Recent Advances in the Optimization of Nucleic Acid Aptamers and Aptasensors
Abstract
1. Introduction
2. Nucleic Acid Aptamer
2.1. Nucleic Acid Aptamer
2.2. SELEX Technology
2.3. Separation of Target-Bonded and Non-Binding Aptamers
2.3.1. Capillary Electrophoresis SELEX Technology
2.3.2. Microfluidic SELEX Technology
2.3.3. Cell-SELEX Technology
3. Aptasensors
3.1. Aptasensors
3.2. Optical Aptasensors
3.2.1. Fluorescent Aptasensors
3.2.2. Colorimetric Aptasensors
3.2.3. Surface Plasmon Resonance Aptasensors
| Analytes | Working Principles | Aptamer Immobilization | Detection Method | Sensitivity (LoD) | Features/Potential Applications | References |
|---|---|---|---|---|---|---|
| Tau441 protein | Changes in the number of target molecules bound to the aptamer-modified biosensor tip cause a shift in the interference pattern that can be measured in real time | Biotin-aptamer was immobilized onto streptavidin (SA) biosensor tips | Biolayer interferometry method | 6.7 nM in vitro | Highly selective, portable, label-free, and real-time monitoring | [135] |
| E. coli O157:H7 | The specific binding between immobilized peptide aptamers on the cavity surface and bacteria’s surface structures (e.g., LPS) induces a shift in the minimum in the optical transmission spectrum | Optical fibre surface was functionalized with extended salinization using APTES and with peptide aptamers immobilized on its surface | Microcavity in-line with Mach–Zehnder interferometer (μIMZI) | 10 CFU/mL for E. coli O157:H7 | Highly specific and sensitive, label-free, and lowest analyte volume | [136] |
| Proteus mirabilis (pathogen) | The amino-modified aptamers against P. mirabilis were conjugated to the surfaces of SiC quantum dots (QDs) for bacteria recognition; the aptamer with an affinity for target protein can bound to P. mirabilis, which causes a decrease in the fluorescence intensity of DNA-SiC QDs (fluorescence quenching strategy) | The carboxyl groups on the surface of SiC QDs and amino-modified DNA aptamers had a condensation reaction by EDC/NHS coupling to form fluorescence aptasensor | Fluorescent detection | 526 CFU/mL with a linear range of 103 to 108 CFU/mL | Potential tool for identification of P. mirabilis in forensic food poisoning cases | [55] |
| Digoxin (DGX) | The PVA hydrogel acted as a fluorescent probe. The fluorescence intensity of the hydrogel was quenched by aptamer-stabilized Au nanoparticles (NPs) as energy acceptors; upon addition of digoxin, aptamer/drug complex was formed, and the fluorescence of the hydrogel was restored because of destabilization and aggregation of AuNPs in the presence of salt | Aptamer of DGX is physically adsorbed on the surface of the 20nm AuNPs and then mixed with solutions of NaCl and PVA with or without DGX for detection | Fluorescent detection with on/off/on strategy | 2.9 ng/L with a linear range of 10–1000 ng/L | High selectivity, eco-friendly, and rapid operation | [137] |
| Zearalenone (ZEN, one of the most prevalent mycotoxins) | The 17-nt fluorophore Cy5.5-labelled complementary DNA strand and ZEN competitively bind with the aptamer immobilized on the fibre, enabling signal-off fluorescent detection | A 40-nt ZEN-specific aptamer (8Z31) is covalently immobilized on the optical fibre by coupling its amino group at the 5′ end with aldehyde group introduced on the fibre surface | Fluorescent detection | 18.4 pM (equivalent to 29.3 ppt) with a semi-log linear range of 10 pM–10 nM | Extremely high specificity, regenerated 28 times, decreased cost | [138] |
| Kanamycin (Kana, an important antibiotic used in veterinary medicine) | The quencher TTQF binds strongly with ssDNA, whereas it exhibits very weak interactions with dsDNA; by subtly designing the structure of the DNA aptamer probes, the target-induced fluorescence signal is amplified to different levels for sensing | Fluorescein labelled at the end of the aptamer as FRET donor (an organic quencher TTQF as acceptor) | Fluorescence spectra (FRET) | 10 nM with a linear range of 10 nM to 3 μM | High specificity, selectivity, and stability for antibiotic determination in food and environment | [139] |
| Endotoxin | Upon binding with endotoxin, the aptamer undergoes conformation changes and decreases the distance between the Au nanoparticle (AuNP) and the Au nanofilm (AuNF), resulting in the red shifting of scattered light spectrum | The aptamer has an amine group at the 5′ end coupling to the SAM layer on the AuNF and a thiol group at the 3′ end, which forms a covalent bond with AuNP | Time-averaged scattering spectra OR colour camera for single-particle mode | 15–31 EU/mL (ensemble mode) OR single-particle resolution | Reusable, highly selective | [140] |
| Lipopolysaccharide (LPS) biomarker of Gram-negative bacteria | Orientation of liquid crystal (LC) molecules at the LC-aqueous interface can be changed upon binding of aptamer with LPS in the mixture solution, inducing optical appearance from bright to dark | LC molecules are immobilized and formed onto glass slides for optical sensing cells | Polarized light microscopy | Ultralow, 0.4 pg/mL for LPS biomarker, and 27 cfu/mL for E. coli bacteria | Simple, label-free, high sensitivity and specificity, and rapid and reliable detection of Gram-negative bacteria | [141] |
| Ochratoxin A (OTA, the most dangerous mycotoxin) | The aptasensing strategy was based on the conformational switch of the immobilized π-shaped DNA aptamer structure on the glass substrate in presence of the target; a shift in the orientation of LCs from random to homeotropic state led to the change in optical appearance of the sensor platform from bright to dark | Two complementary strands of aptamers (CPs) were first immobilized onto glass substrates, subsequently modified with DMOAP/APTES/GA, and then incubated with solution of OTA-specific aptamer for immobilization | Polarized light microscopy | Detecting OTA at the ultra-trace level as low as 0.63 aM | Portable and real-time sensing probes with high performance for food safety control and clinical application | [121] |
| Prostate-specific antigen (PSA, prostate cancer biomarker) | The principle of PSA detection by the aptasensor is based on the changes in the directional regularity of LCs caused by the specific aptamer at the CTAB-arranged LC-aqueous interface; the optical vision of the aptasensor background changes from colourful to dark by forming the aptamer–PSA complex that induces the re-arrangement of LC molecules from disordered to homeotropic direction | The cleaned glass slide was modified by soaking in DMOAP solution, CTAB + 5CB mixture solution, and then aptamer solution, subsequently; afterward, the solution of PSA was pipetted on to the aptasensor for detection | Polarized light microscopy | 0.148 pg/mL with a range of 1–1200 pg/mL | Label-free, simple and low-cost, highly sensitive aptasensor for diagnosis of PSA | [142] |
| Insulin | Insulin-binding DNA aptamer (IGA3) is a guanine (G)-rich DNA that forms an anti-parallel G-quadruplex and binds insulin selectively. Nickel (II) salphen complex modified with piperidine side chain [NI(II)-SP] acted as the ligand to the immobilized aptamer via non-covalent π-π stacking between aromatic rings of folded single-strand IGA3 aptamer and square planar salphen ligand; a colour change from yellow to brownish orange is induced upon the binding of insulin with the immobilized IGA3-Ni(II)SP complex | Insulin-binding DNA aptamer (IGA3) was employed as the synthetic receptor and conjugated to the amine-modified porous silica microparticles (PSiMPs) via imine covalent bonds using glutaraldehyde crosslinker | Fibre optic reflectance spectra | 3.71 μIU/mL with a linear range of 10–50 μIU/mL | High selectivity, rapid response, promising for insulin monitoring simply by observing the aptasensor colour change. | [143] |
| Aflatoxin M1 (AFM1) | Preservation of AuNPs against NaCl-induced aggregation by detaching complementary strand (CS) from dsDNA-modified silica nanoparticles (SNPs) following the addition of target, and the colour of sample remains red; salt-induced aggregation of AuNPs occurs in the lack of AFM1, and the colour of sample becomes purple as dsDNA-modified SNPs are stable and CS cannot bind to AuNPs (as demonstrated in Figure 5) | Biotin-labelled AFM1 aptamer and its complementary strand (CS) immobilized onto the surface of silica nanoparticles (SNPs) through streptavidin–biotin (SA-Bio) interaction. Aptamers as recognition probe and AuNPs as colorimetric indicators | Colorimetric detection | 30 ng/L with a linear range of 300–75,000 ng/L | Simplicity (detected with the naked eye), better selectivity by using CS of aptamer | [120] |
| Retinol-binding protein 4 (RBP4) (a biomarker for the early diagnosis of T2DM) | Retinol-binding protein aptamer (RBP-A) physically adsorbs on the AuNPs surface and stabilizes the AuNPs against NaCl-induced aggregation; upon the addition of RBP4, RBP-A binds to RBP4 and detaches from the AuNPs surface, leaving the AuNPs unprotected. Addition of NaCl causes aggregation of AuNPs, leading to a colour change in the AuNPs solution from red to purple/blue | RBP-A is physically adsorbed on the surface of the AuNPs through van der Waals and hydrophobic interactions | Colorimetric detection | 90.76 nM | Simple yet effective, specific, and rapid detection of RBP4 | [144] |
| Interleukin-6 (IL-6) and C-reactive protein (CRP) (sepsis biomarkers) | Aptamer-functionalized AuNPs used for detection of CRP and IL-6 biomarkers; using colorimetric assay, aggregation value of AuNPs in presence of the target was determined in two phases: in solution and on μPADs; and a competitive reaction between target and AuNPs was performed to bind to the aptamer, bare nanoparticles were aggregated in the presence of salt, and the concentration of the analyte was measured according to the amount of colour change due to aggregation | Aptamer for either CRP or IL-6 protein biomarkers was not modified and physically adsorbed onto the surface of the AuNPs; the aptamer-functionalized AuNPs were mixed with CRP and IL-6 proteins in PBS solution and then incubated for quantification | Colorimetric detection | Linear ranges were 50–1000 mg/L and 1–25 ng/L, and LoD values were 1.9 mg/L and 0.07 ng/L for CRP and IL-6, respectively | Microfluidic paper-based analytical devices (µPAD), simple but highly stable, reproducible, accurate, and recoverable for sepsis diagnosis; determination of two biomarkers with naked eyes and using image analysis | [145] |
| Mammaglobin proteins at the surface of circulating breast cancer cells (CTCs) | The resonance wavelength and amplitude are impacted and shift proportionally to the refractive change induced by the binding between immobilized aptamers and surface proteins of the cells | Thiolated MAMA2 aptamers immobilized on the surface of Au-coated optical tilted fibre Bragg gratings | SPR with tilted fibre Bragg gratings (TFBGs) sensitivity enhancement | 49 cells/mL (down to 10 cells/mL when AuNPs used for signal amplification) | Highly sensitive, selective, and quick response for early-stage metastatic cancer diagnosis | [146,147] |
| Plasmodium falciparum glutamate dehydrogenase (PfGDH, malaria biomarker) | U-bent optical fibres as a LSPR probe creates higher-order modes, resulting in a higher penetration depth of the evanescent field; the probe measures the refractive index changes, initiated by the binding of PfGDH to the aptamer | The U-bent region of plastic optic fibres (POF) was de-cladded and then sputtered with gold for LSPR; the thiolated-PfGDH aptamer NG3 and 6-mercapto 1-hexanol (MCH) were then immobilized on the Au surface via incubation | LSPR detecting refractive index changes | 264 pM with only 175 mL of the sample used | Low-cost, simplistic, robust aptamer-based stable recognition system, and multiplexing capability | [148] |
| SARS-CoV-2 spike protein | A specific aptameric sequence was immobilized on short polyethyleneglycol (PEG) interface on gold nanofilm deposited on a D-shaped plastic optical fibre (POF) probe, and the binding of SARS-CoV-2 spike glycoprotein with the aptamers induced a clear red shift in the resonance wavelength, which is a very sensitive surface plasmon resonance (SPR) phenomenon being monitored for spike protein detection, as demonstrated in Figure 7a. | A mixed layer obtained from a mixture of PEGthiol and BiotinPEGlipo was prepared on Au nanofilm, and a streptavidin coating was next performed; finally, a biotin-modified aptamer was immobilized on the streptavidin layer, as demonstrated in Figure 7b. | SPR | 37 nM, with a range of 25–1000 nM | High specificity, rapid response, low-cost, and compact, portable diagnostic tool | [133] |
| Cancer cells | In the processes of cytosensing, cancer cells were selectively captured onto the T-shaped aptamer-coated Ω-shaped fibre optic (Ω-FO) surface; in the following photothermal therapy (PTT) process, high local temperature induced necrosis of cancer cells, which could be released from the FO surface (as demonstrated in Figure 8) | A sandwich layer of PDA/AuNPs/PDA was designed as a coating around the surface of the Ω-FO to enhance LSPR sensitivity and photothermal conversion efficiency; T-shaped aptamers (AS1411 and MUC1) were immobilized on the coating surface to sensitively capture cancer cells in blood circulation system | Localized surface plasmon resonance (LSPR) detecting refractive index changes | 13 cells/mL | The sandwich layer coating strongly enhanced efficiencies of cytosensor and PTT; significant potential for cancer metastasis inhibition | [134] |
| Helicobacter pylori (H. pylori) | The optical fibre surface was modified with a layer of AuNPs; then, the polyA-tailed aptamer and the block agent of DNA with secondary structure were fixed on AuNPs via the Au-N bond; and when H. pylori is present in the system, the aptamers on the optical fibre surface specifically recognize and capture H. pylori, which caused the intensity change in the LSPR absorption peak due to the refractive index change of the solution. | Original sequence of aptamer was truncated to effectively capture H. pylori; cheaper polyA-tailed aptamers utilized instead of thiolated aptamers; a spacer nucleic acid with short stem-loop structure was adopted to control the aptamer density on AuNPs on the surface of the J-shaped optical fibre probe | Localized surface plasmon resonance (LSPR) | 45 CFU/mL with a linear range of 1.0 × 102–1.0 × 108 CFU/mL | Easy-to-prepare, high sensitivity, and rapid analysis for detection of pathogenic bacteria in environmental monitoring and disease diagnosis | [149] |
| Zearalenone (ZEN, one of the most prevalent mycotoxins) | Target analyte-tuned growth of gold nano-seeds (AuNSs) coated with unmodified aptamers was derived from the DNA-mediated control of the metal nanoparticle shape and optimized to determine concentration of the analyte according to the LSPR peak wavelength of the grown AuNPs | 5 nm AuNSs were modified with both 8Z31 and Z143 aptamers by physical adsorption via incubation together in the mixture solutions | Absorption spectra (LSPR) | ∼72 ppb with a range of 10 ppb to 100 ppm | Rapid detection of small molecules based on analyte-tuned growth of AuNS, combined with machine learning-enhanced spectrum analysis | [150] |
| Four mycotoxins (AFB1, FB1, OTA, and ZEN) | Optical fibres are utilized not only as the carrier to immobilize single-stranded binding protein (SSB) to prepare the sensing region but also as the transducer of the chemiluminescent emission to detect the sensor response; when the fibre sensing probe with SA-Bio-HRP complex on the fibre surface was immersed in the solution containing chemiluminescence substrates, chemiluminescent emission was transmitted through the optical fibre and collected by the photon-counting detector | Firstly, each target and its biotinylated aptamer (Bio-Apt) are mixed in solution for complete specific recognition; then, the fibre sensing probe with immobilized SSB was placed into the mixture solution to make SSB bind to the remaining Bio-Apt molecules that are not recognized by the targets; and afterward, the probe was inserted into a solution to form SA-Bio-HRP complex on the fibre surface | Chemiluminescent emission and photon counting-detector | 0.015–0.423 pg/mL with a range from pg/mL to 105 pg/mL for detecting different mycotoxins | Four mycotoxins (AFB1, FB1, OTA, or ZEN) can be simultaneously determined from one run for on-site analysis of real food samples | [151] |
3.3. Field-Effect Transistor Aptasensors
| Analytes | Aptasensor Architecture | Aptamer Immobilization | FET Type | Sensitivity (LoD) | Feature/Special Findings | References |
|---|---|---|---|---|---|---|
| Dopamine (DA, brain hormone) | Sequential conjugation of carboxylated polypyrrole nanotubes (CPNTs) and DA-specific aptamer molecules on the interdigitated microelectrodes (IMEs), and the modified substrate integrated into liquid-ion gating system surrounded by pH7.4 buffer (as demonstrated in Figure 9). | DNA aptamer specific to DA as a gating modulator was chemically attached to the surface of CPNTs. | Liquid-ion gated FET | 100 pM | Smaller diameter CPNTs of ca. 120 nm achieved over 100 times higher sensitivity than the larger diameter of ca. 200 nm. | [76] |
| Dopamine (DA) | Micron-sized electrolyte-gated graphene field-effect transistors (EG-gFETs) were fabricated with silicon wafer via sputtering, lithographic patterning, and ion milling. Then, single-layer graphene (SLG) grown by thermal chemical vapour deposition (CVD) on Cu foils was transferred onto the wafer, patterned with lithography, etched by plasma, and passivated. | EG-gFET arrays were functionalized firstly with 1-dodecanethiol (DDT) for gate passivation and then with crosslinker 1-pyrenebutyric acid N-hydroxysuccinimide (PBASE). Afterward, a 44nt-long DNA aptamer for high affinity to DA with a 5′ amino-link termination was bonded with PBASE on the sensor surface. | Electrolyte-gated graphene field-effect transistors (EG-gFETs) | 1 aM with dynamic range of 10−18 to 10−8 M | Demonstrated highly pertinent for developing novel point-of-care devices that require stable, high-throughput detection of physiologically and clinically relevant dopamine concentrations. | [169] |
| Hemagglutinin (HA) protein (biomarker for H5N1 avian influenza virus AIV) | The Au electrode of a sensing part as extended gate is functionalized with DNA aptamer specific to HA protein. Surface potential is generated due to a conformational change in DNA aptamer induced by the binding of HA protein. The generated surface potential is transferred with an FET transducer by connecting the sensing part to the gate of a commercial MOS-FET transducer, where the drain current is measured as a function of the gate voltage applied with a reference electrode inserted into the target solution. | The specific DNA aptamer against the HA protein was hybridized with thiol-tagged shorter complimentary DNA strand to enable formation of covalent bonds with the gold electrode using a thiol group. The DNA aptamer was also hybridized with HRP-tagged complimentary DNA strand to create the DNA 3-way junction. | Extended-gate FET | 5.9 pM with a dynamic range of 10 pM to 10 nM | First use of an aptamer-functionalized FET for AIV detection. | [80] |
| Ochratoxin A (OTA, a mycotoxin contaminant) | An array of graphene FETs was integrated on a single silicon chip. A single-layer graphene was transferred onto a Si substrate with 300 nm SiO2 layer by a wet transfer and then patterned to form graphene channels via oxygen plasma etching. E-beam-assisted evaporation was used to deposit Au electrodes. | Specially designed aptamer for OTA with amino-modified at 5′ end was covalently bound onto the sensor surface via a linker of PBASE molecule. The transverse electrical field was used to increase the density of PBASE linker immobilization on graphene by π-π stacking. | Graphene field-effect transistors (gFET) | 1.4 pM in a dynamic range 5–500 pM | Real-time, highly responsive, and rapid detection of OTA is achieved. Ionic strength effect on sensitivity to OTA in buffer is demonstrated. | [165] |
| Ochratoxin A (OTA) | Constructed with high-purity carbon nanotube (CNT) films, svelte yttrium oxide dielectric layers, and extensive gold nanoparticles (AuNPs) to form an ultrathin, active channel. Strategic immobilization of DNA aptamers onto AuNPs confers the sensor with exceptional selectivity. Interactions between the aptamers and the target molecule prompt electrostatic modifications, leading to a reliable biomolecular response. | Thiolated DNA aptamers specific to OTA were immobilized on the sensor surface through forming Au-S bonds with AuNPs distributed along the CNT network, thereby anchoring it firmly to the FET structure. BSA adsorption was subsequently applied to block any remaining active sites, preventing non-specific binding. | CNT FET | 0.2 fM with a range of 8 fM to 80 pM | Label-free, highly sensitive, and rapid detection method for OTA. | [172] |
| Tetracycline | To construct the FET devices, the source and drain electrodes were prepared by depositing 10 nm Ti and 50 nm Au onto silicon-based graphene using a shadow mask and the e-beam evaporation deposition method. The PDMS microfluidic chip was prepared with the aid of photolithography and an inversion process. A Ag/AgCl reference electrode in PBS solution was used as the solution gate through the microfluidic channel. | The ssDNA aptamers were denatured and then injected into the channel and incubated to ensure sufficient fixation of the aptamer, followed by rinsing of the channel with PBS solution and then BSA to remove excessive aptamers and block nonspecific reaction sites. | Camphor−rosin clean transferred graphene FET (CRCT-FET) | 100 fM with a range of 10 pM to 100 nM. | The carrier mobility of the FET made with ultraclean graphene is improved by more than 10 times compared with the FET prepared by conventional PMMA transfer (CPT) method. | [170] |
| Tetracycline | The solution-gated graphene transistor (SGGT)-based sensor with gate, source, and drain electrode and a monolayer graphene channel connecting the source and drain was prepared following the procedure of substrate cleaning, magnetron sputtering, and wet transfer of graphene. The prepared sensor was put into acetone solution to remove PMMA as the organic carrier for the monolayer graphene. The electrode was then encapsulated with PDMS and silver paste to form a reaction tank. | The aptamer against tetracycline (denoted as APT40) with thiol group at 3′ end form covalent Au–S bonds on the Au gate electrode for immobilization. | Solution-gated graphene FET (SGG-FET or SGGT) | 2.073 pM | Demonstrated miniaturized aptamer-SGGT biosensor with appropriate detection strategies could provide an ideal portable sensing platform. | [173] |
| Spike protein of SARS-CoV2 | An aluminum contact was deposited on an intrinsic silicon field-effect transistor sensor and then functionalised with APTES, glutaraldehyde (GA), aptamer, and glycine molecules in sequence. The glycine is aimed at terminating any unbound aldehydes which might create non-specific binding sites. | The specific aptamer against spike protein with an amine group attached to the 5′ end was immobilized by reacting with aldehyde of glutaraldehyde, which was already bound with the amine-terminated silane on the sensor surface. | Schottky field-effect transistor | A linear range of 100 fM to 10 pM (75 pg/mL to 7.5 ng/mL) | Feasibility of an aptamer-FET to detect spike protein. | [174] |
| Hepatitis C virus (HCV) | The graphene solution-gated FET (g-SGFET) was fabricated with silicon (Si) wafer (p-type doped with boron) by multiple steps using optical lithography, ion milling, sputtering, and plasma etching, as well as CVD-grown graphene method. Two-step process of graphene functionalization in ultra-high vacuum (UHV) chamber was performed to create single-atom vacancies in transistor graphene layer using ion-sputtering and then to form covalent bond with p-aminothiophenol (pATP) linker molecules via evaporation of 27 Langmuir of pATP. The functionalization strategy preserves graphene properties and allows for precise control of the density of immobilized aptamer probe molecules. | A 76 nt-long ssDNA aptamer termed AptD-1312 with a thiohexyl motif at 5′ end was selected as an optimal probe molecule for detection of HCV core protein. A 76 nt-long and thiohexyl-modified ssDNA molecule termed D-ACTG was also used as a negative control. The immobilization of thiol-modified AptD-1312 or D-ACTG onto the p-ATP-functionalized surface of the g-SGFET was performed with denaturation and incubation for crosslinking via disulfide bridge. | Gra-phene solution-gated field-effect transistors (g-SGFET or SGGFET) | 15.6 aM with a linear range of 10−14 to 10−18 M in the buffer solution; 90.9 aM in a linear range of 10−13 to 10−18 M in human blood plasma | Unprecedented sensitivity is due to molecular antenna effect of pATP linker, which can capture a subtle charge transfer at the linker/graphene interface driven by local polarization of graphene by the proximity of the aptamer/protein system. The technology is easily scalable in an inexpensive configuration and transportable to the point of care (PoC) in clinical environments. | [171] |
| Thrombin (a model biomarker) | Arrays of silicon nanowire field-effect transistors (SiNW FETs) were fabricated from silicon-on-insulator (SOI) substrates. | EHTES organosilane was used to graft thrombin-binding aptamer (TBA) onto the HfO2 gate oxide of Si nanowires. The TBA probes molecules with an amine termination at the 5′ end. | Silicon nanowire field-effect transistor (SiNW FET) | Tested at a concentration of 2.7 μM only | Feasible to be co-integrated with CMOS readout circuits in the future. | [175] |
| Interleukin 6 (IL-6) | The silicon nanowire field-effect transistor (SiNW FET) devices were commercially available. Each n-type SiNW FET device had two nanowires, and each nanowire had a length of 2 μm and a width of 200 nm. After thoroughly cleaned and activated to form a new oxide layer with the OH groups, three salinization methods with mixed SAM solution, APTES solution, and APS solution were used to perform the subsequent surface modifications. | For the immobilization of the IL-6 aptamer, the mixed SAMs solution was adopted for salinization process for the surface of the Si NWFET device. After the treatment of GA, the SiNW FET device was immersed in the aptamer solution to form covalent bond between the amine group at 5′ end of the aptamer and the aldehyde group on the device surface. | Silicon nanowire field-effect transistor (SiNW FET) | 2.1 pg/mL (100 fM) | The lowest concentration of detection for the aptamer-functionalized SiNW FET was two orders of magnitude lower than the antibody-functionalized SiNW FET. | [176] |
| Troponin I (TnI) | The extended gate electrodes were fabricated with sequential deposition of metal layers using electron beam evaporation and the followed photolithography steps were performed to expose two specific regions on the gold electrodes: one served as the sensing electrode connected to the gate terminal of a metal–oxide–semiconductor field-effect transistor (MOSFET), while the other functioned as a reference electrode for applying the gate bias. | The aptamer was modified with a S-S bond at 5′ end, facilitating its anchoring to the Au electrode surface via thiol–Au chemistry. In the aptamer immobilization process, the aptamer was first combined with SDS to denature, followed by S-S reduction by TCEP to improve the aptamer immobilization onto Au electrode surface. | Extended-gate field-effect transistor (EGFET) | A linear response range of 1 ng/mL to 100 ng/mL | The use of interchangeable extended gate chips greatly improves the modularity and scalability of the system, making it suitable for healthcare applications. | [177] |
| Cardiac troponin I (cTnI, biomarker of acute myocardial infarction, AMI) | Constructed with ion-sensitive field-effect transistors (ISFETs) and an additional gate that connects with the gate of the ISFET and acts as the sensing area with ample space for molecule recognition and substrate exchange. A Prussian blue-gold nanoparticle (PB-AuNP) composite was immobilized on the extended gate of the EGISFET, providing rich active sites and excellent electrocatalytic ability to amplify signal for pH change. ISFETs have a dual gate working mode of a liquid gate coupling with a back gate where concentrations of targets are converted to pH changes during the detection. | Aptamer specific to cTnI was labelled with biotin at 5′ end was reacted with streptavidin and then with biotin-labelled HRP to prepare capture probe through the high affinity of a biotin-streptavidin-biotin structure. Afterward, auxiliary probe of complimentary strand DNA, thiolated at 3′ end, was dropped on the surface of the APTMS-modified EGISFET and covalently bonded with PB-AuNPs via Au–S bonding. | Extended-gate ion-sensitive field-effect transistor (EGISFET) | 0.3 pg/mL with a linear range of 1–1000 pg/mL | A target-induced strand release strategy is demonstrated to shorten the recognition time of cTnI using a complimentary DNA strand for rapid detection and sensor regeneration. Potential for early warning and rapid diagnosis of AMI in emergency treatment. | [178] |
| Kanamycin (KAN) | The PEGFET sensor consists of an n-MOSFET, a TiO2/ITO extended gate electrode, and an Ag/AgCl reference electrode. The photoelectric gate electrode is made of ITO glass as the substrate, on which a photosensitive TiO2 layer (6 mm diameter circular hole) surrounded by a polyimide film works as the sensing area. The gate of the source metre is connected to an Ag/AgCl reference electrode, while its source and drain are connected to the source and drain of a commercial n-MOSFET. The gate terminal of the n-MOSFET is linked to a TiO2/ITO electrode, serving as an extended gate. Both the extended gate and Ag/AgCl reference electrode are immersed in the test solution to form a complete gate source circuit. | During the sensing process, the single-strand DNA (ssDNA) aptamer is linked onto the Au nanoclusters (NCs) for KAN recognition. When target KAN is present, the KAN molecules will bind to the surface of AuNCs and weaken their catalytic ability. Thus, the deposited DAB layer becomes thinner, and the photocurrent increases. Different amounts of DAB precipitate on the photoelectrode surface, leading to gate voltage shift and source-drain current response. | Photoelectrochemical extended-gate field-effect transistor (PEGFET) | At nM level with a linear range of 10 nM to 100 μM | Provide new insights for the development of EGFET sensor with photoelectrochemical process for practical sensing applications. | [179] |
| Brain-derived neurotrophic factor (BDNF) | Constructed on a flexible polyimide (PI) membrane coated with Au/reduced graphene oxide (r-GO) and utilized an extended-gate field-effect transistor (EGFET) as the transducer. The negatively charged DNA aptamers as recognition sites on the surface of the modified electrode enable the specific binding of BDNF within the r-GO/PBASE/APT/BSA modified electrode matrix. Applying a positive bias to the floating gate electrode attracts positively charged BDNF molecules at pH 7 to the modified electrode surface to enhance the binding and induce the surface potential charge for detection. | DNA aptamer specific to BDNF immobilized on sequentially modified PI membrane substrate with Au/r-GO/PBASE via a chemical linker EDC (1-ethyl-3-(dimethylaminopropyl) carbodiimide-NHS (N-hydroxysuccinimide) for coupling reaction with PBASE. The surface was further modified with BSA to enhance sensitivity and selectivity for BDNF. | Extended-gate field-effect transistor (EGFET) | 0.4 nM with a dynamic range of 0.025 to 1000 nM | First electrical aptasensor for BDNF detection, reliable, highly sensitive, wide range, cost effective, fast response, flexible, portable, and early diagnosis. | [166] |
| Vascular endothelial growth factor 165 (VEGF165) | A DNA strand (T1) is to functionalize the graphene channel interface of gFET with PASE as the linker for capture of another DNA strand (T2) containing the sequence complementary to the aptamer (P1) specific to VEGF165, which was released from the double-stranded DNA-aptamer complex (P1-T2), owing to higher affinity of the aptamer P1 to VEGF165. The captured T2 could trigger hybridization chain reaction in the presence of hairpin H1 and H2 on the channel interface; thus, the DNA structure on the interface obviously changed in the presence of target protein. Due to being negatively charged, DNA hybridization enhanced the hole-doping effect on the graphene, which caused a target protein-related negative shift in Dirac voltage of the GFET device, and, thus, a sensitivity method for analysis of target protein could be realized by the DNA hybridization on the GFET-based biosensors. | T1 modified with amino group at 5′ end was covalently bound on the graphene interface using PASE as the linker while PASE could be absorbed on graphene via π–π interaction. P1 was the aptamer specific to VEGF165 and hybridized with T2. After the target VEGF165 was recognized by the aptamer, T2 could be released from P1-T2 complex to hybridize with T1 on the channel interface. | Graphene field-effect transistor (gFET) | 3.24 pg/mL | The proposed biosensing strategy possessed excellent extensibility for other proteins or even nucleic acids by simply changing the specific aptamer. | [167] |
| Vascular endothelial growth factor (VEGF) | The remote dual gates were fabricated by coating poly [3-(3-carboxypropyl)thiophene-2,5-diyl] (PT-COOH) transducer film on SiO2/Si substrates. The probe functionalization was performed with aptamer DNA strand (VEap) either alone (PT-VEap) or in conjunction with its complementary single-stranded DNA (PT-VEap/CS). The remote dual gates were integrated with commercial MOS-FET for detection (as illustrated in Figure 10). | Amino-modified aptamer DNA strand VEap for VEGF121 were immobilized onto the PT-COOH film via acylation using EDC/NHS chemistry to form PT-VEap or PT-VEap/CS sensing surface on the remote gates. Charged backbones of the aptamers facilitated specific recognition in complex surroundings and maintained high sensitivity to VEGF across a broad pH range (5–9) and ionic strength (0.05–1.0 × PBS). | Remote dual-gate organic field-effect transistors (OFETs) | 0.1 ng/mL | The PT-VEap and (PT-VEap/CS) aptamer sensors exhibited opposite changes in threshold voltage in response to VEGF, and the CS pairing helped PT-VEap/CS sensors achieve 10 times lower detection limit. | [168] |
| Escherichia coli (E. coli) | FET sensor was built based on rGO, gold nanoparticles (AuNPs), and an amino ssDNA aptamer to detect whole cells of E. coli. At first, source and drain electrodes were formed on silicon wafer using photolithography and lift-off process. Then, rGO was coated onto the sensor surface with GO solution via thermal reduction, followed by AuNPs sputtering. | Amine terminated amino ssDNA aptamer specific to E. coli was immobilized on the rGO surface by EDC/NHS chemistry. | rGO-AuNPs field-effect transistor | 3 CFU/mL with a linear range of 3–3 × 106 CFU/mL | Amine terminated ssDNA aptamers are less expensive than thiolated aptamers (attached to AuNPs) and provide a strong covalent bond with rGO; thus, FET performance was enhanced but its cost reduced. | [180] |
| Cortisol (a key stress biomarker) | To fabricate FETs on flexible substrates for conformal skin contact, thin-film In2O3 was formed on polyimide via spin coating In2O3 precursor followed by solution-processed sol-gel chemistry. The In2O3 layer was then patterned by photolithography and reactive ion etching to form the channel regions. Interdigitated Au/Ti electrodes were patterned to form source and drain contacts. | The In2O3 was functionalized with APTES and PTMS (1:9 v/v ratio) via self-assembly using vapour-phase deposition. Cortisol aptamer with a thiol modification at the 5′ end was covalently immobilized on amino-silanized In2O3 FET channels using MBS as a crosslinker. | Flexible field-effect transistor (FET) | 1 pM with a dynamic range of 1 pM to 1 µM | Demonstrated the flexible FET array system can be straightforwardly adapted in wearable and mobile formats for additional physiological biomarkers, inc. targets at low concentrations in sweat. | [181] |
| Cortisol (a key depression marker) | ISEFT sensor was fabricated on n-type silicon wafer with multiple steps, including wet oxidation for SiO2 layer, boron implantation for contact interconnects, lithography lift-off process for source and drain contact patches with an Al/Ti/Au metal stack, another wet oxidation process to produce SiO2 layer to serve as a device passivation, and another lithography process to open window contacts for the source/drain/gate. Afterward, a dry oxidation process to grown thin-layer SiO2 to passivate the sensing area. Finally, another lift-off process to produce outer source/drain contacts using the similar Al/Ti/Au stack. | The ISFET surface was functionalized with APTES, self-assembled through vapour-phase deposition via salinization process. The APTES molecule features a group that covalently binds to gate oxide and an amine group at the other end, which can react further with linker MBS. The thiol group at the 5′ end of the aptamers cross-linked with the amine-terminated silanes via MBS. | Ion-sensitive field-effect transistor (ISFET) | 1 fM with a dynamic range of 1 fM to 1 µM | Suitable for integration into portable and wearable devices. A great potential for the development of accurate point-of-care monitoring system for early detection of depression disorders. | [182] |
3.4. Electrochemical Aptasensors
3.4.1. Electrochemical Impedance Spectroscopy Aptasensors
3.4.2. Voltametric Aptasensors
3.4.3. Amperometric Aptasensors
4. Post-SELEX Optimization Process of Aptamers
4.1. Truncation
4.2. Other Post-SELEX Optimization Methods
5. Conclusions and Outlook
- (1)
- It is more difficult to screen aptamers for small molecule targets than for large ones. It is worth highlighting a new type of SELEX technology, named Capture-SELEX, can be used in the field of screening aptamers for small molecule targets, despite that few SELEX technologies applied in this field [265].
- (2)
- Point-of-care (POC) diagnostic systems have become increasingly demanded in healthcare and clinical diagnosis; aptamer-based biosensing systems have proven their feasibility, but they are still in their infancy. There is still a significant gap in affordability, standardization, and commercialization [266,267,268].
- (3)
- Wearable aptasensors are a brand-new field that combines flexible materials, artificial intelligence, machine learning, and aptasensors, but it is still in its infancy at present. In the future, there are still huge challenges ahead in improving the consumption of wearable devices, the collection of detection data, and the storage of wearable aptasensors under various physiological conditions and in complex external environments [269].
- (4)
- Besides aptamers, some emerging nanomaterials also demonstrated promise for the enhanced performance of aptasensors, despite some condition limitations observed [270,271]. For example, the sustainability and toxicity of nanomaterials in sensor applications have been insufficiently investigated. The fabrication of nanomaterials and nano-biosensors is usually complicated and investment-intensive, which could also cause affordability and accessibility issues for nanomaterial-based aptasensors. Thus, more research should be performed to address these issues.
- (5)
- The discovery of aptamers and their applications in sensing have become an interdisciplinary research field across physics, chemistry, biology, materials science, and computer science, and several recent aptasensor designs have demonstrated that deep learning and predictive models can effectively enhance the performance of aptasensors, while significantly shortening the discovery time of the aptamers as well as running costs [272,273,274,275,276,277,278,279,280,281,282,283,284,285]. In the future, the development and adoption of advanced predictive algorithms and computational tools are expected to have a significant impact on the development of high-performance and low-cost aptasensors. Especially, hybrid in silico approaches combining machine learning (ML) and structure-driven modelling will be increasingly common for the balanced benefits in scalability (ML) and physical realism (structural modelling).
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Farid, S.; Ghosh, S.; Dutta, M.; Stroscio, M.A. Aptamer-Based Optical and Electrochemical Sensors: A Review. Chemosensors 2023, 11, 569. [Google Scholar] [CrossRef]
- Lan, Y.; Farid, S.; Meshik, X.; Xu, K.; Choi, M.; Ranginwala, S.; Wang, Y.; Burke, P.; Dutta, M.; Stroscio, M.A. Detection of immunoglobulin E with a graphene-based field-effect transistor aptasensor. J. Sens. 2018, 2018, 1–8. [Google Scholar] [CrossRef]
- Famulok, M.; Mayer, G.; Blind, M. Nucleic acid aptamers from selection in vitro to applications in vivo. Acc. Chem. Res. 2000, 33, 591–599. [Google Scholar] [CrossRef]
- Wang, Y.; Ye, Z.; Ping, J.; Jing, S.; Ying, Y. Development of an aptamer-based impedimetric bioassay using microfluidic system and magnetic separation for protein detection. Biosens. Bioelectron. 2014, 59, 106–111. [Google Scholar] [CrossRef]
- Lamberti, I.; Scarano, S.; Esposito, C.L.; Antoccia, A.; Antonini, G.; Tanzarella, C.; Franciscis, V.D.; Minunni, M. In vitro selection of RNA aptamers against CA125 tumor marker in ovarian cancer and its study by optical biosensing. Methods 2016, 97, 58–68. [Google Scholar] [CrossRef]
- Villalonga, A.; Pérez-Calabuig, A.M.; Villalonga, R. Electrochemical biosensors based on nucleic acid aptamers. Anal. Bioanal. Chem. 2020, 412, 55–72. [Google Scholar] [CrossRef]
- Zhao, Y.; Yavari, K.; Liu, J. Critical evaluation of aptamer binding for biosensor designs. TrAC Trends Anal. Chem. 2022, 146, 116480. [Google Scholar] [CrossRef]
- Sanford, A.A.; Rangel, A.E.; Feagin, T.A.; Lowery, R.G.; Argueta-Gonzalez, H.S.; Heemstra, J.M. RE-SELEX: Restriction enzyme-based evolution of structure-switching aptamer biosensors. Chem. Sci. 2021, 12, 11692–11702. [Google Scholar] [CrossRef]
- Mok, W.; Li, Y. Recent progress in nucleic acid aptamer-based biosensors and bioassays. Sensors 2008, 8, 7050–7084. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Yang, G.; Zhang, X.; Qu, F. Development of aptamer screening against proteins and its applications. Chin. J. Anal. Chem. 2020, 48, 560–572. [Google Scholar] [CrossRef]
- Zheng, G.; Zhao, L.; Yuan, D.; Li, J.; Guang, Y.; Song, D.; Miao, H.; Shu, L.; Mo, X.; Xu, X.; et al. A genetically encoded fluorescent biosensor for monitoring ATP in living cells with heterobifunctional aptamers. Biosens. Bioelectron. 2022, 198, 113827. [Google Scholar] [CrossRef]
- Kleinjung, F.; Klussmann, S.; Erdmann, V.A.; Scheller, F.W.; Fürste, J.P.; Bier, F.F. High-affinity RNA as a recognition element in a biosensor. Anal. Chem. 1998, 70, 328–331. [Google Scholar] [CrossRef]
- Eriksson, E.S.E.; Joshi, L.; Billeter, M.; Eriksson, L.A. De novo tertiary structure prediction using RNA123—Benchmarking and application to Macugen. J. Mol. Model. 2014, 20, 2389. [Google Scholar] [CrossRef]
- Hayashi, T.; Oshima, H.; Tsukasa, M.; Nagata, T.; Masato, K.; Kinoshita, M. Binding of an RNA aptamer and a partial peptide of a prion protein: Crucial importance of water entropy in molecular recognition. Nucleic Acids Res. 2014, 42, 6861–6875. [Google Scholar] [CrossRef]
- Kwame, S.; Phillips, J.A.; Xiong, X.; Meng, L.; Simaeys, D.V.; Chen, H.; Maetin, J.; Tan, W. Nucleic acid aptamers for biosensors and bio-analytical applications. Analyst 2009, 134, 1765–1775. [Google Scholar] [CrossRef]
- Byun, J. Recent progress and opportunities for nucleic acid aptamers. Life 2021, 11, 193. [Google Scholar] [CrossRef]
- Fischer, M.J.M.; Schmidt, J.; Stanislav Koulchitskym, S.; Klussmann, S.; Vater, A.; Meßlinger, K. Effect of a calcitonin gene-related peptide-binding L-RNA aptamer on neuronal activity in the rat spinal trigeminal nucleus. J. Headache Pain. 2018, 19, 3. [Google Scholar] [CrossRef]
- Ruckman, J.; Green, L.S.; Beeson, J.; Waugh, S.; Gillette, W.L.; Henninger, D.D.; Claesson-Welsh, L.; Janjić, N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998, 273, 20556–20567. [Google Scholar] [CrossRef] [PubMed]
- Savla, R.; Taratula, O.; Garbuzenko, O.; Minko, T. Tumor targeted quantum dot-mucin 1 aptamer-doxorubicin conjugate for imaging and treatment of cancer. J. Control. Release 2011, 153, 16–22. [Google Scholar] [CrossRef]
- Mann, D.; Reinemann, C.; Stoltenburg, R.; Strehlitz, B. In vitro selection of DNA aptamers binding ethanolamine. Biochem. Biophys. Res. Commun. 2005, 338, 1928–1934. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, J.; Hirofumi, I.; Yokota, Y.; Sugimoto, N. In vitro selection of aptamers that act with Zn2+. J. Inorg. Biochem. 2000, 82, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Hamula, C.; Le, X.C.; Li, X. DNA aptamers binding to multiple prevalent M-types of Streptococcus pyogenes. Anal. Chem. 2011, 83, 3640–3647. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Parekh, P.; Turner, P.; Moyer, R.W.; Tan, W. Generating aptamers for recognition of virus-infected cells. Clin. Chem. 2009, 55, 813–822. [Google Scholar] [CrossRef]
- Chen, F.; Hu, Y.; Li, D.; Chen, H.; Zhang, X.-L. CS-SELEX generates high-affinity ssDNA aptamers as molecular probes for Hepatitis C virus envelope glycoprotein E2. PLoS ONE 2009, 4, e8142. [Google Scholar] [CrossRef]
- Gold, L. Oligonucleotides as research, diagnostic, and therapeutic agents. J. Biol. Chem. 1995, 270, 13581–13584. [Google Scholar] [CrossRef]
- Zhang, Y.; Lai, B.; Juhas, M. Recent advances in aptamer discovery and applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef]
- Fraser, L.A.; Cheung, Y.W.; Kinghorn, A.B.; Guo, W.; Shui, S.C.-C.; Jianta, C.; Liu, M.; Bhuyan, S.; Nsn, L.; Shum, H.C.; et al. Microfluidic technology for nucleic acid aptamer evolution and application. Adv. Biosyst. 2019, 3, 1900012. [Google Scholar] [CrossRef]
- Gopinath, S.C.B. Methods developed for SELEX. Anal. Bioanal. Chem. 2006, 387, 171–182. [Google Scholar] [CrossRef]
- Binkley, J.; Allen, P.; Brown, D.M.; Green, L.; Tuerk, C.; Gold, L. RNA ligands to human nerve growth factor. Nucleic Acids Res. 1995, 23, 3198–3205. [Google Scholar] [CrossRef][Green Version]
- Marshall, K.A.; Ellington, A.D. In vitro selection of RNA aptamers. Methods Enzymol. 2000, 318, 193–214. [Google Scholar] [CrossRef]
- Uphoff, K.W.; Bell, S.D.; Ellington, A.D. In vitro selection of aptamers: The dearth of pure reason. Curr. Opin. Struct. Biol. 1996, 6, 281–288. [Google Scholar] [CrossRef]
- Famulok, M.; Szostak, J.W. In vitro selection of specific ligand-binding nucleic acids. Angew. Chem. Int. Ed. Engl. 1992, 31, 979–988. [Google Scholar] [CrossRef]
- Mendonsa, S.D.; Bowser, M.T. In vitro evolution of functional DNA using capillary electrophoresis. J. Am. Chem. Soc. 2004, 126, 20–21. [Google Scholar] [CrossRef]
- Zhu, C.; Yang, G.; Ghulam, M.; Li, L.; Qu, F. Evolution of multi-functional capillary electrophoresis for high-efficiency selection of Aptamers. Biotechnol. Adv. 2019, 37, 107432. [Google Scholar] [CrossRef]
- Yu, X.; Yu, Y. A mathematical analysis of the selective enrichment of NECEEM-Based Non-SELEX. Appl. Biochem. Biotechnol. 2014, 17, 2019–2027. [Google Scholar] [CrossRef] [PubMed]
- Tok, J.; Lai, J.; Leung, T.; Li, S.F.Y. Selection of aptamers for signal transduction proteins by capillary electrophoresis. Electrophoresis 2010, 31, 2055–2062. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Zhu, C.; Zhao, L.; Li, L.; Huang, Y.; Zhang, Y.; Qu, F. Pressure controllable aptamers picking strategy by targets competition. Chin. Chem. Lett. 2020, 32, 218–220. [Google Scholar] [CrossRef]
- Zhu, C.; Feng, Z.; Qin, H.; Chen, L.; Yan, M.; Li, L.; Qu, F. Recent progress of SELEX methods for screening nucleic acid aptamers. Talanta 2014, 266, 124998. [Google Scholar] [CrossRef]
- Chung, Y.-D.; Tsai, Y.-C.; Wang, C.-H.; Lee, G.-B. Aptamer selection via versatile microfluidic platforms and their diverse applications. Lab. A Chip 2025, 25, 1047–1080. [Google Scholar] [CrossRef] [PubMed]
- Gotrik, M.R.; Feagin, T.A.; Csordas, A.T.; Nakamoto, M.A.; Soh, H.T. Advancements in aptamer discovery technologies. Acc. Chem. Res. 2016, 49, 1903–1910. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-Y.; Hsieh, I.-S.; Tung, C.-H.; Weng, C.-H.; Wu, J.-E.; Yu, J.-S.; Hong, T.-M.; Chen, Y.-L. A novel DNA aptamer targeting lung cancer stem cells exerts a therapeutic effect by binding and neutralizing Annexin A2. Mol. Ther. Nucleic Acids 2022, 27, 956–968. [Google Scholar] [CrossRef]
- Sun, X.; Xie, L.; Qiu, S.; Li, H.; Zhou, L.; Zhang, Y.; Zhang, L.; Xie, T.; Chen, Y.; Zhang, L.; et al. Elucidation of CKAP4-remodeled cell mechanics in driving metastasis of bladder cancer through aptamer-based target discovery. Proc. Natl. Acad. Sci. USA 2022, 119, e2110500119. [Google Scholar] [CrossRef]
- Ren, M.; Zhou, J.; Song, Z.; Mei, H.; Zhou, M.; Fu, Z.F.; Han, H.; Zhao, L. Aptamer and RVG functionalized gold nanorods for targeted photothermal therapy of neurotropic virus infection in the mouse brain. Chem. Eng. J. 2021, 411, 128557. [Google Scholar] [CrossRef]
- Zhu, H.; Wu, E.; Pan, Z.; Zhang, C.; Zhang, Y.; Liao, Q.; Wang, Y.; Sun, Y.; Ye, M.; Wu, W. Development of an aptamer-based molecular tool for specifically targeting Microglia via the CD64 Protein. Anal. Chem. 2023, 95, 3238–3246. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Liu, H.; Hu, Y.; Wang, Y.; Zhao, M.; Yuan, Y.; Han, Y.; Jing, Y.; Cui, J.; Ren, X.; et al. Selection and identification of a novel ssDNA aptamer targeting human skeletal muscle. Bioact. Mater. 2023, 20, 166–178. [Google Scholar] [CrossRef]
- Pleiko, K.; Saulite, L.; Parfejevs, V.; Miculis, K.; Vjaters, E.; Riekstina, U. Differential binding cell-SELEX method to identify cell-specific aptamers using high-throughput sequencing. Sci. Rep. 2019, 9, 8142. [Google Scholar] [CrossRef]
- Khan, N.I.; Song, E. Lab-on-a-Chip Systems for Aptamer-Based Biosensing. Micromachines 2020, 11, 220. [Google Scholar] [CrossRef]
- O’Sullivan, C.K. Aptasensors—The future of biosensing? Anal. Bioanal. Chem. 2002, 372, 44–48. [Google Scholar] [CrossRef]
- Hosseinzadeh, L.; Mazloum-Ardakani, M. Advances in aptasensor technology. Adv. Clin. Chem. 2020, 99, 237–279. [Google Scholar] [CrossRef]
- Sassolas, A.; Blum, L.J.; Leca-Bouvier, B.D. Electrochemical aptasensors. Electroanalysis 2009, 21, 1237–1250. [Google Scholar] [CrossRef]
- Sequeira-Antunes, B.; Ferreira, H.A. Nucleic acid aptamer-based biosensors: A Review. Biomedicines 2023, 11, 3201. [Google Scholar] [CrossRef] [PubMed]
- Radi, A.-E. Electrochemical aptamer-based biosensors: Recent Advances and Perspectives. Int. J. Electrochem. 2011, 1, 863196. [Google Scholar] [CrossRef]
- Yao, W.; Shi, J.; Ling, J.; Guo, Y.; Ding, C.; Ding, Y. SiC-functionalized fluorescent aptasensor for determination of Proteus mirabilis. Microchim. Acta 2020, 187, 406. [Google Scholar] [CrossRef]
- Song, S.; Wang, L.; Li, J.; Fan, C.; Zhao, J. Aptamer-based biosensors. TrAC Trends Anal. Chem. 2008, 27, 108–117. [Google Scholar] [CrossRef]
- Hong, P.; Li, W.; Li, J. Applications of aptasensors in clinical diagnostics. Sensors 2012, 12, 1181–1193. [Google Scholar] [CrossRef]
- Wang, Z.; Wilkop, T.; Xu, D.; Dong, Y.; Ma, G.; Cheng, Q. Surface plasmon resonance imaging for affinity analysis of aptamer–protein interactions with PDMS microfluidic chips. Anal. Bioanal. Chem. 2007, 389, 819–825. [Google Scholar] [CrossRef]
- So, H.-M.; Won, K.; Yong, H.K.; Kim, B.-K.; Beyong, H.R.; Pil, S.N.; Kim, H.; Lee, J.O. Single-walled carbon nanotube biosensors using aptamers as molecular recognition elements. J. Am. Chem. Soc. 2005, 127, 11906–11907. [Google Scholar] [CrossRef] [PubMed]
- Polsky, R.; Gill, R.; Kaganovsky, L.; Willner, I. Nucleic acid-functionalized Pt nanoparticles: Catalytic labels for the amplified electrochemical detection of biomolecules. Anal. Chem. 2006, 78, 2268–2271. [Google Scholar] [CrossRef]
- Zayats, M.; Huang, Y.; Gill, R.; Ma, C.; Willner, I. Label-free and reagentless aptamer-based sensors for small molecules. J. Am. Chem. Soc. 2006, 128, 13666–13667. [Google Scholar] [CrossRef]
- Phillips, J.A.; Xu, Y.; Xia, Z.; Fan, Z.H.; Tan, W. Enrichment of cancer cells using aptamers immobilized on a microfluidic channel. Anal. Chem. 2008, 81, 1033–1039. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, S.; Obubuafo, A.; Soper, S.A.; Spivak, D.A. Surface immobilization methods for aptamer diagnostic applications. Anal. Bioanal. Chem. 2007, 390, 1009–1021. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, H.; Wrenger, C. Disease-specific biomarker discovery by aptamers. Cytom. Part A 2009, 75A, 727–733. [Google Scholar] [CrossRef]
- Jayasena, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [CrossRef]
- Yildirim, N.; Long, F.; Gao, C.; He, M.; Shi, H.-C.; Gu, A.Z. Aptamer-based optical biosensor for rapid and sensitive detection of 17β-Estradiol in water samples. Environ. Sci. Technol. 2012, 46, 3288–3294. [Google Scholar] [CrossRef]
- Dittmer, W.U.; Reuter, A.; Simmel, F.C. A DNA-based machine that can cyclically bind and release Thrombin. Angew. Chem. Int. Ed. 2004, 43, 3550–3553. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, V.; Xiao, Y.; Shlyahovsky, B.; Willner, I. Aptamer-functionalized Au nanoparticles for the amplified optical detection of Thrombin. J. Am. Chem. Soc. 2004, 126, 11768–11769. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.-A.; Leclerc, M. Optical sensors based on hybrid aptamer/conjugated polymer complexes. J. Am. Chem. Soc. 2004, 126, 1384–1387. [Google Scholar] [CrossRef]
- Feng, C.; Dai, S.; Wang, L. Optical aptasensors for quantitative detection of small biomolecules: A review. Biosens. Bioelectron. 2014, 59, 64–74. [Google Scholar] [CrossRef]
- Zahra, Q.; Khan, Q.A.; Luo, Z. Advances in optical aptasensors for early detection and diagnosis of various cancer types. Front. Oncol. 2021, 11, 632165. [Google Scholar] [CrossRef]
- Ng, S.; Lim, H.S.; Ma, Q.; Gao, Z. Optical Aptasensors for adenosine triphosphate. Theranostics 2016, 6, 1683–1702. [Google Scholar] [CrossRef]
- Mao, S.; Yu, K.; Lu, G.; Chen, J. Highly sensitive protein sensor based on thermally-reduced graphene oxide field-effect transistor. Nano Res. 2011, 4, 921–930. [Google Scholar] [CrossRef]
- Ghosh, S.; Khan, N.I.; Tsavalas, J.G.; Song, E. Selective detection of lysozyme biomarker utilizing large area chemical vapor deposition-grown graphene-based field-effect transistor. Front. Bioeng. Biotechnol. 2018, 6, 29. [Google Scholar] [CrossRef]
- Ohno, Y.; Maehashi, K.; Matsumoto, K. Label-free biosensors based on aptamer-modified graphene field-effect transistors. J. Am. Chem. Soc. 2010, 132, 18012–18013. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Lee, J.; Seo, S.E.; Kim, K.H.; Park, C.S.; Lee, S.H.; Ban, H.S.; Lee, B.D.; Song, H.C.; Kim, J.; et al. High-performance conducting polymer nanotube-based liquid-ion gated field-effect transistor aptasensor for dopamine exocytosis. Sci. Rep. 2020, 10, 3772. [Google Scholar] [CrossRef]
- Wang, J.; Chen, D.; Huang, W.; Yang, N.; Yuan, Q.; Yang, Y. Aptamer-functionalized field-effect transistor biosensors for disease diagnosis and environmental monitoring. Exploration 2023, 3, 20210027. [Google Scholar] [CrossRef]
- Nguyen, T.T.-H.; Nguyen, C.M.; Huynh, M.A.; Vu, H.H.; Nguyen, T.K.; Nguyen, N.T. Field effect transistor based wearable biosensors for healthcare monitoring. J. Nanobiotechnol. 2023, 21, 411. [Google Scholar] [CrossRef]
- Vu, C.-A.; Chen, W.-Y. Predicting future prospects of aptamers in field-effect transistor biosensors. Molecules 2020, 25, 680. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.; Lee, Y.; Lee, T.; Ahn, J.H. Aptamer-based field-effect transistor for detection of Avian Influenza Virus in chicken serum. Anal. Chem. 2020, 92, 5524–5531. [Google Scholar] [CrossRef]
- Tran, T.-T.; Mulchandani, A. Carbon nanotubes and graphene nano field-effect transistor-based biosensors. TrAC Trends Anal. Chem. 2016, 79, 222–232. [Google Scholar] [CrossRef]
- Kim, S.G.; Lee, J.S.; Jun, J.; Shin, D.H.; Jang, J. Ultrasensitive bisphenol A field-effect transistor sensor using an aptamer-modified multichannel carbon nanofiber transducer. ACS Appl. Mater. Interfaces 2016, 8, 6602–6610. [Google Scholar] [CrossRef] [PubMed]
- Kwon, O.S.; Park, S.J.; Hong, J.-Y.; Han, A.-R.; Lee, J.S.; Lee, J.S.; Oh, J.H.; Jang, J. Flexible FET-type VEGF aptasensor based on nitrogen-doped graphene converted from conducting polymer. ACS Nano 2012, 6, 1486–1493. [Google Scholar] [CrossRef] [PubMed]
- Grabowska, I.; Sharma, N.; Vasilescu, A.; Iancu, M.; Badea, G.; Boukherroub, R.; Ogale, S.; Szunerits, S. Electrochemical aptamer-based biosensors for the detection of cardiac biomarkers. ACS Omega 2018, 3, 12010–12018. [Google Scholar] [CrossRef]
- Khan, N.I.; Maddaus, A.G.; Song, E. A low-cost inkjet-printed aptamer-based electrochemical biosensor for the selective detection of lysozyme. Biosensors 2018, 8, 7. [Google Scholar] [CrossRef]
- Crulhas, B.P.; Karpik, A.E.; Delella, F.K.; Castro, G.R.; Pedrosa, V.A. Electrochemical aptamer-based biosensor developed to monitor PSA and VEGF released by prostate cancer cells. Anal. Bioanal. Chem. 2017, 409, 6771–6780. [Google Scholar] [CrossRef]
- Rohrbach, F.; Karadeniz, H.; Erdem, A.; Famulok, M.; Mayer, G. Label-free impedimetric aptasensor for lysozyme detection based on carbon nanotube-modified screen-printed electrodes. Anal. Biochem. 2012, 421, 454–459. [Google Scholar] [CrossRef]
- Cheng, A.; Ge, B.; Yu, H.-Z. Aptamer-based biosensors for label-free voltametric detection of lysozyme. Anal. Chem. 2007, 79, 5158–5164. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-D.; Chen, Z.-B.; Zhao, H.-T.; Guo, L.; Mu, X. An aptamer-based biosensor for the detection of lysozyme with gold nanoparticles amplification. Sens. Actuators B 2010, 149, 110–115. [Google Scholar] [CrossRef]
- Lian, Y.; He, F.; Mi, X.; Tong, F.; Shi, X. Lysozyme aptamer biosensor based on electron transfer from SWCNTs to SPQC-IDE. Sens. Actuators B Chem. 2014, 199, 377–383. [Google Scholar] [CrossRef]
- Liang, G.; Man, Y.; Jin, X.; Pan, L.; Liu, X. Aptamer-based biosensor for label-free detection of ethanolamine by electrochemical impedance spectroscopy. Anal. Chim. Acta 2016, 936, 222–228. [Google Scholar] [CrossRef]
- Ikebukuro, K.; Kiyohara, C.; Sode, K. Electrochemical detection of protein using a double aptamer sandwich. Anal. Lett. 2004, 37, 2901–2909. [Google Scholar] [CrossRef]
- Rodriguez, M.C.; Kawde, A.-N.; Wang, J. Aptamer biosensor for label-free impedance spectroscopy detection of proteins based on recognition-induced switching of the surface charge. Chem. Commun. 2005, 34, 4267. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, W.; Wang, J.; Yang, C.; Yang, F.; Yang, X. A sensitive impedimetric thrombin aptasensor based on polyamidoamine dendrimer. Talanta 2009, 78, 1240–1245. [Google Scholar] [CrossRef]
- Wei, Y.; Li, B.; Wang, X.; Duan, Y. Magnified fluorescence detection of silver(I) ion in aqueous solutions by using nano-graphite-DNA hybrid and DNase, I. Biosens. Bioelectron. 2014, 58, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Ji, G.; Tian, J.; Xing, F.; Feng, Y. Optical biosensor based on graphene and its derivatives for detecting biomolecules. Int. J. Mol. Sci. 2022, 23, 10838. [Google Scholar] [CrossRef] [PubMed]
- Lafleur, J.P.; Jönsson, A.; Senkbeil, S.; Kutter, J.P. Recent advances in lab-on-a-chip for biosensing applications. Biosens. Bioelectron. 2016, 76, 213–233. [Google Scholar] [CrossRef] [PubMed]
- Kong, R.-M.; Zhang, X.; Ding, L.; Yang, D.; Qu, F. Label-free fluorescence turn-on aptasensor for prostate-specific antigen sensing based on aggregation-induced emission–silica nanospheres. Anal. Bioanal. Chem. 2017, 409, 5757–5765. [Google Scholar] [CrossRef]
- Zhang, M.; Guo, X. Emerging strategies in fluorescent aptasensor toward food hazard aflatoxins detection. Trends Food Sci. Technol. 2022, 129, 621–633. [Google Scholar] [CrossRef]
- Sahoo, H. Förster resonance energy transfer—A spectroscopic nanoruler: Principle and applications. J. Photochem. Photobiol. C: Photochem. Rev. 2011, 12, 20–30. [Google Scholar] [CrossRef]
- Szöllosi, J.; Damjanovich, S.; Mátyus, L. Application of fluorescence resonance energy transfer in the clinical laboratory: Routine and Research. Cytometry 1998, 34, 159–179. [Google Scholar] [CrossRef]
- Kocjan, B.J.; Seme, K.; Poljak, M. Detection and differentiation of human papillomavirus genotypes HPV-6 and HPV-11 by FRET-based real-time PCR. J. Virol. Methods 2008, 153, 245–249. [Google Scholar] [CrossRef]
- Lu, C.-H.; Li, J.; Qi, X.-J.; Song, X.-R.; Yang, H.-H.; Chen, X.; Chen, G.-N. Multiplex detection of nucleases by a graphene-based platform. J. Mater. Chem. 2011, 21, 10915. [Google Scholar] [CrossRef]
- Park, Y.; Dang, T.V.; Jeong, U.; Kim, M.I.; Kim, J. Comparison of optical and electrical sensor characteristics for efficient analysis of attachment and detachment of aptamer. Biosensors 2022, 12, 979. [Google Scholar] [CrossRef]
- He, S.; Song, B.; Li, D.; Zhu, C.; Qi, W.; Wen, Y.; Wang, L.; Song, S.; Fang, H.; Fan, C. A graphene nanoprobe for rapid, sensitive, and multicolor fluorescent DNA analysis. Adv. Funct. Mater. 2010, 20, 453–459. [Google Scholar] [CrossRef]
- Chou, S.S.; De, M.; Luo, J.; Rotello, V.M.; Huang, J.; Dravid, V.P. Nanoscale graphene oxide (nGO) as artificial receptors: Implications for biomolecular interactions and sensing. J. Am. Chem. Soc. 2012, 134, 16725–16733. [Google Scholar] [CrossRef] [PubMed]
- Xi, G.; Chen, T.; Wang, X. A reduced graphene oxide-based fluorescence resonance energy transfer sensor for highly sensitive detection of matrix metalloproteinase 2. Int. J. Nanomed. 2016, 2016, 1537. [Google Scholar] [CrossRef]
- Vermisoglou, E.; Panáček, D.; Jayaramulu, K.; Pykal, M.; Frébort, I.; Kolář, M.; Hajdúch, M.; Zbořil, R.; Otyepka, M. Human virus detection with graphene-based materials. Biosens. Bioelectron. 2020, 166, 112436. [Google Scholar] [CrossRef]
- Shaban, S.M.; Kim, D.-H. Recent advances in aptamer sensors. Sensors 2021, 21, 979. [Google Scholar] [CrossRef] [PubMed]
- Marín, M.J.; Schofield, C.L.; Field, R.A.; Russell, D.A. Glyconanoparticles for colorimetric bioassays. Analyst 2014, 140, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Shayesteh, O.H.; Ghavami, R. A novel label-free colorimetric aptasensor for sensitive determination of PSA biomarker using gold nanoparticles and a cationic polymer in human serum. Spectrochim. Acta Part A 2019, 226, 117644. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, X.; Li, J.; Qiang, W.; Sun, L.; Li, H.; Xu, D. Microfluidic chip-based silver nanoparticles aptasensor for colorimetric detection of thrombin. Talanta 2015, 150, 81–87. [Google Scholar] [CrossRef]
- Cate, D.M.; Adkins, J.A.; Mettakoonpitak, J.; Henry, C.S. Recent developments in paper-based microfluidic devices. Anal. Chem. 2014, 87, 19–41. [Google Scholar] [CrossRef]
- Doeven, E.H.; Barbante, G.J.; Kerr, E.; Hogan, C.F.; Endler, J.A.; Francis, P.S. Red–green–blue electrogenerated chemiluminescence utilizing a digital camera as detector. Anal. Chem. 2014, 86, 2727–2732. [Google Scholar] [CrossRef]
- Zhou, W.; Liang, W.; Li, X.; Chai, Y.; Yuan, R.; Xiang, Y. MicroRNA-triggered, cascaded and catalytic self-assembly of functional ‘DNAzyme ferris wheel’ nanostructures for highly sensitive colorimetric detection of cancer cells. Nanoscale 2015, 7, 9055–9061. [Google Scholar] [CrossRef]
- Lou, B.; Zhou, Z.; Du, Y.; Dong, S. Resistance-based logic aptamer sensor for CCRF-CEM and Ramos cells integrated on microfluidic chip. Electrochem. Commun. 2015, 59, 64–67. [Google Scholar] [CrossRef]
- Yue, F.; Li, F.; Kong, Q.; Guo, Y.; Sun, X. Recent advances in aptamer-based sensors for aminoglycoside antibiotics detection and their applications. Sci. Total Environ. 2021, 762, 143129. [Google Scholar] [CrossRef]
- Zhang, N.; Liu, B.; Cui, X.; Li, Y.; Tang, J.; Wang, H.; Zhang, D.; Li, Z. Recent advances in aptasensors for mycotoxin detection: On the surface and in the colloid. Talanta 2020, 223, 121729. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, F.; Abbaszadeh, H.; Dolatabadi, J.E.N.; Aghebati-Maleki, L.; Yousefi, M. Application of various optical and electrochemical aptasensors for detection of human prostate specific antigen: A review. Biosens. Bioelectron. 2019, 142, 111484. [Google Scholar] [CrossRef] [PubMed]
- Jalalian, S.H.; Lavaee, P.; Ramezani, M.; Danesh, N.M.; Alibolandi, M.; Abnous, K.; Taghdisi, S.M. An optical aptasensor for aflatoxin M1 detection based on target-induced protection of gold nanoparticles against salt-induced aggregation and silica nanoparticles. Spectrochim. Acta Part. A 2021, 246, 119062. [Google Scholar] [CrossRef]
- Khoshbin, Z.; Abnous, K.; Taghdisi, S.M.; Verdian, A. A novel liquid crystal-based aptasensor for ultra-low detection of ochratoxin a using a π-shaped DNA structure: Promising for future on-site detection test strips. Biosens. Bioelectron. 2021, 191, 113457. [Google Scholar] [CrossRef]
- Wei, X.; Yin, M.; Zhang, L.; Lin, H.; Wang, J.; Xie, W.; Xu, D. Surface plasmon resonance (SPR) biosensor for detection of mycotoxins: A review. J. Immunol. Methods 2022, 510, 113349. [Google Scholar] [CrossRef] [PubMed]
- Wijaya, E.; Lenaerts, C.; Maricot, S.; Hastanin, J.; Habraken, S.; Vilcot, J.P.; Boukherroub, R.; Szunerits, S. Surface plasmon resonance-based biosensors: From the development of different SPR structures to novel surface functionalization strategies. Curr. Opin. Solid State Mater. Sci. 2011, 15, 208–224. [Google Scholar] [CrossRef]
- Cooper, M.A. Optical biosensors in drug discovery. Nat. Rev. Drug Discov. 2002, 1, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Canoa, P.; Simón-Vázquez, R.; Popplewell, J.; González-Fernández, Á. A quantitative binding study of fibrinogen and human serum albumin to metal oxide nanoparticles by surface plasmon resonance. Biosens. Bioelectron. 2015, 74, 376–383. [Google Scholar] [CrossRef]
- Omar, N.; Fen, Y.; Saleviter, S.; Daniyal, W.; Anas, N.; Ramdzan, N.; Roshidi, M. Development of a graphene-based surface plasmon resonance optical sensor chip for potential biomedical application. Materials 2019, 12, 1928. [Google Scholar] [CrossRef]
- Green, R.J.; Frazier, R.A.; Shakesheff, K.M.; Davies, M.C.; Roberts, C.J.; Tendler, S.J.B. Surface plasmon resonance analysis of dynamic biological interactions with biomaterials. Biomaterials 2000, 21, 1823–1835. [Google Scholar] [CrossRef]
- HomolaJ, Y.S.S.; Gauglitz, G. Surface plasmon resonance sensors: Review. Sens. Actuators B 1999, 54, 3–15. [Google Scholar] [CrossRef]
- Inamori, K.; Kyo, M.; Nishiya, Y.; Inoue, Y.; Sonoda, T.; Kinoshita, E.; Koike, T.; Katayama, Y. Detection and quantification of on-chip phosphorylated peptides by surface plasmon resonance imaging techniques using a phosphate capture molecule. Anal. Chem. 2005, 77, 3979–3985. [Google Scholar] [CrossRef]
- Spadavecchia, J.; Manera, M.G.; Quaranta, F.; Siciliano, P.; Rella, R. Surface plasmon resonance imaging of DNA based biosensors for potential applications in food analysis. Biosens. Bioelectron. 2005, 21, 894–900. [Google Scholar] [CrossRef]
- Kanoh, N.; Kyo, M.; Inamori, K.; Ando, A.; Asami, A.; Nakao, A.; Osada, H. SPR imaging of photo-cross-linked small-molecule arrays on gold. Anal. Chem. 2006, 78, 2226–2230. [Google Scholar] [CrossRef] [PubMed]
- Myers, F.B.; Lee, L.P. Innovations in optical microfluidic technologies for point-of-care diagnostics. Lab. A Chip 2008, 8, 2015. [Google Scholar] [CrossRef] [PubMed]
- Cennamo, N.; Pasquardini, L.; Arcadio, F.; Lunelli, L.; Vanzetti, L.; Carafa, V.; Altucci, L.; Zeni, L. SARS-CoV-2 spike protein detection through a plasmonic D-shaped plastic optical fiber aptasensor. Talanta 2022, 233, 122532. [Google Scholar] [CrossRef]
- Kong, X.; He, X.; He, F.; Li, Y.; Feng, Y.; Li, Y.; Luo, Z.; Shen, J.; Duan, Y. Sandwich layer-modified Ω-shaped fiber-optic LSPR enables the development of an aptasensor for a Cytosensing–Photothermal Therapy Circuit. ACS Sens. 2024, 9, 4637–4645. [Google Scholar] [CrossRef]
- Ziu, I.; Laryea, E.T.; Fayza, A.; Wu, C.G.; Sanela, M. A dip-and-read optical aptasensor for detection of tau protein. Anal. Bioanal. Chem. 2020, 412, 1193–1201. [Google Scholar] [CrossRef]
- Janik, M.; Brzozowska, E.; Czyszczoń, P.; Celebańska, A.; Koba, M.; Gamian, A.; Bock, W.J.; Śmietana, M. Optical fiber aptasensor for label-free bacteria detection in small volumes. Sens. Actuators B 2021, 330, 129316. [Google Scholar] [CrossRef]
- Shirani, M.; Kalantari, H.; Khodayar, M.J.; Kouchak, M.; Rahbar, N. An ultra-sensitive optical aptasensor based on gold nanoparticles/poly vinyl alcohol hydrogel as acceptor/emitter pair for fluorometric detection of digoxin with on/off/on strategy. Spectrochim. Acta Part. A 2020, 250, 119345. [Google Scholar] [CrossRef]
- Zhao, H.; Ren, S.; Wei, Z.; Lou, X. Evanescent wave optical-fiber aptasensor for rapid detection of zearalenone in corn with unprecedented sensitivity. Biosensors 2022, 12, 438. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Chen, P.; Zhang, L.; Sun, P.; Huang, Y.; Liu, X.; Fan, Q. A universal optical aptasensor for antibiotics determination based on a new high-efficiency Förster resonance energy transfer pair. Microchim. Acta 2024, 191, 561. [Google Scholar] [CrossRef]
- Zhu, P.; Papadimitriou, V.A.; van Dongen, J.E.; Cordeiro, J.; Neeleman, Y.; Santoso, A.; Chen, S.; Eijkel, J.C.T.; Peng, H.; Segerink, L.I.; et al. An optical aptasensor for real-time quantification of endotoxin: From ensemble to single-molecule resolution. Sci. Adv. 2023, 9, eadf5509. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, M.; Chen, J.; Yang, R.; Huang, Z.; Liu, Z.; Li, N.; Shui, L. Liquid crystal-based optical aptasensor for the sensitive and selective detection of Gram-negative bacteria. Sci. China Chem. 2022, 65, 2023–2030. [Google Scholar] [CrossRef]
- Khoshbin, Z.; Zahraee, H.; Ramezani, M.; Alibolandi, M.; Verdian, A.; Abnous, K.; Taghdisi, S.M. Surfactant-regularized liquid crystal aptasensor for optical monitoring of prostate-specific antigen: Appropriate for health care monitoring. Microchem. J. 2023, 193, 109002. [Google Scholar] [CrossRef]
- Taib, M.; Tan, L.L.; Abd Karim, N.H.; Ta, G.C.; Heng, L.Y.; Khalid, B. Reflectance aptasensor based on metal salphen label for rapid and facile determination of insulin. Talanta 2020, 207, 120321. [Google Scholar] [CrossRef]
- Moabelo, K.L.; Lerga, T.M.; Jauset-Rubio, M.; Sibuyi, N.R.S.; O’Sullivan, C.K.; Meyer, M.; Madiehe, A.M. A label-free gold nanoparticles-based optical aptasensor for the detection of retinol binding protein 4. Biosensors 2022, 12, 1061. [Google Scholar] [CrossRef]
- Marjan, M.; Mirzaei, S.; Rezayan, A.H.; Abbasi, V.; Mehrizi, A.A. µPAD-based colorimetric nanogold aptasensor for CRP and IL-6 detection as sepsis biomarkers. Microchem. J. 2023, 197, 109744. [Google Scholar] [CrossRef]
- Loyez, M.; Hassan, E.M.; Lobry, M.; Liu, F.; Caucheteur, C.; Wattiez, R.; DeRosa, M.C.; Willmore, M.G.; Albert, J. Rapid detection of circulating breast cancer cells using a multiresonant optical fiber aptasensor with plasmonic amplification. ACS Sens. 2020, 5, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Lobry, M.; Loyez, M.; Hassan, E.M.; Chah, K.; DeRosa, M.C.; Goormaghtigh, E.; Wattiez, R.; Caucheteur, C. Multimodal plasmonic optical fiber grating aptasensor. Opt. Express 2020, 28, 7539. [Google Scholar] [CrossRef]
- Sanjay, M.; Singh, N.K.; Ngashangva, L.; Goswami, P. A smartphone-based fiber-optic aptasensor for label-free detection of Plasmodium falciparum glutamate dehydrogenase. Anal. Methods 2020, 12, 1333–1341. [Google Scholar] [CrossRef]
- Ning, W.; Hu, S.; Zhou, C.; Luo, J.; Li, Y.; Zhang, C.; Luo, Z.; Li, Y. An ultrasensitive J-shaped optical fiber LSPR aptasensor for the detection of Helicobacter pylori. Anal. Chim. Acta 2023, 1278, 341733. [Google Scholar] [CrossRef] [PubMed]
- Hua, M.Z.; Liu, J.; Roopesh, M.S.; Lu, X. Microfluidic Optical Aptasensor for Small Molecules Based on Analyte-Tuned Growth of Gold Nanoseeds and Machine Learning-Enhanced Spectrum Analysis: Rapid Detection of Mycotoxins. ACS Sens. 2024, 9, 6299–6308. [Google Scholar] [CrossRef]
- Jia, Y.; Zhao, S.; Li, D.; Yang, J.; Yang, L. Portable chemiluminescence optical fiber aptamer-based biosensors for analysis of multiple mycotoxins. Food Control 2022, 144, 109361. [Google Scholar] [CrossRef]
- Lu, G.; Ocola, L.E.; Chen, J. Reduced graphene oxide for room-temperature gas sensors. Nanotechnology 2009, 20, 445502. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-W.; Wu, C.-S.; Chuang, C.-K.; Pang, S.-T.; Pan, T.-M.; Yang, Y.-S.; Ko, F.-H. Real-time and label-free detection of the prostate-specific antigen in human serum by a polycrystalline silicon nanowire field-effect transistor biosensor. Anal. Chem. 2013, 85, 7912–7918. [Google Scholar] [CrossRef]
- Zhou, G.; Chang, J.; Cui, S.; Pu, H.; Wen, Z.; Chen, J. Real-time, selective detection of Pb2+ in water using a reduced graphene oxide/gold nanoparticle field-effect transistor device. ACS Appl. Mater. Interfaces 2014, 6, 19235–19241. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Chen, J. Graphene-based electronic biosensors. J. Mater. Res. 2017, 32, 2954–2965. [Google Scholar] [CrossRef]
- Mao, S.; Chang, J.; Pu, H.; Lu, G.; He, Q.; Zhang, H.; Chen, J. Two-dimensional nanomaterial-based field-effect transistors for chemical and biological sensing. Chem. Soc. Rev. 2017, 46, 6872–6904. [Google Scholar] [CrossRef] [PubMed]
- Wadhera, T.; Kakkar, D.; Wadhwa, G.; Raj, B. Recent advances and progress in development of the field effect transistor biosensor: A review. J. Electron. Mater. 2019, 48, 7635–7646. [Google Scholar] [CrossRef]
- Sedki, M.; Chen, Y.; Mulchandani, A. Non-carbon 2D materials-based field-effect transistor biosensors: Recent advances, challenges, and future perspectives. Sensors 2020, 20, 4811. [Google Scholar] [CrossRef]
- Vu and Chen Field-effect transistor biosensors for biomedical applications: Recent advances and future prospects. Sensors 2019, 19, 4214. [CrossRef]
- Nehra, A; Krishna Nand Singh Current trends in nanomaterial embedded field effect transistor-based biosensor. Biosens. Bioelectron. 2015, 74, 731–743. [CrossRef]
- Poghossian, A.; Yoshinobu, T.; Simonis, A.; Ecken, H.; Lüth, H.; Schöning, M.J. Penicillin detection by means of field-effect based sensors: EnFET, capacitive EIS sensor or LAPS? Sens. Actuators B 2001, 78, 237–242. [Google Scholar] [CrossRef]
- Zhou, Y.; Feng, T.; Li, Y.; Ao, X.; Liang, S.; Yang, X.; Wang, L.; Xu, X.; Zhang, W. Recent advances in enhancing the sensitivity of biosensors based on field effect transistors. Adv. Electron. Mater. 2024, 11, 2400712. [Google Scholar] [CrossRef]
- Douaki, A.; Stuber, A.; Hengsteler, J.; Momotenko, D.; Rogers, D.M.; Rocchia, W.; Hirst, J.D.; Nakatsuka, N.; Garoli, D. Theoretical analysis of divalent cation effects on aptamer recognition of neurotransmitter targets. Chem. Commun. 2023, 59, 14713. [Google Scholar] [CrossRef] [PubMed]
- Stuber, A.; Douaki, A.; Hengsteler, J.; Buckingham, D.; Momotenko, D.; Garoli, D.; Nakatsuka, N. Aptamer conformational dynamics modulate neurotransmitter sensing in nanopores. ACS Nano 2023, 17, 19168–19179. [Google Scholar] [CrossRef] [PubMed]
- Nekrasov, N.; Jaric, S.; Kireev, D.; Emelianov, A.V.; Orlov, A.V.; Gadjanski, I.; Nikitin, P.I.; Akinwande, D.; Bobrinetskiy, I. Real-time detection of ochratoxin A in wine through insight of aptamer conformation in conjunction with graphene field-effect transistor. Biosens. Bioelectron. 2022, 200, 113890. [Google Scholar] [CrossRef]
- Salehirozveh, M.; Bonne, R.; Kumar, P.; Abazar, P.; Dehghani, P.; Mijakovic, I.; Roy, V.A. Enhanced detection of brain-derived neurotrophic factor (BDNF) using a reduced graphene oxide field-effect transistor aptasensor. Nanoscale 2025, 17, 4543. [Google Scholar] [CrossRef]
- Chen, L.; Li, G.; Yang, A.; Wu, J.; Yan, F.; Ju, H. A DNA-functionalized graphene field-effect transistor for quantitation of vascular endothelial growth factor. Sens. Actuators B 2022, 351, 130964. [Google Scholar] [CrossRef]
- Chen, X.; Li, M.; Yang, X.; Cui, J.; Ge, J.; Liu, Y.; Ma, M. Reliable and specific biosensing on single- and double-stranded aptamer functionalized remote dual-gate organic field-effect transistors: A comparison. Talanta 2025, 287, 127634. [Google Scholar] [CrossRef]
- Abrantes, M.; Alexandra, D.; Domingues, T.; Nemala, S.S.; Monteiro, P.; Borme, J.; Alpuim, P.; Jacinto, L. Ultrasensitive dopamine detection with graphene aptasensor multitransistor arrays. J. Nanobiotechnol. 2022, 20, 495. [Google Scholar] [CrossRef]
- Wang, S.; Sun, M.; Zhang, Y.; Ji, H.; Gao, J.; Song, S.; Sun, J.; Liu, H.; Zhang, Y.; Han, L. Ultrasensitive antibiotic perceiving based on aptamer-functionalized ultraclean graphene field-effect transistor biosensor. Anal. Chem. 2022, 94, 14785–14793. [Google Scholar] [CrossRef]
- Palacio, I.; Moreno, M.; Náñez, A.; Purwidyantri, A.; Domingues, T.; Cabral, P.D.; Borme, J.; Marciello, M.; Mendieta-Moreno, I.; Torres-Vázquez, B.; et al. Attomolar detection of hepatitis C virus core protein powered by molecular antenna-like effect in a graphene field-effect aptasensor. Biosens. Bioelectron. 2023, 222, 115006. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Xiao, M.; Feng, X.; Li, T.; Li, P.; Cao, X.; Zhang, J.; Liu, Y.; Wang, L. Real-time, ultra-sensitive and label-free detection of OTA based on DNA aptamer functionalized carbon nanotube field-effect transistor. Sens. Actuators B 2024, 413, 135883. [Google Scholar] [CrossRef]
- Dou, X.; Wu, Q.; Luo, S.; Yang, J.; Dong, B.; Wang, L.; Qu, H.; Zheng, L. A miniaturized biosensor for rapid detection of tetracycline based on a graphene field-effect transistor with an aptamer modified gate. Talanta 2024, 271, 125702. [Google Scholar] [CrossRef]
- Farrow, T.; Laumier, S.; Sandall, I.; van Zalinge, H. An aptamer-functionalised Schottky-field effect transistor for the detection of proteins. Biosensors 2022, 12, 347. [Google Scholar] [CrossRef]
- Rony, M.; Valérie, S.; Fontelaye, C.; Guillaume, N.; Spinelli, N.; Barraud, S. Aptasensors based on silicon nanowire field-effect transistors for electrical detection of thrombin. Microelectron. Eng. 2023, 286, 112130. [Google Scholar] [CrossRef]
- Hu, W.-P.; Wu, Y.-M.; Vu, C.-A.; Chen, W.-Y. Ultrasensitive detection of Interleukin 6 by using silicon nanowire field-effect transistors. Sensors 2023, 23, 625. [Google Scholar] [CrossRef]
- Hung, S.-C.; Lee, Y.-H. Aptamer-based planar electric double-layer field-effect transistor: A novel approach for sensitive Troponin I sensing. Biosensors 2025, 15, 285. [Google Scholar] [CrossRef]
- Zhang, W.; Jiang, J.; Liu, T.; Wang, X.; Zhang, W.; Wang, Y.; Chu, Z.; Jin, W. A rapid and ultrasensitive cardiac troponin I aptasensor based on an ion-sensitive field-effect transistor with extended gate. Talanta 2024, 277, 126364. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Qin, H.; Wei, X.; Tao, T.; Li, Q.; Mao, S. Antibiotic residue detection by novel photoelectrochemical extended-gate field-effect transistor sensor. J. Hazard. Mater. 2024, 485, 136897. [Google Scholar] [CrossRef]
- Alvandi, H.; Hossein, A.; Hajghassem, H.; Rahimi, F.; Moghadam, R.A.; Firoozbakhtian, A. Aptasensor based on rGO-AuNPs field-effect transistor for selective detection of Escherichia coli in river water. Anal. Biochem. 2025, 700, 115796. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhao, C.; Wang, Z.; Yang, K.A.; Cheng, X.; Liu, W.; Yu, W.; Lin, S.; Zhao, Y.; Cheung, K.M.; et al. Wearable aptamer-field-effect transistor sensing system for noninvasive cortisol monitoring. Sci. Adv. 2022, 8, eabk0967. [Google Scholar] [CrossRef]
- Thanh, T.; Nguyen, C.M.; Huynh, M.A.; Trinh, Q.T.; Tanner, P.; Sven Ingebrandt, V.X.T.; Nguyen, T.-K.; Nguyen, N.-T. Ultra-sensitive aptameric field-effect transistor for cortisol sensing. Sens. Actuators Rep. 2025, 9, 100324. [Google Scholar] [CrossRef]
- Feng, X.; Li, P.; Xiao, M.; Li, T.; Chen, B.; Wang, X.; Wang, L. Recent advances in the detection of pathogenic microorganisms and toxins based on field-effect transistor biosensors. Crit. Rev. Food Sci. Nutr. 2023, 64, 9161–9190. [Google Scholar] [CrossRef]
- Syedmoradi, L.; Ahmadi, A.; Norton, M.L.; Omidfar, K. A review on nanomaterial-based field effect transistor technology for biomarker detection. Mikrochim. Acta 2019, 186, 739. [Google Scholar] [CrossRef]
- Luo, X.; Davis, J.J. Electrical biosensors and the label free detection of protein disease biomarkers. Chem. Soc. Rev. 2013, 42, 5944. [Google Scholar] [CrossRef]
- Klinghammer, S.; Voitsekhivska, T.; Licciardello, N.; Kim, K.; Baek, C.-K.; Cho, H.; Wolter, K.-J.; Kirschbaum, C.; Baraban, L.; Cuniberti, G. Nanosensor-based real-time monitoring of stress biomarkers in human saliva using a portable measurement system. ACS Sens. 2020, 5, 4081–4091. [Google Scholar] [CrossRef]
- Estrela, P.; Paul, D.; Song, Q.; Stadler, L.K.J.; Wang, L.; Huq, E.; Davis, J.J.; Ferrigno, P.K.; Migliorato, P. Label-free sub-picomolar protein detection with field-effect transistors. Anal. Chem. 2010, 82, 3531–3536. [Google Scholar] [CrossRef] [PubMed]
- Costa, L.; Brissos, V.; Lemos, F.; Ribeiro, F.R.; Cabral, J.M.S. Comparing the effect of immobilization methods on the activity of lipase biocatalysts in ester hydrolysis. Bioprocess. Biosyst. Eng. 2008, 31, 323–327. [Google Scholar] [CrossRef]
- Sadighbayan, D.; Hasanzadeh, M.; Ghafar-Zadeh, E. Biosensing based on field-effect transistors (FET): Recent progress and challenges. TrAC Trends Anal. Chem. 2020, 133, 116067. [Google Scholar] [CrossRef] [PubMed]
- Asai, K.; Yamamoto, T.; Nagashima, S.; Ogata, G.; Hibino, H.; Einaga, Y. An electrochemical aptamer-based sensor prepared by utilizing the strong interaction between a DNA aptamer and diamond. Analyst 2020, 145, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Xie, M.; Zhao, F.; Han, S. Application of nanomaterial modified aptamer-based electrochemical sensor in detection of heavy metal ions. Foods 2022, 11, 1404. [Google Scholar] [CrossRef] [PubMed]
- Willner, I.; Zayats, M. Electronic aptamer-based sensors. Angew. Chem. Int. Ed. 2007, 46, 6408–6418. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-O.; So, H.-M.; Jeon, E.-K.; Chang, H.; Won, K.; Kim, Y.H. Aptamers as molecular recognition elements for electrical nanobiosensors. Anal. Bioanal. Chem. 2007, 390, 1023–1032. [Google Scholar] [CrossRef]
- Bott, A.W. Electrochemical techniques for the characterization of redox polymers. Curr. Sep. 2001, 19, 71–75. [Google Scholar]
- Katz, E.; Willner, I. Probing biomolecular interactions at conductive and semiconductive surfaces by impedance spectroscopy: Routes to impedimetric immunosensors, DNA-sensors, and enzyme biosensors. Electroanalysis 2003, 15, 913–947. [Google Scholar] [CrossRef]
- Farzin, L.; Shamsipur, M.; Samandari, L.; Sheibani, S. Recent advances in designing nanomaterial based biointerfaces for electrochemical biosensing cardiovascular biomarkers. J. Pharm. Biomed. Anal. 2018, 161, 344–376. [Google Scholar] [CrossRef]
- Guan, J.-G.; Miao, Y.-Q.; Zhang, Q.-J. Impedimetric biosensors. J. Biosci. Bioeng. 2004, 97, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Lisdat, F.; Schäfer, D. The use of electrochemical impedance spectroscopy for biosensing. Anal. Bioanal. Chem. 2008, 391, 1555–1567. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, J.; Cai, Y.; Xu, S.; Wu, H.; Chen, X.; Huang, Y.; Li, F. Integrated electrochemical aptasensor array toward monitoring anticancer drugs in sweat. Anal. Chem. 2024, 96, 4997–5005. [Google Scholar] [CrossRef]
- Leva-Bueno, J.; Peyman, S.A.; Millner, P.A. A review on impedimetric immunosensors for pathogen and biomarker detection. Med. Microbiol. Immunol. 2020, 209, 343–362. [Google Scholar] [CrossRef]
- Magar, H.S.; Hassan, R.Y.A.; Mulchandani, A. Electrochemical impedance spectroscopy (EIS): Principles, construction, and biosensing applications. Sensors 2021, 21, 6578. [Google Scholar] [CrossRef]
- Prodromidis, M.I. Impedimetric immunosensors—A review. Electrochim. Acta 2010, 55, 4227–4233. [Google Scholar] [CrossRef]
- Kokkinos, C.; Economou, A.; Prodromidis, M.I. Electrochemical immunosensors: Critical survey of different architectures and transduction strategies. TrAC Trends Anal. Chem. 2016, 79, 88–105. [Google Scholar] [CrossRef]
- Randviir, E.P.; Banks, C.E. A review of electrochemical impedance spectroscopy for bioanalytical sensors. Anal. Methods 2022, 14, 4602–4624. [Google Scholar] [CrossRef] [PubMed]
- Edmonds, T.E. Voltammetric and amperometric transducers. In Chemical Sensors; Springer: Dordrecht, The Netherlands, 1988. [Google Scholar] [CrossRef]
- Chen, A.; Shah, B. Electrochemical sensing and biosensing based on square wave voltammetry. Anal. Methods 2013, 5, 2158. [Google Scholar] [CrossRef]
- Lu, Y.; Liang, X.; Niyungeko, C.; Zhou, J.; Xu, J.; Tian, G. A review of the identification and detection of heavy metal ions in the environment by voltammetry. Talanta 2018, 178, 324–338. [Google Scholar] [CrossRef]
- Kaur, H.; Shorie, M. Nanomaterial based aptasensors for clinical and environmental diagnostic applications. Nanoscale Adv. 2019, 1, 2123–2138. [Google Scholar] [CrossRef]
- Singh, A.; Sharma, A.; Ahmed, A.; Sundramoorthy, A.K.; Furukawa, H.; Arya, S.; Khosla, A. Recent advances in electrochemical biosensors: Applications, Challenges, and Future scope. Biosensors 2021, 11, 336. [Google Scholar] [CrossRef]
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, Y.; Liu, S.; Yu, S.; Yu, Z.-R.; Low, S.S. Application and progress of chemometrics in voltammetric biosensing. Biosensors 2022, 12, 494. [Google Scholar] [CrossRef]
- Kissinger, P.T.; Heineman, W.R. Cyclic voltammetry. J. Chem. Educ. 1983, 60, 702. [Google Scholar] [CrossRef]
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical biosensors—Sensor principles and architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef]
- Soni, D.K.; Ahmad, R.; Dubey, S.K. Biosensor for the detection of Listeria monocytogenes: Emerging trends. Crit. Rev. Microbiol. 2018, 44, 590–608. [Google Scholar] [CrossRef]
- Xu, Q.; Pan, Y.; Li, W.; Yang, Z. Amperometric sensors. In Fundamentals of Sensor Technology; Barhoum, A., Altintas, Z., Eds.; Woodhead Publishing Series in Electronic and Optical Materials; Woodhead Publishing: London, UK, 2023; pp. 123–145. [Google Scholar] [CrossRef]
- Baracu, A; Livia Alexandra Gugoaşă Review—Recent advances in microfabrication, design and applications of amperometric sensors and biosensors. J. Electrochem. Soc. 2021, 168, 037503. [CrossRef]
- Ricci, F.; Vallée-Bélisle, A.; Simon, A.; Porchetta, A.; Plaxco, K.W. Using Nature’s ‘Tricks’ to rationally tune the binding properties of biomolecular receptors. Acc. Chem. Res. 2016, 49, 1884–1892. [Google Scholar] [CrossRef]
- Hasegawa, H.; Savory, N.; Abe, K.; Ikebukuro, K. Methods for improving aptamer binding affinity. Molecules 2016, 21, 421. [Google Scholar] [CrossRef]
- Pan, J.; Xu, W.; Li, W.; Chen, S.; Dai, Y.; Yu, S.; Zhou, Q.; Xia, F. Electrochemical aptamer-based sensors with tunable detection range. Anal. Chem. 2023, 95, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xie, Y.; Zhang, Y.; Li, C.; Xu, W. Programmable 3D rigid clathrate hydrogels based on self-assembly of tetrahedral DNA and linker PCR products. Chem. Commun. 2020, 56, 13181–13184. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.; Xu, J.; Tan, X.; Liu, Z.; Xu, L.; Peng, R. Dual-aptamer modification generates a unique interface for highly sensitive and specific electrochemical detection of tumor cells. ACS Appl. Mater. Interfaces 2014, 6, 7309–7315. [Google Scholar] [CrossRef]
- Jolly, P.; Formisano, N.; Tkáč, J.; Kasák, P.; Frost, C.G.; Estrela, P. Label-free impedimetric aptasensor with antifouling surface chemistry: A prostate specific antigen case study. Sens. Actuators B 2015, 209, 306–312. [Google Scholar] [CrossRef]
- Duan, N.; Ding, X.; He, L.; Wu, S.; Wei, Y.; Wang, Z. Selection, identification and application of a DNA aptamer against Listeria monocytogenes. Food Control 2013, 33, 239–243. [Google Scholar] [CrossRef]
- Lai, Y.-T.; DeStefano, J.J. DNA aptamers to human immunodeficiency virus reverse transcriptase selected by a primer-free SELEX method: Characterization and comparison with other aptamers. Nucleic Acid. Ther. 2012, 22, 162–176. [Google Scholar] [CrossRef]
- Chamorro-Garcia, A.; Parolo, C.; Ortega, G.; Idili, A.; Green, J.; Ricci, F.; Plaxco, K.W. The sequestration mechanism as a generalizable approach to improve the sensitivity of biosensors and bioassays. Chem. Sci. 2022, 13, 12219–12228. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Hu, J.; Lu, F. Aptamers used for biosensors and targeted therapy. Biomed. Pharmacother. 2020, 132, 110902. [Google Scholar] [CrossRef] [PubMed]
- Clark, V.; Waters, K.; Orsburn, B.; Bumpus, N.N.; Kundu, N.; Sczepanski, J.T.; Ray, P.; Arroyo-Currás, N. Human Cyclophilin B Nuclease activity revealed via nucleic acid-based electrochemical sensors. Angewandte Chemie 2022, 134, e202211292. [Google Scholar] [CrossRef]
- Yoo, H.; Jo, H.; Oh, S.S. Detection and beyond: Challenges and advances in aptamer-based biosensors. Mater. Adv. 2020, 1, 2663–2687. [Google Scholar] [CrossRef]
- Jarczewska, M.; Malinowska, E. The application of antibody–aptamer hybrid biosensors in clinical diagnostics and environmental analysis. Anal. Methods 2020, 12, 3183–3199. [Google Scholar] [CrossRef]
- Oliveira, R.; Pinho, E.; Sousa, A.L.; DeStefano, J.J.; Azevedo, N.F.; Almeida, C. Improving aptamer performance with nucleic acid mimics: De novo and post-SELEX approaches. Trends Biotechnol. 2022, 40, 549–563. [Google Scholar] [CrossRef]
- Yu, H.; Zhu, J.; Shen, G.; Deng, Y.; Geng, X.; Wang, L. Improving aptamer performance: Key factors and strategies. Mikrochimica Acta 2023, 190, 255. [Google Scholar] [CrossRef]
- Wang, T.; Chen, C.; Larcher, L.M.; Barrero, R.A.; Veedu, R.N. Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnol. Adv. 2019, 37, 28–50. [Google Scholar] [CrossRef]
- Chushak, Y.; Stone, M.O. In silico selection of RNA aptamers. Nucleic Acids Res. 2009, 37, e87. [Google Scholar] [CrossRef]
- Soon, S.; Nordin, N.A. In silico predictions and optimization of aptamers against Streptococcus agalactiae surface protein using computational docking. Mater. Today 2019, 16, 2096–2100. [Google Scholar] [CrossRef]
- Bell, D.R.; Weber, J.K.; Yin, W.; Huynh, T.; Duan, W.; Zhou, R. In silico design and validation of high-affinity RNA aptamers targeting epithelial cellular adhesion molecule dimers. Proc. Natl. Acad. Sci. USA 2020, 117, 8486–8493. [Google Scholar] [CrossRef]
- Fallah, A.; Havaei, S.A.; Sedighian, H.; Kachuei, R.; Fooladi, A.A.I. Prediction of aptamer affinity using an artificial intelligence approach. J. Mater. Chem. B 2024, 12, 8825–8842. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Cho, J.; Lee, B.-H.; Hwang, D.; Park, J.-W. Design and prediction of aptamers assisted by In Silico methods. Biomedicines 2023, 11, 356. [Google Scholar] [CrossRef] [PubMed]
- Le, T.T.; Chumphukam, O.; Cass, A.E.G. Determination of minimal sequence for binding of an aptamer. A comparison of truncation and hybridization inhibition methods. RSC Adv. 2014, 4, 47227–47233. [Google Scholar] [CrossRef]
- Nguyen, M.-D.; Osborne, M.T.; Prevot, G.T.; Churcher, Z.R.; Johnson, P.E.; Simine, L.; Dauphin-Ducharme, P. Truncations and in silico docking to enhance the analytical response of aptamer-based biosensors. Biosens. Bioelectron. 2024, 265, 116680. [Google Scholar] [CrossRef]
- Radom, F.; Jurek, P.M.; Mazurek, M.P.; Otlewski, J.; Jeleń, F. Aptamers: Molecules of great potential. Biotechnol. Adv. 2013, 31, 1260–1274. [Google Scholar] [CrossRef]
- Bing, T.; Yang, X.; Mei, H.; Cao, Z.; Shangguan, D. Conservative secondary structure motif of streptavidin-binding aptamers generated by different laboratories. Bioorganic Med. Chem. 2010, 18, 1798–1805. [Google Scholar] [CrossRef]
- Akitomi, J.; Kato, S.; Yoshida, Y.; Horii, K.; Furuichi, M.; Waga, I. ValFold: Program for the aptamer truncation process. Bioinformation 2011, 7, 38–40. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinformatics 2010, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Kilgour, M.; Liu, T.; Walker, B.D.; Ren, P.; Simine, L. E2EDNA: Simulation protocol for DNA aptamers with ligands. J. Chem. Inf. Model. 2021, 61, 4139–4144. [Google Scholar] [CrossRef]
- Shangguan, D.; Tang, Z.; Mallikaratchy, P.; Xiao, Z.; Tan, W. Optimization and Modifications of Aptamers Selected from Live Cancer Cell Lines. ChemBioChem 2007, 8, 603–606. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Guo, L.; He, J.; Xu, H.; Xie, J. Stepping library-based Post-SELEX strategy approaching to the minimized aptamer in SPR. Anal. Chem. 2017, 89, 6559–6566. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Ye, H.; Guo, H.; Ma, X.; Yue, L.; Wang, Z. Aptamer truncation strategy assisted by molecular docking and sensitive detection of T-2 toxin using SYBR Green I as a signal amplifier. Food Chem. 2022, 381, 132171. [Google Scholar] [CrossRef]
- DeRosa, M.C.; Lin, A.; Mallikaratchy, P.; McConnell, E.M.; McKeague, M.; Patel, R.; Shigdar, S. In vitro selection of aptamers and their applications. Nat. Rev. Methods Primers 2023, 3, 54. [Google Scholar] [CrossRef]
- Zheng, X.; Hu, B.; Gao, S.X.; Liu, D.J.; Sun, M.J.; Jiao, B.H.; Wang, L.H. A saxitoxin-binding aptamer with higher affinity and inhibitory activity optimized by rational site-directed mutagenesis and truncation. Toxicon 2015, 101, 41–47. [Google Scholar] [CrossRef]
- Kimoto, M.; Nakamura, M.; Hirao, I. Post-ExSELEX stabilization of an unnatural-base DNA aptamer targeting VEGF165 toward pharmaceutical applications. Nucleic Acids Res. 2016, 44, 7487–7494. [Google Scholar] [CrossRef][Green Version]
- Zhao, L.; Qi, X.; Yan, X.; Huang, Y.; Liang, X.; Zhang, L.; Wang, S.; Tan, W. Engineering aptamer with enhanced affinity by triple helix-based terminal fixation. J. Am. Chem. Soc. 2019, 141, 17493–17497. [Google Scholar] [CrossRef]
- Gao, S.; Zheng, X.; Jiao, B.; Wang, L. Post-SELEX optimization of aptamers. Anal. Bioanal. Chem. 2016, 408, 4567–4573. [Google Scholar] [CrossRef]
- Mallikaratchy, P.R.; Ruggiero, A.; Gardner, J.R.; Kuryavyi, V.; Maguire, W.F.; Heaney, M.L.; McDevitt, M.R.; Patel, D.J.; Scheinberg, D.A. A multivalent DNA aptamer specific for the B-cell receptor on human lymphoma and leukemia. Nucleic Acids Res. 2010, 39, 2458–2469. [Google Scholar] [CrossRef]
- Sheng, W.; Chen, T.; Tan, W.; Fan, Z.H. Multivalent DNA nanospheres for enhanced capture of cancer cells in microfluidic devices. ACS Nano 2013, 7, 7067–7076. [Google Scholar] [CrossRef]
- Kim, Y.-M.; Cao, Z.; Tan, W. Molecular assembly for high-performance bivalent nucleic acid inhibitor. Proc. Natl. Acad. Sci. USA 2008, 105, 5664–5669. [Google Scholar] [CrossRef]
- Kwon, P.S.; Ren, S.; Kwon, S.-J.; Kizer, M.E.; Kuo, L.; Xie, M.; Zhu, D.; Zhou, F.; Zhang, F.; Kim, D.; et al. Designer DNA architecture offers precise and multivalent spatial pattern-recognition for viral sensing and inhibition. Nat. Chem. 2019, 12, 26–35. [Google Scholar] [CrossRef]
- Qin, W.; Chen, L.; Wang, Z.; Li, Q.; Fan, C.; Wu, M.; Zhang, Y. Bioinspired DNA nanointerface with anisotropic aptamers for accurate capture of circulating tumor cells. Adv. Sci. 2020, 7, 2000647. [Google Scholar] [CrossRef]
- Dou, B.; Xu, L.; Jiang, B.; Yuan, R.; Xiang, Y. Aptamer-functionalized and gold nanoparticle array-decorated magnetic graphene nanosheets enable multiplexed and sensitive electrochemical detection of rare circulating tumor cells in whole blood. Anal. Chem. 2019, 91, 10792–10799. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Liu, S.; Jiang, Y.; Lin, J.-M.; Yu, L.; Hu, Q. Simultaneous detection of multiple tumor markers in blood by functional liquid crystal sensors assisted with target-induced dissociation of aptamer. Anal. Chem. 2020, 92, 3867–3873. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Gao, Q.; Cheung, M.C.; Leung, H.M.; Chi, T.; Sleiman, H.F.; Wai, K.; Lo, P.K. A highly versatile platform based on geometrically well-defined 3D DNA nanostructures for selective recognition and positioning of multiplex targets. Nanoscale 2016, 8, 18291–18295. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Zhang, C.; Wang, Y.; Chen, G. Research progress of whole-cell-SELEX selection and the application of cell-targeting aptamer. Mol. Biol. Rep. 2022, 49, 7979–7993. [Google Scholar] [CrossRef]
- Ma, P.; Duan, N.; Ye, H.; Xia, Y.; Ding, Z.; Wang, Z. Selection, truncation and fluorescence polarization based aptasensor for Weissella viridescens detection. Talanta 2022, 246, 123499. [Google Scholar] [CrossRef]
- Kim, Y.S.; Chung, J.; Song, M.Y.; Jurng, J.; Kim, B.C. Aptamer cocktails: Enhancement of sensing signals compared to single use of aptamers for detection of bacteria. Biosens. Bioelectron. 2014, 54, 195–198. [Google Scholar] [CrossRef]
- Lam, S.Y.; Lau, H.L.; Kwok, C.K. Capture-SELEX: Selection strategy, aptamer identification, and biosensing application. Biosensors 2022, 12, 1142. [Google Scholar] [CrossRef] [PubMed]
- Villalonga, A.; Mayol, B.; Villalonga, R.; Vilela, D. Electrochemical aptasensors for clinical diagnosis. A review of the last five years. Sens. Actuators B 2022, 369, 132318. [Google Scholar] [CrossRef]
- Ansari, M.A.; Verma, D.; Hamizan, M.A.; Mukherjee, M.D.; Mohd-Naim, N.F.; Ahmed, M.U. Trends in aptasensing and the enhancement of diagnostic efficiency and accuracy. ACS Synth. Biol. 2025, 14, 21–40. [Google Scholar] [CrossRef]
- Liu, L.S.; Wang, F.; Ge, Y.; Lo, P.K. Recent developments in aptasensors for diagnostic applications. ACS Appl. Mater. Interfaces 2020, 13, 9329–9358. [Google Scholar] [CrossRef] [PubMed]
- Downs, A.M.; Plaxco, K.W. Real-time, in vivo molecular monitoring using electrochemical aptamer based sensors: Opportunities and challenges. ACS Sens. 2022, 7, 2823–2832. [Google Scholar] [CrossRef]
- Rabiee, N.; Rabiee, M. Wearable aptasensors. Anal. Chem. 2024, 96, 19160–19182. [Google Scholar] [CrossRef] [PubMed]
- Varnakavi, N.; Lee, N. A Review on biosensors and recent development of nanostructured materials-enabled biosensors. Sensors 2021, 21, 1109. [Google Scholar] [CrossRef]
- Lino, C.; Barrias, S.; Chaves, R.; Adega, F.; Martins-Lopes, P.; Fernandes, J.R. Biosensors as diagnostic tools in clinical applications. Biochim. Et. Biophys. Acta (BBA)—Rev. Cancer 2022, 1877, 188726. [Google Scholar] [CrossRef]
- Khoshbin, Z.; Housaindokht, M.R.; Izadyar, M.; Bozorgmehr, M.R.; Verdian, A. Recent advances in computational methods for biosensor design. Biotechnol. Bioeng. 2021, 118, 555–578. [Google Scholar] [CrossRef]
- Douaki, A.; Garoli, D.; Inam, A.K.M.S.; Angeli, M.A.C.; Cantarella, G.; Rocchia, W.; Wang, J.; Petti, L.; Lugli, P. Smart approach for the design of highly selective aptamer-based biosensors. Biosensors 2022, 12, 574. [Google Scholar] [CrossRef]
- Darmostuk, M.; Rimpelova, S.; Gbelcova, H.; Ruml, T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2015, 33, 1141–1161. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, N.A.; Mohamed Zulkifli, R.; Hussin, H.; Nadri, M.H. In silico approach for Post-SELEX DNA aptamers: A mini-review. J. Mol. Graph. Model. 2021, 105, 107872. [Google Scholar] [CrossRef]
- Emami, N.; Pakchin, P.S.; Ferdousi, R. Computational predictive approaches for interaction and structure of aptamers. J. Theor. Biol. 2020, 497, 110268. [Google Scholar] [CrossRef]
- Cataldo, R.; Leuzzi, M.; Alfinito, E. Modelling and development of electrical aptasensors: A short review. Chemosensors 2018, 6, 20. [Google Scholar] [CrossRef]
- Wei, X.; Ma, P.; Mahmood, K.I.; Zhang, Y.; Wang, Z. A review: Construction of aptamer screening methods based on improving the screening rate of key steps. Talanta 2022, 253, 124003. [Google Scholar] [CrossRef]
- Bashir, A.; Yang, Q.; Wang, J.; Hoyer, S.; Chou, W.; McLean, C.; Davis, G.; Gong, Q.; Armstrong, Z.; Jang, J.; et al. Machine learning guided aptamer refinement and discovery. Nat. Commun. 2021, 12, 2366. [Google Scholar] [CrossRef] [PubMed]
- Tobia, J.P.; Huang, P.-J.J.; Ding, Y.; Narayan, R.S.A.; Liu, J. Machine learning directed aptamer search from conserved primary sequences and secondary structures. ACS Synth. Biol. 2023, 12, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Chan, C.H.; Yao, S.; Chu, H.Y.; Lyu, M.; Chen, Z.; Xiao, H.; Ma, Y.; Yu, S.; Li, F.; et al. DeepAptamer: Advancing high-affinity aptamer discovery with a hybrid deep learning model. Mol. Ther. Nucleic Acids 2025, 36, 102436. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, Y.; Kuster, D.; Ye, Q.; Huang, W.; Fürbacher, S.; Zhang, J.; Doll, P.; Lin, W.; Dong, S.; et al. Single-step discovery of high-affinity RNA ligands by UltraSELEX. Nat. Chem. Biol. 2025, 21, 1118–1126. [Google Scholar] [CrossRef] [PubMed]
- Cossettini, A.; Pasquardini, L.; Romani, A.; Feriani, A.; Pinamonti, D.; Manzano, M. Computational aptamer design for spike glycoprotein (S) (SARS-CoV-2) detection with an electrochemical aptasensor. Appl. Microbiol. Biotechnol. 2024, 108, 259. [Google Scholar] [CrossRef] [PubMed]
- Fasogbon, I.V.; Ondari, E.N.; Tusubira, D.; Rangasamy, L.; Venkatesan, J.; Musyoka, A.M.; Aja, P.M. Recent focus in non-SELEX-computational approach for de novo aptamer design: A mini review. Anal. Biochem. 2025, 699, 115756. [Google Scholar] [CrossRef] [PubMed]
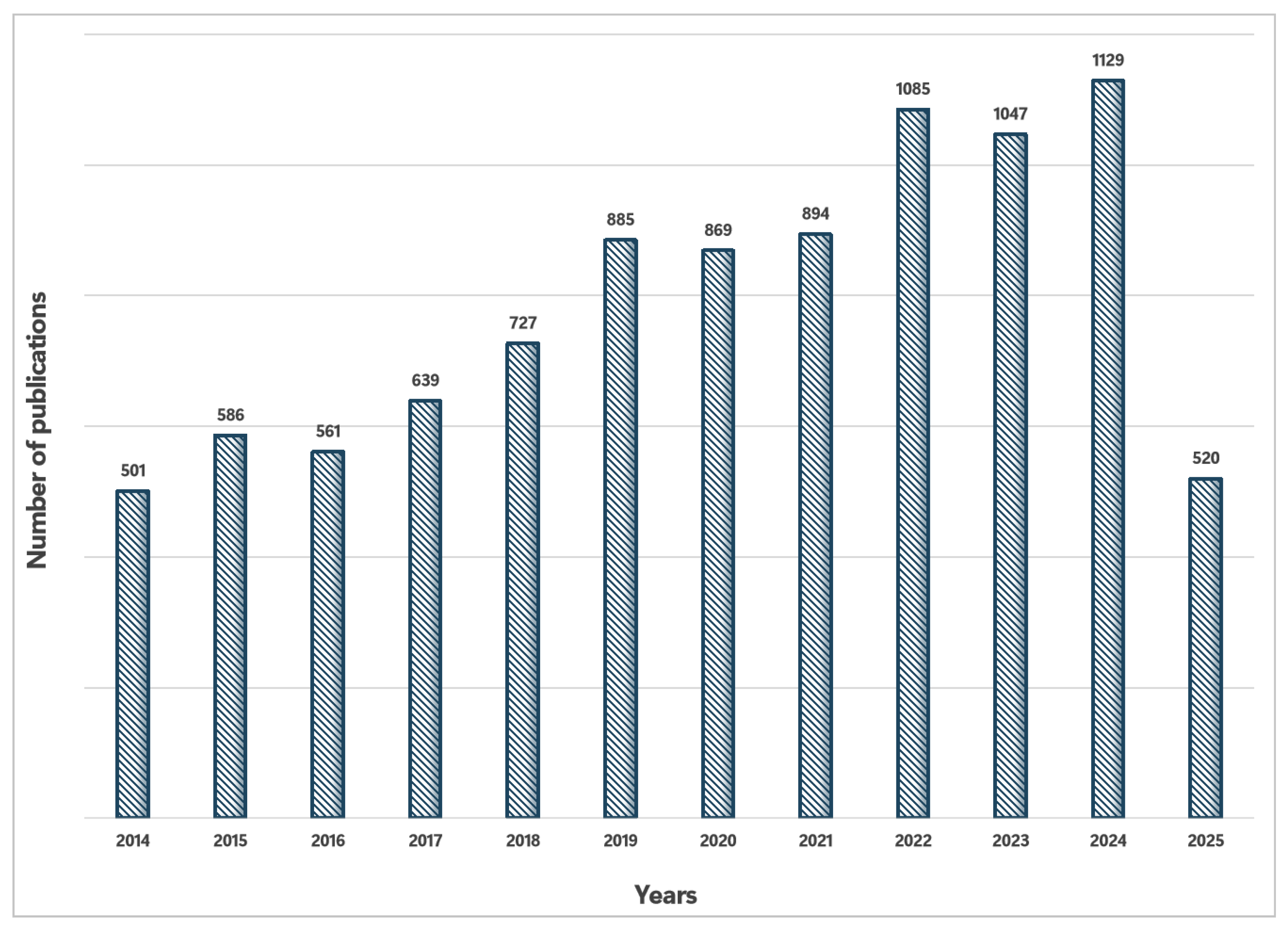
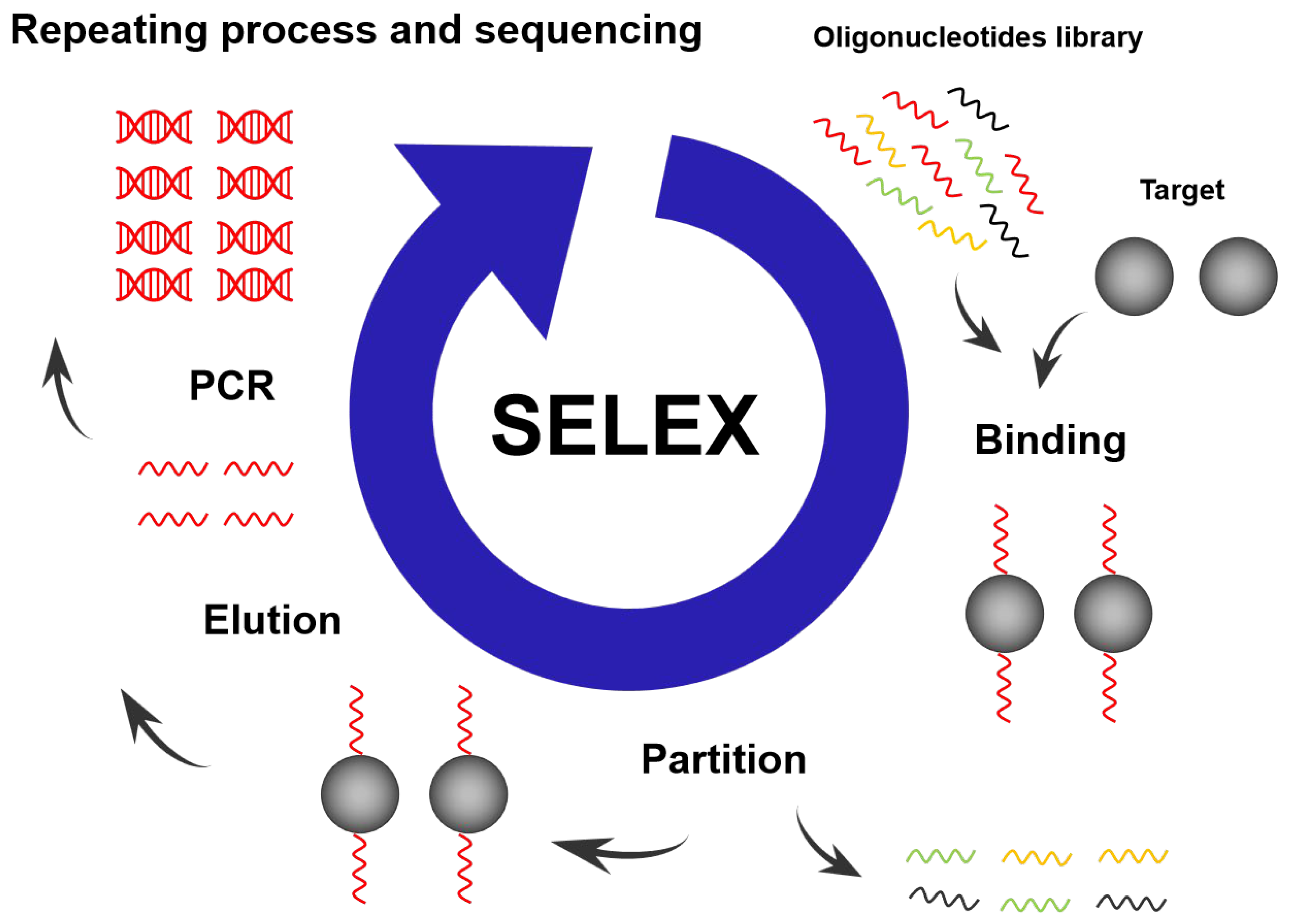
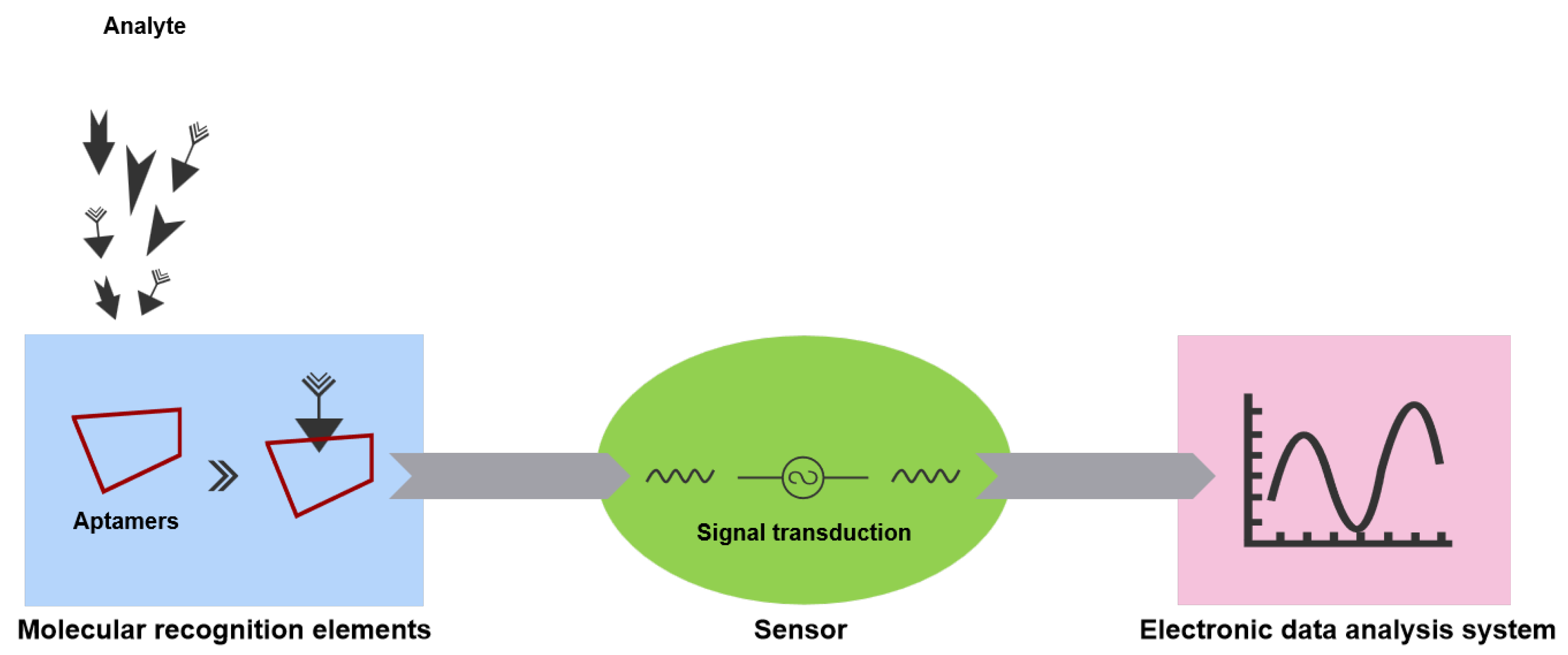
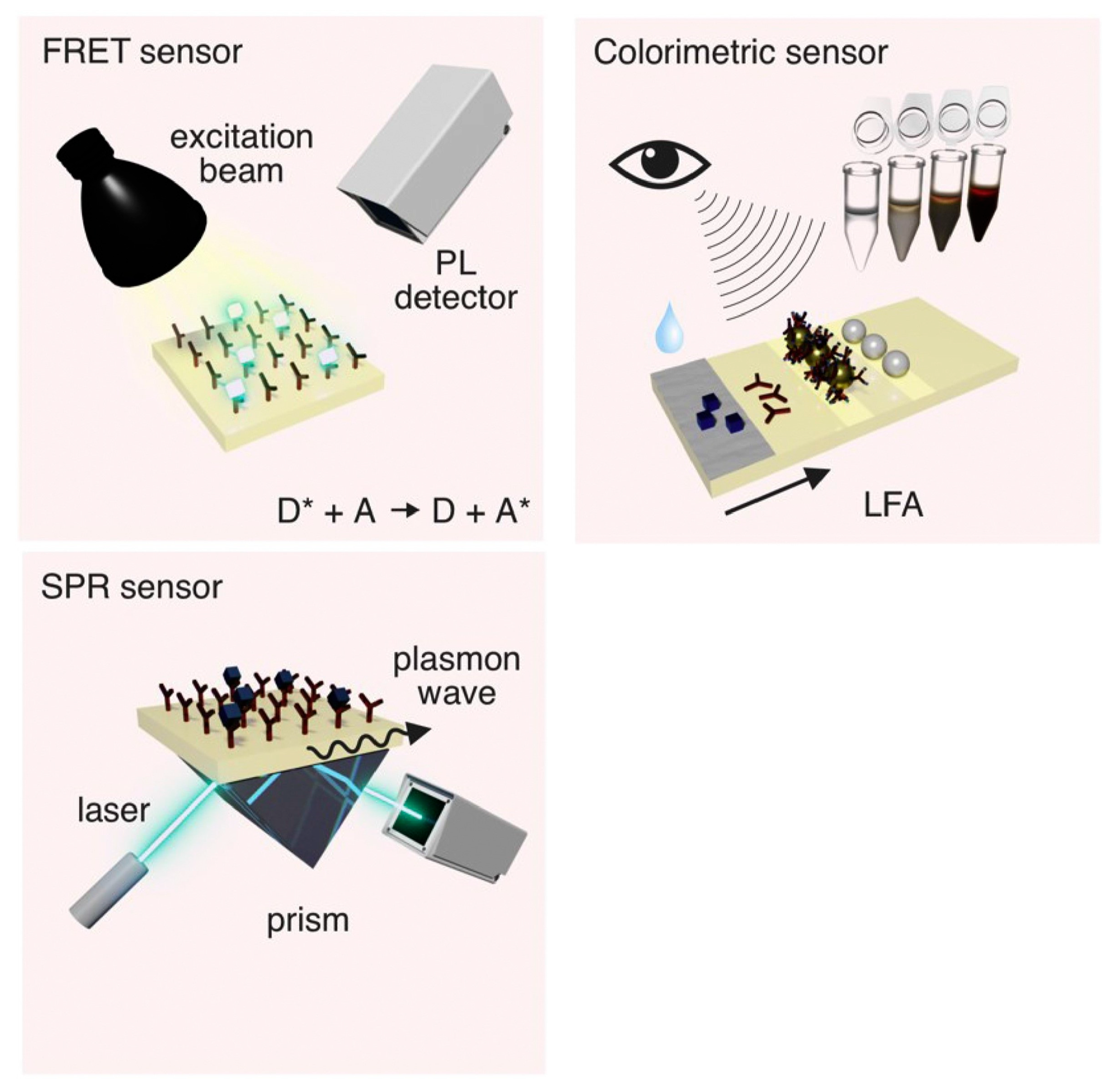
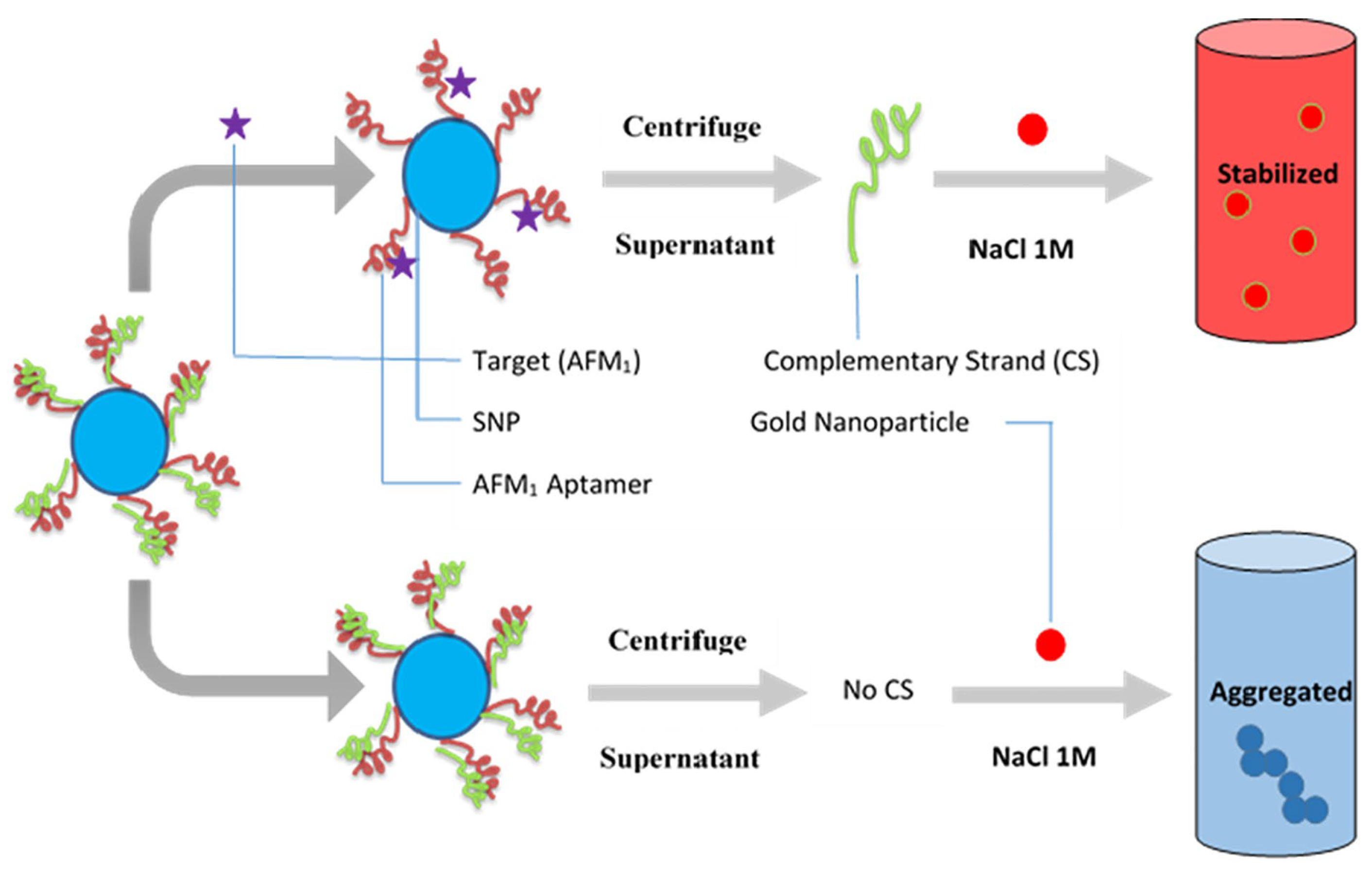
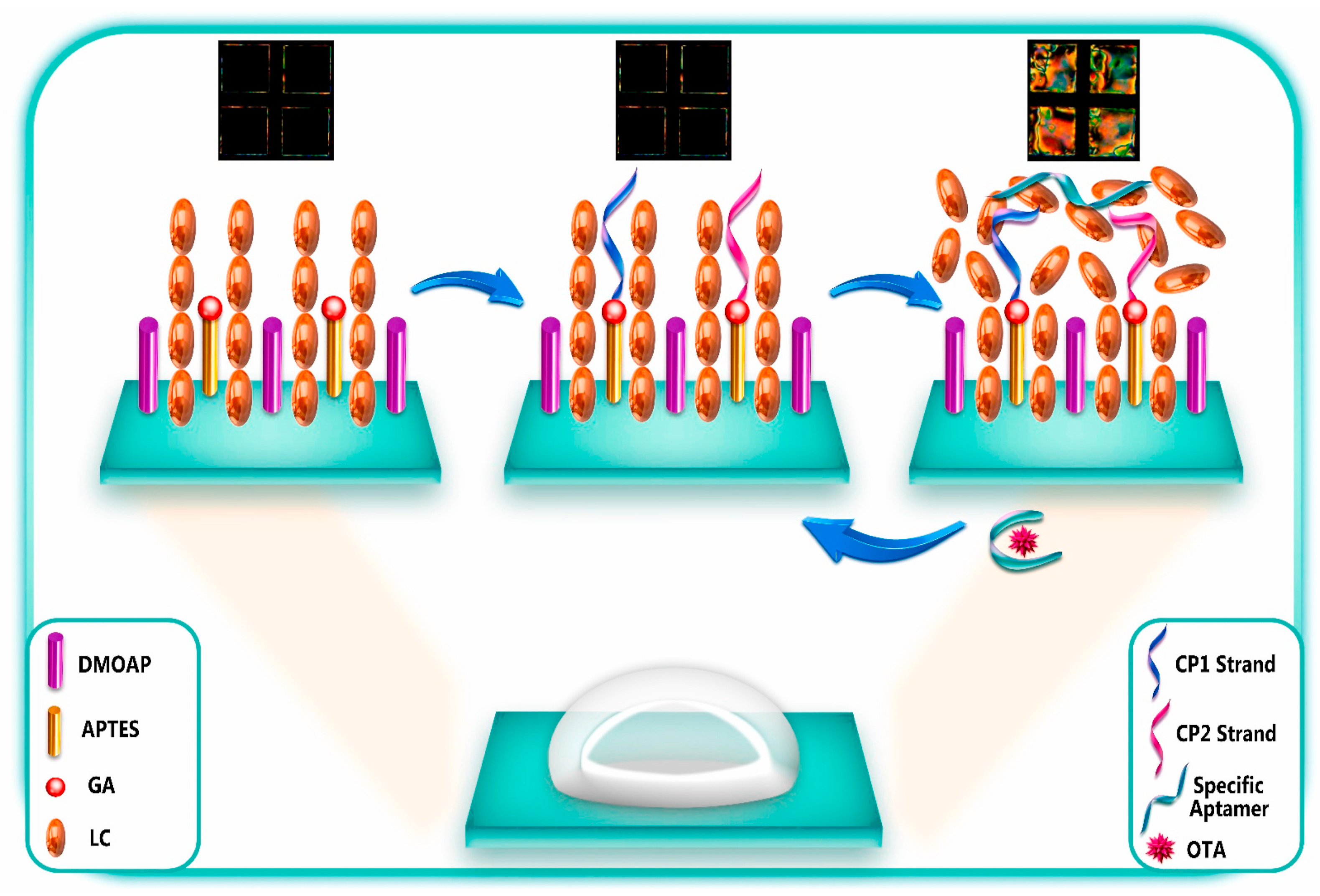
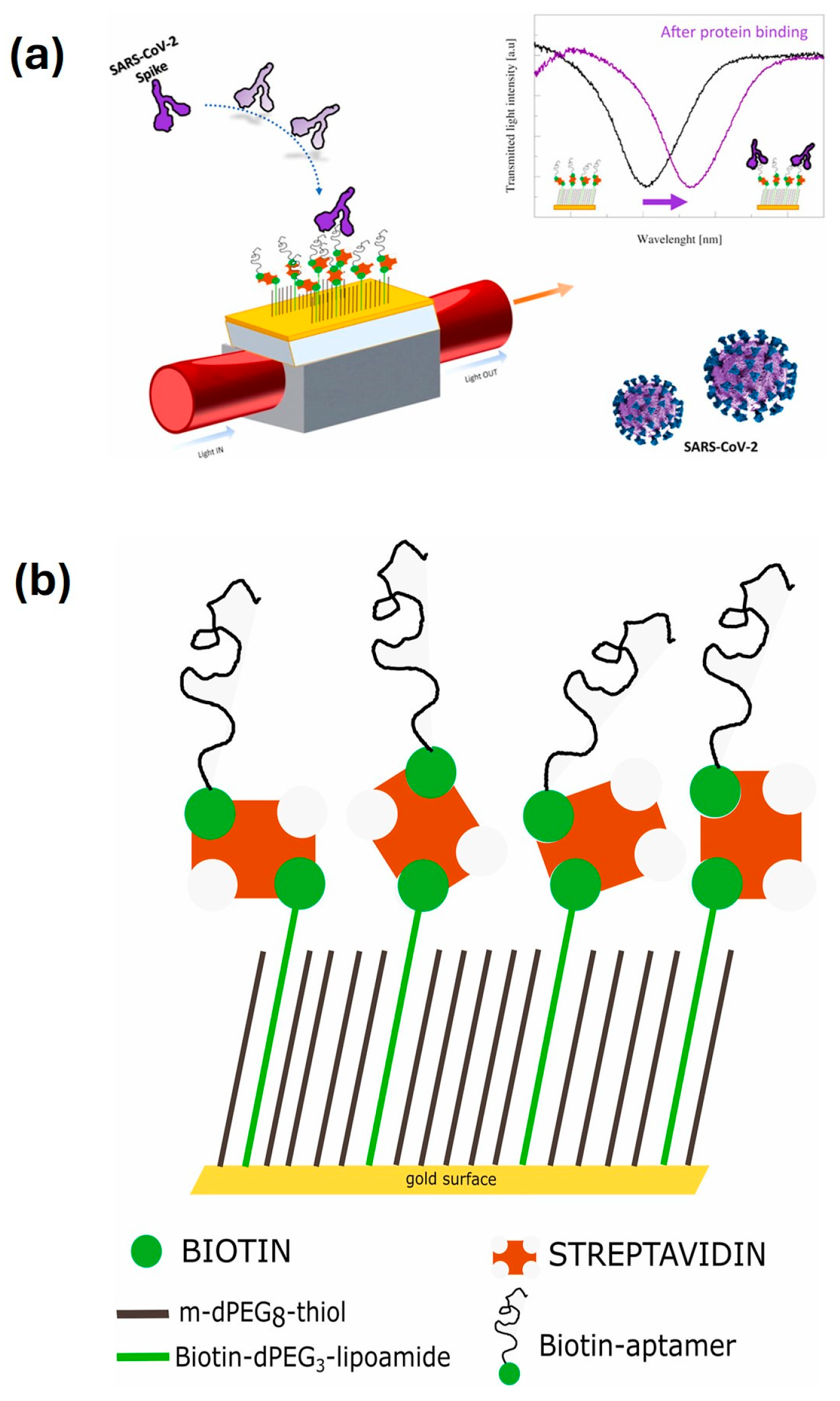
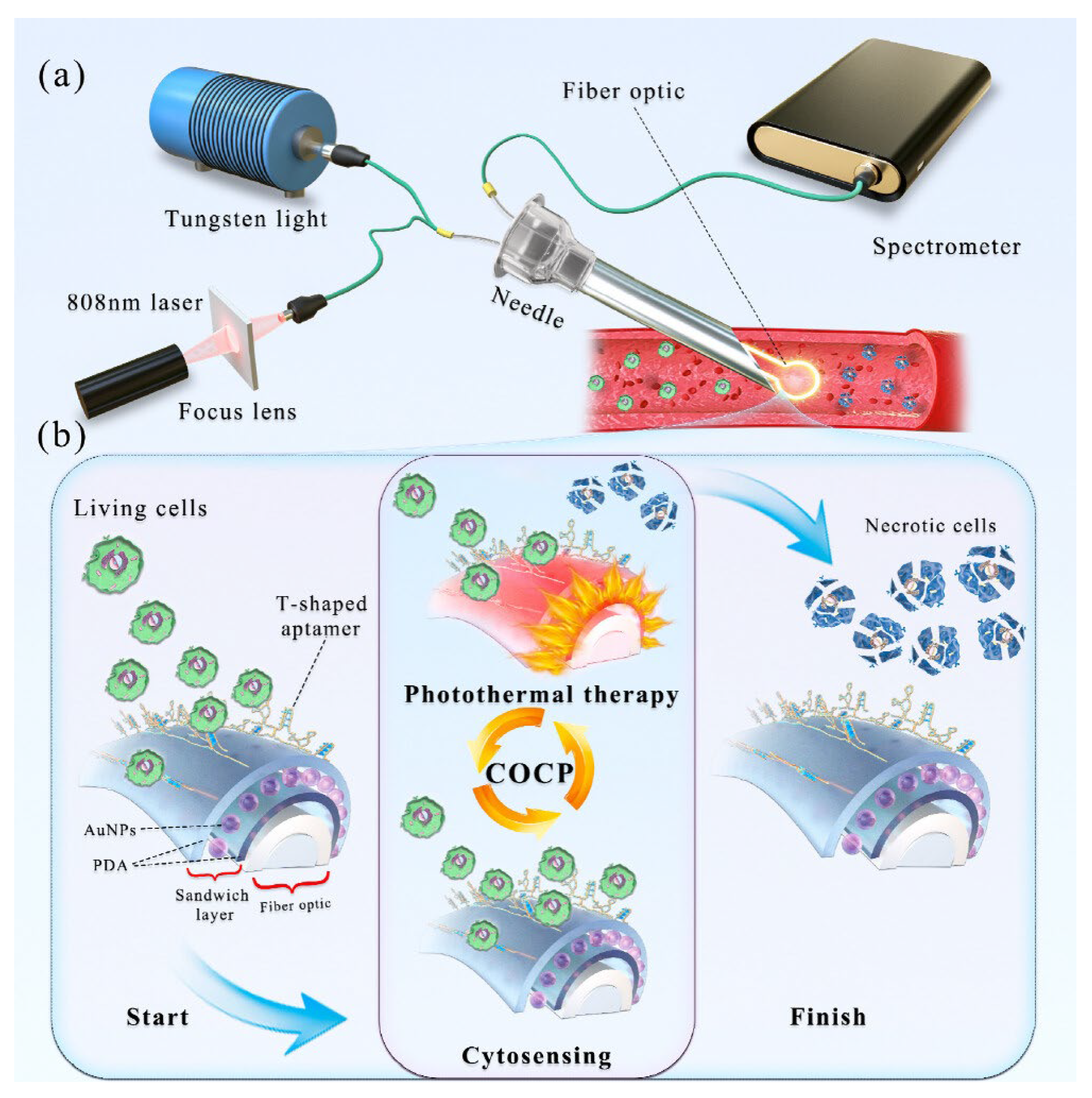
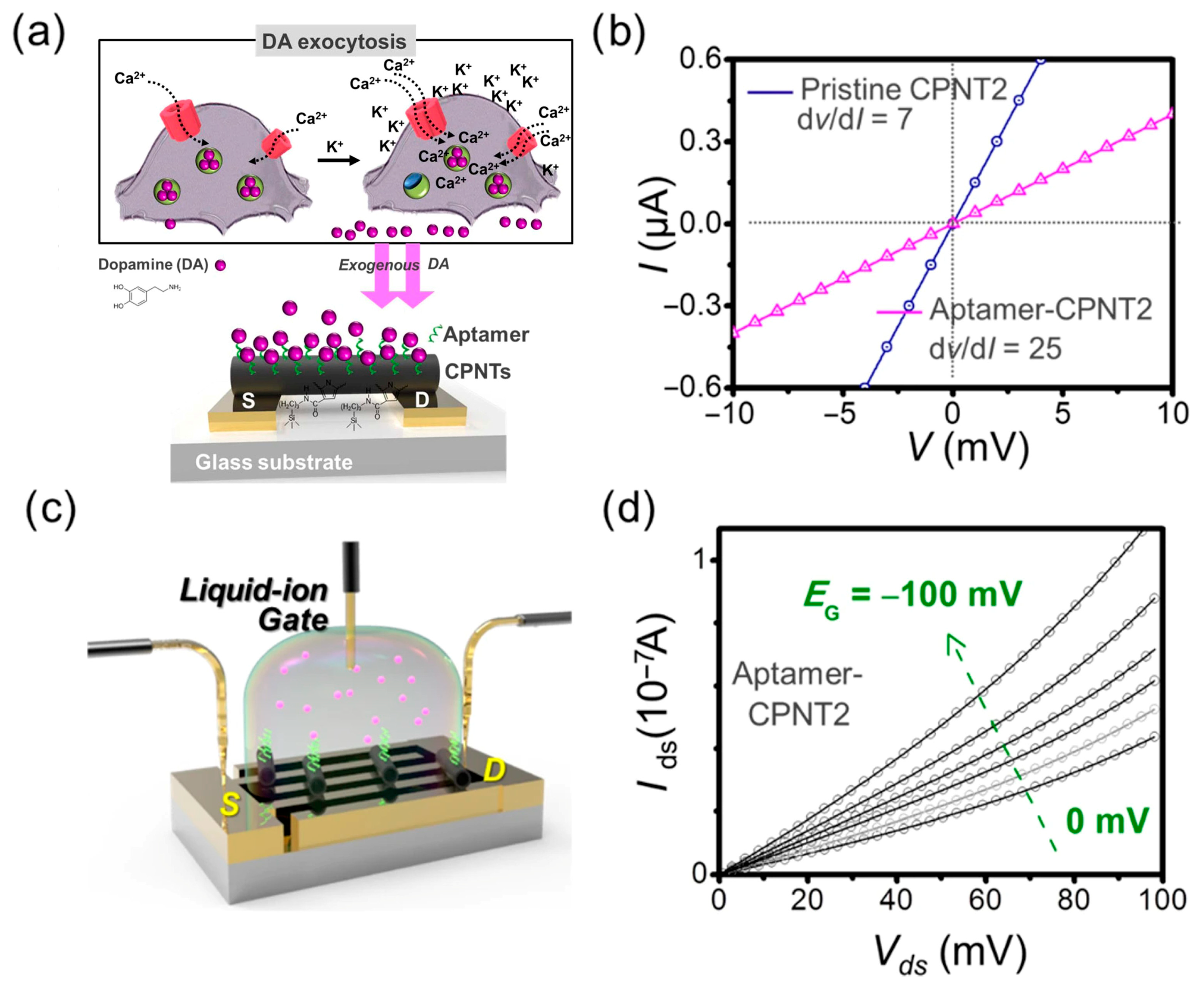

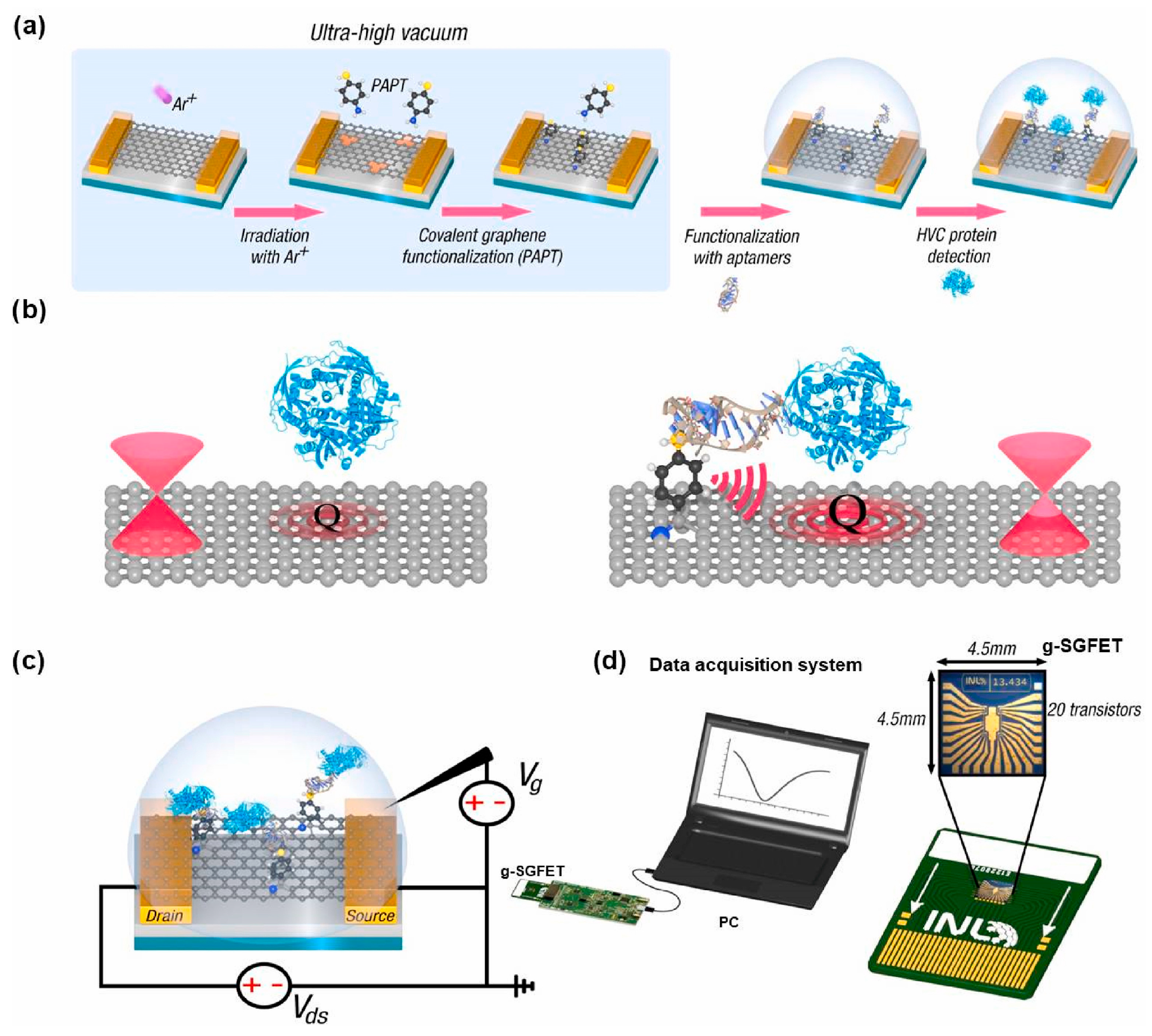


| Feature | Aptamers | Antibodies |
|---|---|---|
| Nature | Short ssDNA or RNA oligonucleotides | Large protein molecules (~150 kDa) |
| Production | Fully synthetic via SELEX | Biological (immunization, hybridoma, and cell culture) |
| Time to Develop | Weeks | Months |
| Batch Consistency | High (chemical synthesis) | Variable (biological expression) |
| Size | Small (5–15 kDa) | Large (~150 kDa) |
| Target Range | Proteins, small molecules, toxins, ions, and even non-immunogenic targets | Mostly proteins and larger antigens |
| Stability | Stable to pH, heat; reversible folding | Sensitive to temperature, pH; irreversible denaturation |
| Modification | Easily and precisely modified (labels, drugs, and nanomaterials) | Modifications more limited and complex |
| Tissue Penetration | Better (small size) | Limited (large size) |
| Immunogenicity | Very low | May trigger immune responses |
| Cost | Relatively low (chemical synthesis) | Higher (animal/cell-based production) |
| Regulatory Approval | Fewer approved (e.g., Pegaptanib) | Many approved therapeutics and diagnostics |
| Degradation | Susceptible to nucleases (can be stabilized chemically) | Proteolytic degradation but generally longer half-life in vivo |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Nie, M. Recent Advances in the Optimization of Nucleic Acid Aptamers and Aptasensors. Biosensors 2025, 15, 641. https://doi.org/10.3390/bios15100641
Wang Y, Nie M. Recent Advances in the Optimization of Nucleic Acid Aptamers and Aptasensors. Biosensors. 2025; 15(10):641. https://doi.org/10.3390/bios15100641
Chicago/Turabian StyleWang, Yuan, and Mengyan Nie. 2025. "Recent Advances in the Optimization of Nucleic Acid Aptamers and Aptasensors" Biosensors 15, no. 10: 641. https://doi.org/10.3390/bios15100641
APA StyleWang, Y., & Nie, M. (2025). Recent Advances in the Optimization of Nucleic Acid Aptamers and Aptasensors. Biosensors, 15(10), 641. https://doi.org/10.3390/bios15100641





