.-G.; Deng, W.-J. Antibiotic Residues in Food: Extraction, Analysis, and Human Health Concerns. J. Agric. Food Chem. 2019, 67, 7569–7586. [Google Scholar] [CrossRef]
- Ames, B.N.; McCann, J.; Yamasaki, E. Methods for Detecting Carcinogens and Mutagens with the Salmonella/Mammalian-Microsome Mutagenicity Test. Mutat. Res./Environ. Mutagen. Relat. Subj. 1975, 31, 347–363. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, Y.; Yang, C. Evaluating In Vitro DNA Damage Using Comet Assay. J. Vis. Exp. 2017, 128, 56450. [Google Scholar]
- Clare, G. The in vitro mammalian chromosome aberration test. Methods Mol. Biol. 2012, 817, 69–91. [Google Scholar]
- Allen, M.; Millett, P.; Dawes, E.; Rushton, N. Lactate Dehydrogenase Activity as a Rapid and Sensitive Test for the Quantification of Cell Numbers in Vitro. Clin. Mater. 1994, 16, 189–194. [Google Scholar] [CrossRef]
- van Meerloo, J.; Kaspers, G.J.L.; Cloos, J. Cell Sensitivity Assays: The MTT Assay. In Cancer Cell Culture: Methods and Protocols; Cree, I.A., Ed.; Humana Press: Totowa, NJ, USA, 2011; pp. 237–245. [Google Scholar]
- Louzao, M.C.; Costas, C. 4 Toxicological Studies with Animals. In Environmental Toxicology: Non-Bacterial Toxins; Botana, L.M., Ed.; De Gruyter: Berlin, Germany, 2024; pp. 103–134. [Google Scholar]
- Bauer, B.; Mally, A.; Liedtke, D. Zebrafish Embryos and Larvae as Alternative Animal Models for Toxicity Testing. Int. J. Mol. Sci. 2021, 22, 13417. [Google Scholar] [CrossRef]
- Nguyen, X.-H. Current Status and Future Prospects of Toxicity Assessment Using Organoids. Toxicol. Res. 2025, 41, 325–333. [Google Scholar] [CrossRef]
- Hu, J.; Wang, W.; Zhu, Z.; Chang, H.; Pan, F.; Lin, B. Quantitative Structure−Activity Relationship Model for Prediction of Genotoxic Potential for Quinolone Antibacterials. Environ. Sci. Technol. 2007, 41, 4806–4812. [Google Scholar] [CrossRef]
- Valko, M.; Morris, H.; Cronin, M.T. Metals, toxicity and oxidative stress. Curr. Med. Chem. 2005, 12, 1161–1208. [Google Scholar] [CrossRef]
- Tamás, M.J.; Sharma, S.K.; Ibstedt, S.; Jacobson, T.; Christen, P. Heavy Metals and Metalloids as a Cause for Protein Misfolding and Aggregation. Biomolecules 2014, 4, 252–267. [Google Scholar] [CrossRef] [PubMed]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.D.; Mazur, M.; Telser, J. Free Radicals and Antioxidants in Normal Physiological Functions and Human Disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef] [PubMed]
- Dias, C.; Nylandsted, J. Plasma Membrane Integrity in Health and Disease: Significance and Therapeutic Potential. Cell Discov. 2021, 7, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Alhmoud, J.F.; Woolley, J.F.; Al Moustafa, A.-E.; Malki, M.I. DNA Damage/Repair Management in Cancers. Cancers 2020, 12, 1050. [Google Scholar] [CrossRef]
- Krewski, D.; Acosta, D., Jr.; Andersen, M. Toxicity testing in the 21st century: A vision and a strategy. J. Toxicol. Environ. Health B Crit. Rev. 2010, 13, 51–138. [Google Scholar] [CrossRef]
- Gao, B.; Liang, L.; Su, L.; Wen, A.; Zhou, C.; Feng, Y. Structural Basis for Regulation of SOS Response in Bacteria. Proc. Natl. Acad. Sci. USA 2023, 120, e2217493120. [Google Scholar] [CrossRef]
- Chiang, S.M.; Schellhorn, H.E. Regulators of Oxidative Stress Response Genes in Escherichia Coli and Their Functional Conservation in Bacteria. Arch. Biochem. Biophys. 2012, 525, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Yura, T. Regulation of the Heat Shock Response in Escherichia Coli: History and Perspectives. Genes Genet. Syst. 2019, 94, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Parsons, J.B.; Rock, C.O. Bacterial Lipids: Metabolism and Membrane Homeostasis. Prog. Lipid Res. 2013, 52, 249–276. [Google Scholar] [CrossRef] [PubMed]
- Rhodius, V.A.; Suh, W.C.; Nonaka, G.; West, J.; Gross, C.A. Conserved and Variable Functions of the σE Stress Response in Related Genomes. PLoS Biol. 2005, 4, e2. [Google Scholar] [CrossRef]
- Kotova, V.Y.; Manukhov, I.V.; Zavilgelskii, G.B. Lux-Biosensors for Detection of SOS-Response, Heat Shock, and Oxidative Stress. Appl. Biochem. Microbiol. 2010, 46, 781–788. [Google Scholar] [CrossRef]
- Padilla-Martínez, F.; Carrizosa-Villegas, L.A.; Rangel-Serrano, Á.; Paramo-Pérez, I.; Mondragón-Jaimes, V.; Anaya-Velázquez, F.; Padilla-Vaca, F.; Franco, B. Cell Damage Detection Using Escherichia Coli Reporter Plasmids: Fluorescent and Colorimetric Assays. Arch. Microbiol. 2015, 197, 815–821. [Google Scholar] [CrossRef]
- Niazi, J.H.; Kim, B.C.; Ahn, J.-M.; Gu, M.B. A Novel Bioluminescent Bacterial Biosensor Using the Highly Specific Oxidative Stress-Inducible Pgi Gene. Biosens. Bioelectron. 2008, 24, 670–675. [Google Scholar] [CrossRef]
- Niazi, J.H.; Kim, B.C.; Gu, M.B. Characterization of Superoxide-Stress Sensing Recombinant Escherichia Coli Constructed Using Promoters for Genes Zwf and Fpr Fused to Lux Operon. Appl. Microbiol. Biotechnol. 2007, 74, 1276–1283. [Google Scholar] [CrossRef]
- Belkin, S.; Smulski, D.R.; Dadon, S.; Vollmer, A.C.; Van Dyk, T.K.; Larossa, R.A. A Panel of Stress-Responsive Luminous Bacteria for the Detection of Selected Classes of Toxicants. Water Res. 1997, 31, 3009–3016. [Google Scholar] [CrossRef]
- Elad, T.; Seo, H.B.; Belkin, S.; Gu, M.B. High-Throughput Prescreening of Pharmaceuticals Using a Genome-Wide Bacterial Bioreporter Array. Biosens. Bioelectron. 2015, 68, 699–704. [Google Scholar] [CrossRef]
- Ben-Israel, O.; Ben-Israel, H.; Ulitzur, S. Identification and Quantification of Toxic Chemicals by Use of Escherichia coli Carrying Lux Genes Fused to Stress Promoters. Appl. Environ. Microbiol. 1998, 64, 4346–4352. [Google Scholar] [CrossRef] [PubMed]
- Hui, C.; Hu, S.; Yang, X.; Guo, Y. A Panel of Visual Bacterial Biosensors for the Rapid Detection of Genotoxic and Oxidative Damage: A Proof of Concept Study. Mutat. Res.-Gen. Tox. En. 2023, 888, 503639. [Google Scholar] [CrossRef] [PubMed]
- Bindels, D.S.; Haarbosch, L.; Van Weeren, L.; Postma, M.; Wiese, K.E.; Mastop, M.; Aumonier, S.; Gotthard, G.; Royant, A.; Hink, M.A.; et al. mScarlet: A Bright Monomeric Red Fluorescent Protein for Cellular Imaging. Nat. Methods 2017, 14, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Bryksin, A.V.; Matsumura, I. Overlap Extension PCR Cloning: A Simple and Reliable Way to Create Recombinant Plasmids. Bio. Tech. 2010, 48, 463–465. [Google Scholar] [CrossRef]
- Wang, B.; Barahona, M.; Buck, M. A Modular Cell-Based Biosensor Using Engineered Genetic Logic Circuits to Detect and Integrate Multiple Environmental Signals. Biosens. Bioelectron. 2013, 40, 368–376. [Google Scholar] [CrossRef]
- Kobets, T.; Smith, B.P.C.; Williams, G.M. Food-Borne Chemical Carcinogens and the Evidence for Human Cancer Risk. Foods 2022, 11, 2828. [Google Scholar] [CrossRef]
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy metal toxicity and the environment. Exp. Suppl. 2012, 101, 133–164. [Google Scholar]
- Butala, M.; Žgur-Bertok, D.; Busby, S.J.W. The Bacterial LexA Transcriptional Repressor. Cell. Mol. Life. Sci. 2008, 66, 82–93. [Google Scholar] [CrossRef]
- Maslowska, K.H.; Makiela-Dzbenska, K.; Fijalkowska, I.J. The SOS System: A Complex and Tightly Regulated Response to DNA Damage. Environ. Mol. Mutagen. 2019, 60, 368–384. [Google Scholar] [CrossRef]
- Podlesek, Z.; Žgur Bertok, D. The Escherichia coli SOS Response: Much More Than DNA Damage Repair; IntechOpen: London, UK, 2023. [Google Scholar]
- Michalowski, C.B.; Giese, K.C. RecA-Dependent Cleavage of LexA Dimers. Mol. Biol. 2008, 377, 148–161. [Google Scholar]
- Chen, J.X.; Lim, B.; Steel, H.; Song, Y.; Ji, M.; Huang, W.E. Redesign of Ultrasensitive and Robust RecA Gene Circuit to Sense DNA Damage. Microb. Biotechnol. 2021, 14, 2481–2496. [Google Scholar] [CrossRef]
- Qu, F.; Zheng, W. Cadmium Exposure: Mechanisms and Pathways of Toxicity and Implications for Human Health. Toxics 2024, 12, 388. [Google Scholar] [CrossRef]
- Rather, I.A.; Koh, W.Y.; Paek, W.K.; Lim, J. The Sources of Chemical Contaminants in Food and Their Health Implications. Front. Pharmacol. 2017, 8, 830. [Google Scholar] [CrossRef]
- Prajapati, J.D.; Kleinekathöfer, U.; Winterhalter, M. How to Enter a Bacterium: Bacterial Porins and the Permeation of Antibiotics. Chem. Rev. 2021, 121, 5158–5192. [Google Scholar] [CrossRef]
- Mahendran, K.R.; Kreir, M.; Weingart, H.; Fertig, N.; Winterhalter, M. Permeation of Antibiotics through Escherichia Coli OmpF and OmpC Porins: Screening for Influx on a Single-Molecule Level. J. Biomol. Screen. 2010, 15, 302–307. [Google Scholar] [CrossRef]
- Hossain, S.T.; Mallick, I.; Mukherjee, S.K. Cadmium Toxicity in Escherichia Coli: Cell Morphology, Z-Ring Formation and Intracellular Oxidative Balance. Ecotox. Environ. Safe 2012, 86, 54–59. [Google Scholar]
- Zhang, G.; Hu, S.; Jia, X. Highly Sensitive Whole-Cell Biosensor for Cadmium Detection Based on a Negative Feedback Circuit. Front. Bioeng. Biotechnol. 2021, 9, 799781. [Google Scholar] [CrossRef] [PubMed]
- Chavakula, R.; Chintala, R.; Tadanki, B. Application of Validated Stability Indicating HPLC Method in Stability Testing of Nor-Metrogyl Tablets. J. Pharm. Res. 2013, 6, 499–503. [Google Scholar] [CrossRef]
- Singh, B.; Bhat, A.; Dutta, L.; Pati, K.R.; Korpan, Y.; Dahiya, I. Electrochemical Biosensors for the Detection of Antibiotics in Milk: Recent Trends and Future Perspectives. Biosensors 2023, 13, 867. [Google Scholar] [CrossRef]
- Itoh, T.; Mitsumori, K.; Kawaguchi, S.; Sasaki, Y.F. Genotoxic Potential of Quinolone Antimicrobials in the in Vitro Comet Assay and Micronucleus Test. Mutat. Res./Genet. Toxicol. Environ. Mutagen. 2006, 603, 135–144. [Google Scholar] [CrossRef]
- Mamber, S.W.; Kolek, B.; Brookshire, K.W.; Bonner, D.P.; Fung-Tomc, J. Activity of Quinolones in the Ames Salmonella TA102 Mutagenicity Test and Other Bacterial Genotoxicity Assays. Antimicrob. Agents Chemother. 1993, 37, 213–217. [Google Scholar] [CrossRef]
- Hansch, C.; McKarns, S.C.; Smith, C.J.; Doolittle, D.J. Comparative QSAR Evidence for a Free-Radical Mechanism of Phenol-Induced Toxicity. Chem.-Biol. Interact. 2000, 127, 61–72. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Lou, M.; He, W.; Quan, S. Sensing Cellular Damages Induced by Food Safety Hazards Using Bacterial Stress-Responsive Biosensors. Biosensors 2025, 15, 695. https://doi.org/10.3390/bios15100695
Li R, Lou M, He W, Quan S. Sensing Cellular Damages Induced by Food Safety Hazards Using Bacterial Stress-Responsive Biosensors. Biosensors. 2025; 15(10):695. https://doi.org/10.3390/bios15100695
Chicago/Turabian StyleLi, Ruiqi, Manzhuan Lou, Wei He, and Shu Quan. 2025. "Sensing Cellular Damages Induced by Food Safety Hazards Using Bacterial Stress-Responsive Biosensors" Biosensors 15, no. 10: 695. https://doi.org/10.3390/bios15100695
APA StyleLi, R., Lou, M., He, W., & Quan, S. (2025). Sensing Cellular Damages Induced by Food Safety Hazards Using Bacterial Stress-Responsive Biosensors. Biosensors, 15(10), 695. https://doi.org/10.3390/bios15100695




