Detecting Nanotopography Induced Changes in Cell Migration Directions Using Oxygen Sensors
Abstract
1. Introduction
2. Materials and Methods
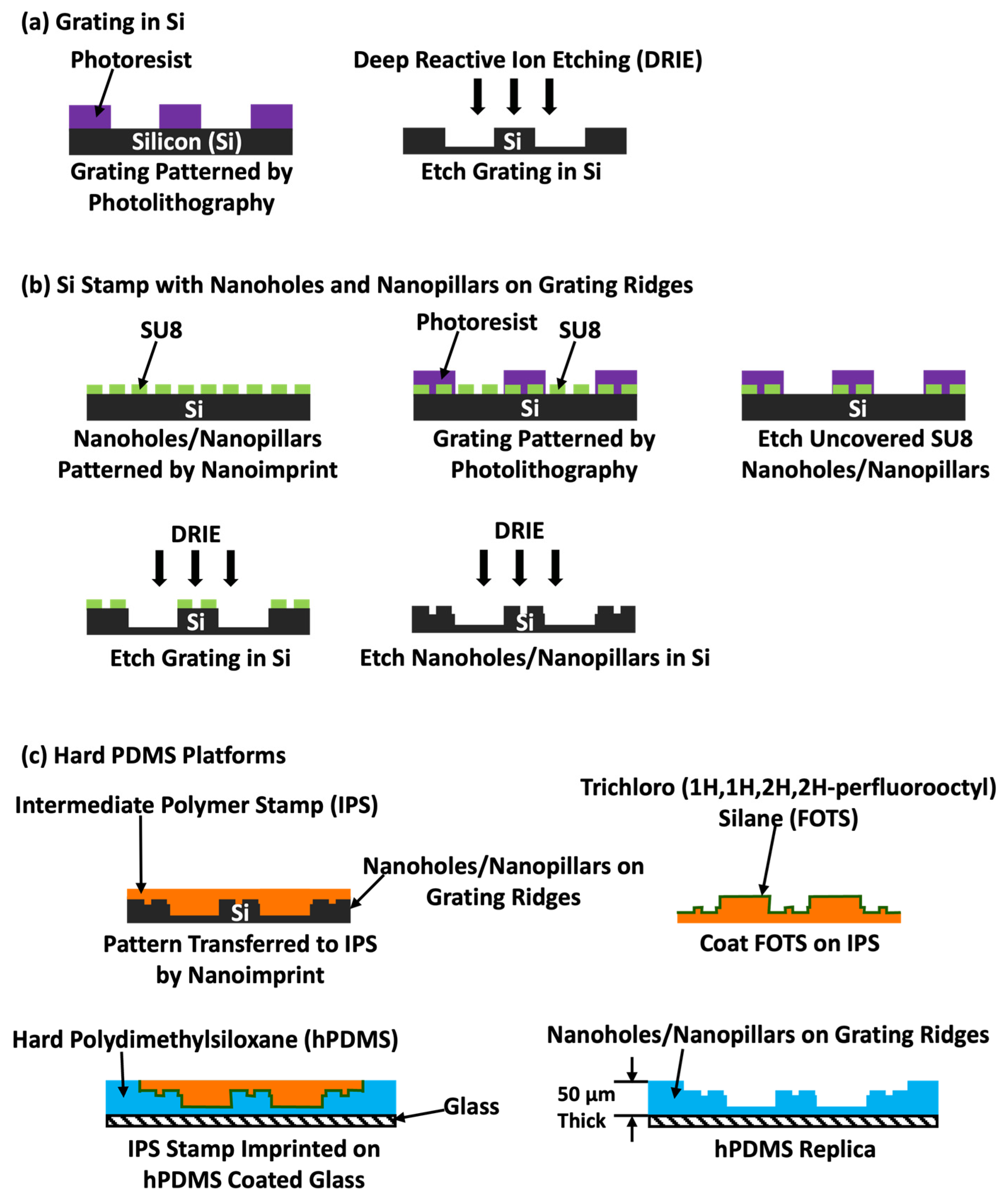
2.1. Nanostructures Patterning on Grating Ridges by Nanoimprint Lithography
2.2. Cell Preparation and Time-Lapse Imaging
2.3. Fixing Cells on Platforms with Gratings and Nanotopography
2.4. F-Actin Structures in MC3T3-E1 Cells
2.5. Platforms with PtOEPK Dye and hPDMS for O2 Sensing
2.6. Live Cell Tracking of Mitochondria and PtOEPK Dye Signals
3. Results and Discussion
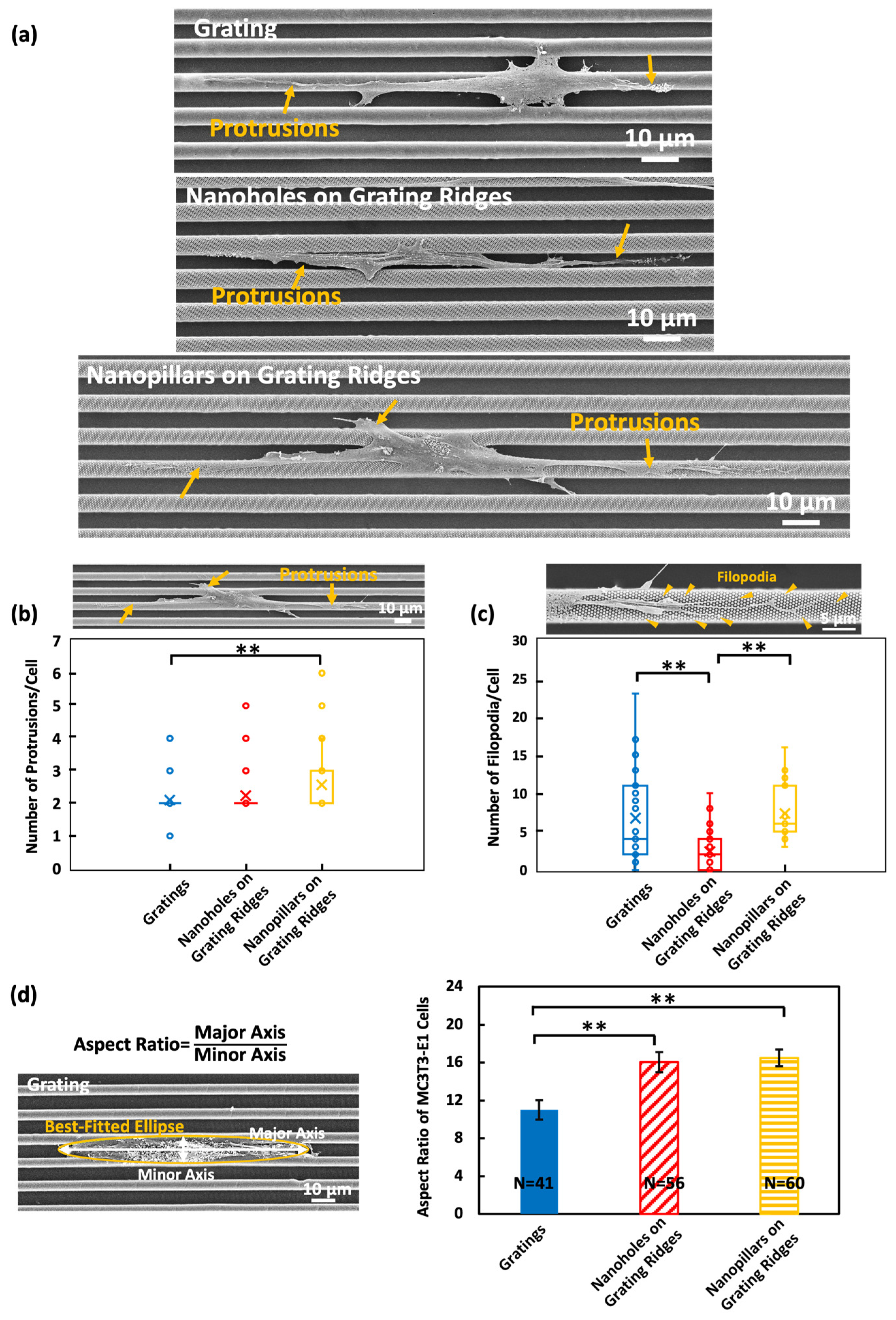
3.1. Protrusion and Filopodium Formation on Platforms with Nanotopography
3.2. Dotted F-Actin Structures in MC3T3-E1 Cells on Nanohole and Nanopillar Surfaces
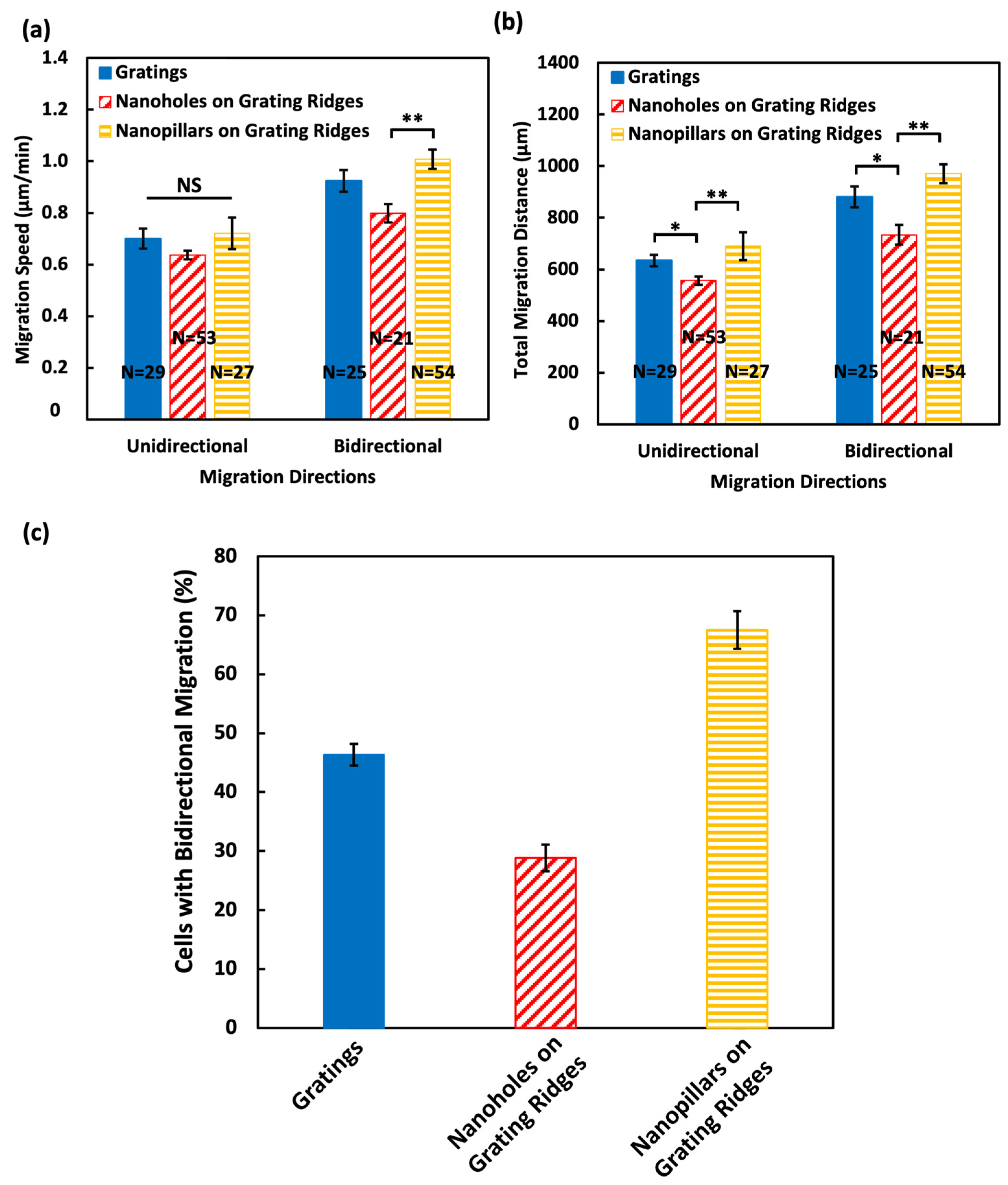
3.3. Nanopillars Enhanced Cell Motility and Induced Migration Directional Changes
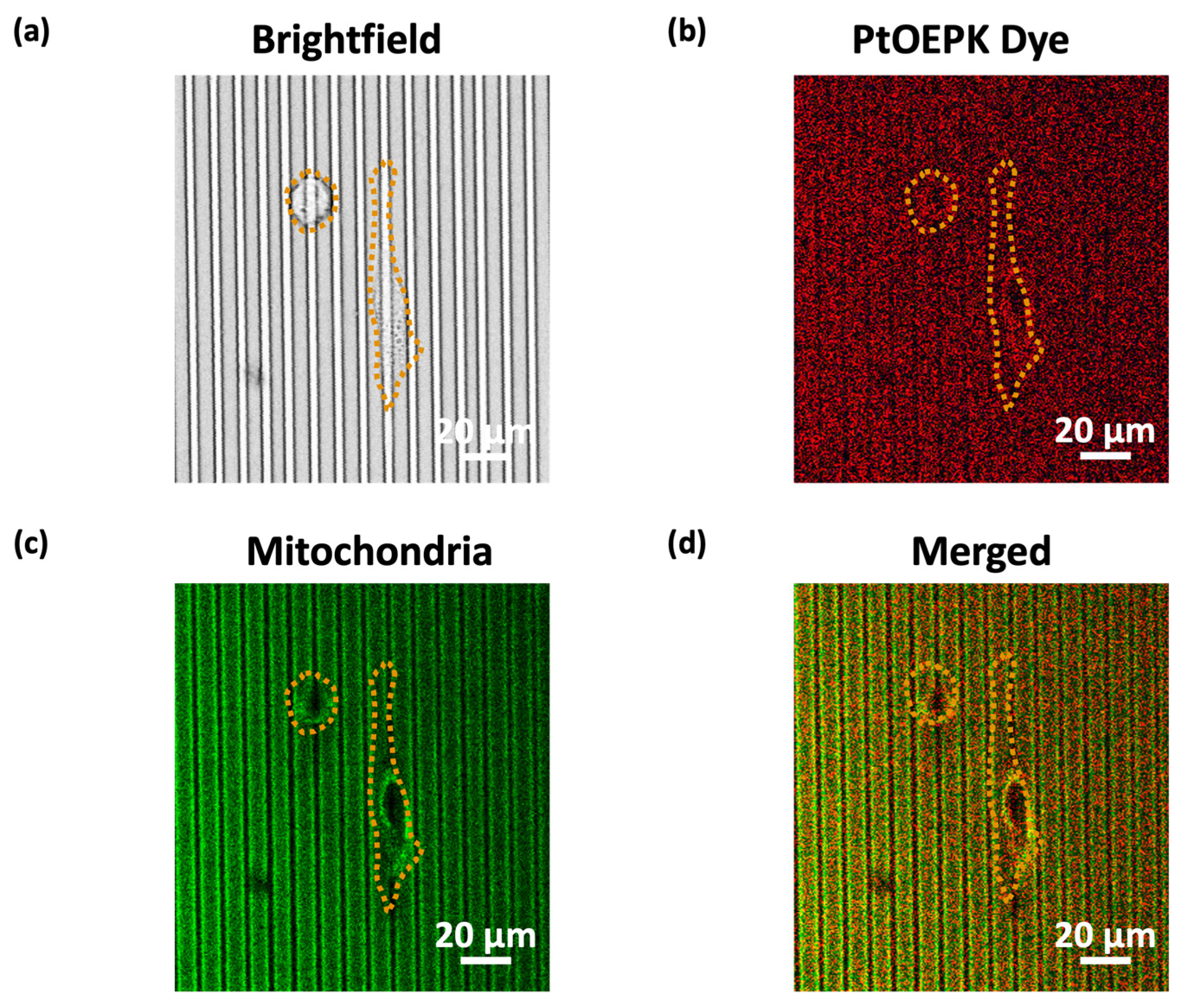
3.4. Correlation between Mitochondria and PtOEPK Dye Signals
3.5. O2 sensing of Single Cells with Unidirectional and Bidirectional Migrations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doyle, A.D.; Yamada, K.M. Mechanosensing via cell-matrix adhesions in 3D microenvironments. Exp. Cell Res. 2016, 343, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, P.; Schweller, R.M.; Khademhosseini, A.; West, J.L.; Bashir, R. 3D biofabrication strategies for tissue engineering and regenerative medicine. Annu. Rev. Biomed. Eng. 2014, 16, 247–276. [Google Scholar] [CrossRef] [PubMed]
- Friedl, P.; Alexander, S. Cancer invasion and the microenvironment: Plasticity and reciprocity. Cell 2011, 147, 992–1009. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, A.I.; Abrams, G.A.; Bertics, P.J.; Murphy, C.J.; Nealey, P.F. Epithelial contact guidance on well-defined micro-and nanostructured substrates. J. Cell Sci. 2003, 116, 1881–1892. [Google Scholar] [CrossRef] [PubMed]
- SenGupta, S.; Parent, C.A.; Bear, J.E. The principles of directed cell migration. Nat. Rev. Mol. Cell Biol. 2021, 22, 529–547. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.Y.; Tong, W.Y.; Shi, J.; Shi, P.; Lam, Y.W.; Pang, S.W. Influence of engineered surface on cell directionality and motility. Biofabrication 2014, 6, 015011. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Pang, S.W. Enhancing Nasopharyngeal Carcinoma Cell Separation with Selective Fibronectin Coating and Topographical Modification on Polydimethylsiloxane Scaffold Platforms. Int. J. Mol. Sci. 2023, 24, 12409. [Google Scholar] [CrossRef] [PubMed]
- Refaaq, F.; Chen, X.; Pang, S.W. Effects of topographical guidance cues on osteoblast cell migration. Sci. Rep. 2020, 10, 20003. [Google Scholar] [CrossRef]
- Wang, M.; Pang, S.W. Controlled Scaffold Platform Designs on Nasopharyngeal Carcinoma Cell Separation. IEEE Access 2021, 9, 113813–113822. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Khademhosseini, A. Advances in engineering hydrogels. Science 2017, 356, eaaf3627. [Google Scholar] [CrossRef]
- Umer, A.; Ghouri, M.D.; Muyizere, T.; Aqib, R.M.; Muhaymin, A.; Cai, R.; Chen, C. Engineered Nano–Bio Interfaces for Stem Cell Therapy. Precis. Chem. 2023, 1, 341–356. [Google Scholar] [CrossRef] [PubMed]
- Esfahani, S.N.; Irizarry, A.M.R.; Xue, X.; Lee, S.B.-D.; Shao, Y.; Fu, J. Micro/nanoengineered technologies for human pluripotent stem cells maintenance and differentiation. Nano Today 2021, 41, 101310. [Google Scholar] [CrossRef]
- Yang, C.Y.; Huang, W.Y.; Chen, L.H.; Liang, N.W.; Wang, H.C.; Lu, J.; Wang, T.W. Neural tissue engineering: The influence of scaffold surface topography and extracellular matrix microenvironment. J. Mater. Chem. B 2021, 9, 567–584. [Google Scholar] [CrossRef]
- Dalby, M.J.; Gadegaard, N.; Oreffo, R.O. Harnessing nanotopography and integrin–matrix interactions to influence stem cell fate. Nat. Mater. 2014, 13, 558–569. [Google Scholar] [CrossRef]
- Saha, K.; Keung, A.J.; Irwin, E.F.; Li, Y.; Little, L.; Schaffer, D.V.; Healy, K.E. Substrate modulus directs neural stem cell behavior. Biophys. J. 2008, 95, 4426–4438. [Google Scholar] [CrossRef] [PubMed]
- Li-Beisson, Y.; Pollard, M.; Sauveplane, V.; Pinot, F.; Ohlrogge, J.; Beisson, F. Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc. Natl. Acad. Sci. USA 2009, 106, 22008–22013. [Google Scholar] [CrossRef]
- Rey-Suarez, B.A.W.; Hourwitz, I.M.T.; Kerr, S.; Shroff, H.; Fourkas, J.T.; Upadhyaya, A. Nanotopography modulates cytoskeletal organization and dynamics during T cell activation. Mol. Biol. Cell 2022, 33, ar88. [Google Scholar]
- Luo, J.; Walker, M.; Xiao, Y.; Donnelly, H.; Dalby, M.J.; Salmeron-Sanchez, M. The influence of nanotopography on cell behaviour through interactions with the extracellular matrix—A review. Bioact. Mater. 2022, 15, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Chien, F.C.; Dai, Y.H.; Kuo, C.W.; Chen, P. Flexible nanopillars to regulate cell adhesion and movement. Nanotechnology 2016, 27, 475101. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhu, S.; Pang, S.W. Directing osteoblastic cell migration on arrays of nanopillars and nanoholes with different aspect ratios. Lab Chip 2021, 21, 2206–2216. [Google Scholar] [CrossRef]
- Herr, C.; Winkler, B.; Ziebert, F.; Aranson, I.S.; Fourkas, J.T.; Losert, W. Spontaneous polarization and cell guidance on asymmetric nanotopography. Commun. Phys. 2022, 5, 114. [Google Scholar] [CrossRef]
- Dabare, P.R.L.; Bachhuka, A.; Visalakshan, R.M.; Shirazi, H.S.; Ostriko, K.; Smith, L.E.; Vasilev, K. Mechanistic insight in surface nanotopography driven cellular migration. ACS Biomater. Sci. Eng. 2021, 7, 4921–4932. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.W.; Kim, D.S.; Suhito, I.R.; Han, D.K.; Lee, T.; Kim, T.H. Enhancing neurogenesis of neural stem cells using homogeneous nanohole pattern-modified conductive platform. Int. J. Mol. Sci. 2019, 21, 191. [Google Scholar] [CrossRef] [PubMed]
- Solaini, G.; Baracca, A.; Lenaz, G.; Sgarbi, G. Hypoxia and mitochondrial oxidative metabolism. Biochim. Biophys. Acta 569 Bioenerg. 2010, 1797, 1171–1177. [Google Scholar] [CrossRef] [PubMed]
- Azimzadeh, M.; Khashayar, P.; Amereh, M.; Tasnim, N.; Hoorfar, M.; Akbari, M. Microfluidic-based oxygen (O2) sensors for on-chip monitoring of cell, tissue and organ metabolism. Biosensors 2022, 12, 6. [Google Scholar] [CrossRef] [PubMed]
- Palacio-Castañeda, V.; Velthuijs, N.; Le Gac, S.; Verdurmen, W.P. Oxygen control: The often overlooked but essential piece to create better in vitro systems. Lab Chip 2022, 22, 1068–1092. [Google Scholar] [CrossRef]
- Ramamoorthy, R.; Dutta, P.; Akbar, S. Oxygen sensors: Materials, methods, designs and applications. J. Mater. Sci. 2003, 38, 4271–4282. [Google Scholar] [CrossRef]
- Godet, I.; Doctorman, S.; Wu, F.; Gilkes, D.M. Detection of hypoxia in cancer models: Significance, challenges; advances. Cells 2022, 11, 686. [Google Scholar] [CrossRef] [PubMed]
- Grist, S.M.; Bennewith, K.L.; Cheung, K.C. Oxygen Measurement in Microdevices. Annu. Rev. Anal. Chem. 2022, 15, 221–246. [Google Scholar] [CrossRef]
- Wang, X.; Wolfbeis, O.S. Optical methods for sensing and imaging oxygen: Materials, spectroscopies and applications. Chem. Soc. Rev. 2014, 43, 3666–3761. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, H.; Jin, Q.; Jia, D.; Liu, T. Ratiometric optical fiber dissolved oxygen sensor based on fluorescence quenching principle. Sensors 2022, 22, 4811. [Google Scholar] [CrossRef]
- Marchetti, M.; Ronda, L.; Cozzi, M.; Bettati, S.; Bruno, S. Genetically encoded biosensors for the fluorescence detection of O2 and reactive O2 species. Sensors 2023, 23, 8517. [Google Scholar] [CrossRef] [PubMed]
- Sáez, P.J.; Vargas, P.; Shoji, K.F.; Harcha, P.A.; Lennon-Duménil, A.-M.; Sáez, J.C. ATP promotes the fast migration of dendritic cells through the activity of pannexin 1 channels and P2 × 7 receptors. Sci. Signal. 2017, 10, eaah7107. [Google Scholar] [CrossRef]
- Barclay, C. Energy demand and supply in human skeletal muscle. J. Muscle Res. Cell Motil. 2017, 38, 143–155. [Google Scholar] [CrossRef]
- Yoon, S.; Fuwad, A.; Jeong, S.; Cho, H.; Jeon, T.J.; Kim, S.M. Surface Deformation of Biocompatible Materials: Recent Advances in Biological Applications. Biomimetics 2024, 9, 395. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Driscoll, M.K.; Guven, C.; Losert, W. Asymmetric nanotopography biases cytoskeletal dynamics and promotes unidirectional cell guidance. Proc. Natl. Acad. Sci. USA 2015, 112, 12557–12562. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Eldeeb, M.A.; Pang, S.W. 3D nanoplasmonic biosensor for detection of filopodia in cells. Lab Chip 2020, 20, 2188–2196. [Google Scholar] [CrossRef]
- Humphrey, J.D.; Dufresne, E.R.; Schwartz, M.A. Mechanotransduction and extracellular matrix homeostasis. Nat. Rev. 589 Mol. Cell Biol. 2014, 15, 802–812. [Google Scholar] [CrossRef]
- Zhang, J.C.; Chen, M.K.; Liang, Y.; Hong, X.; Wang, M.T.; Cheng, Y.; Liu, X.; Tsai, D.P.; Pang, S.W. Nanoimprint meta-device for chiral imaging. Adv. Funct. Mater. 2023, 33, 2306422. [Google Scholar] [CrossRef]
- Grist, S.M.; Chrostowski, L.; Cheung, K.C. Optical oxygen sensors for applications in microfluidic cell culture. Sensors 2010, 10, 9286–9316. [Google Scholar] [CrossRef]
- Ungerböck, B.; Charwat, V.; Ertl, P.; Mayr, T. Microfluidic oxygen imaging using integrated optical sensor layers and a color camera. Lab Chip 2013, 13, 1593–1601. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-Y.; Sadhu, A.S.; Karmakar, R.; Chu, C.-S.; Lin, Y.-N.; Chang, S.-H.; Dalapati, G.K.; Biring, S. Strongly improving the sensitivity of phosphorescence-based optical oxygen sensors by exploiting nano-porous substrates. Biosensors 2022, 12, 774. [Google Scholar] [CrossRef] [PubMed]
- Capitani, N.; Baldari, C.T. F-actin dynamics in the regulation of endosomal recycling and immune synapse assembly. Front. Cell Dev. Biol. 2021, 9, 670882. [Google Scholar] [CrossRef] [PubMed]
- Triantopoulou, N.; Vidaki, M. Local mRNA translation and cytoskeletal reorganization: Mechanisms that tune neuronal responses. Front. Mol. Neurosci. 2022, 15, 949096. [Google Scholar] [CrossRef]
- Kuo, C.W.; Chueh, D.Y.; Chen, P. Investigation of size–dependent cell adhesion on nanostructured interfaces. J. Nanobiotechnol. 2014, 12, 54. [Google Scholar] [CrossRef] [PubMed]
- Tilokani, L.; Nagashima, S.; Paupe, V.; Prudent, J. Mitochondrial dynamics: Overview of molecular mechanisms. Essays Biochem. 2018, 62, 341–360. [Google Scholar] [PubMed]
- Denisenko, T.V.; Gorbunova, A.S.; Zhivotovsky, B. Mitochondrial involvement in migration, invasion and metastasis. Front. Cell Dev. Biol. 2019, 7, 355. [Google Scholar] [CrossRef] [PubMed]
- Bonora, M.; Patergnani, S.; Rimessi, A.; De Marchi, E.; Suski, J.M.; Bononi, A.; Giorgi, C.; Marchi, S.; Missiroli, S.; Poletti, F.; et al. ATP synthesis and storage. Purinergic Signal. 2012, 8, 343–357. [Google Scholar] [CrossRef]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef]
- Duanghathaipornsuk, S.; Farrell, E.J.; Alba-Rubio, A.C.; Zelenay, P.; Kim, D.-S. Detection technologies for reactive oxygen species: Fluorescence and electrochemical methods and their applications. Biosensors 2021, 11, 30. [Google Scholar] [CrossRef]
- Duval, K.; Grover, H.; Han, L.H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Habanjar, O.; Diab-Assaf, M.; Caldefie-Chezet, F.; Delort, L. 3D cell culture systems: Tumor application, advantages; disadvantages. Int. J. Mol. Sci. 2021, 22, 12200. [Google Scholar] [CrossRef] [PubMed]
- Griffith, C.M.; Huang, S.A.; Cho, C.; Khare, T.M.; Rich, M.; Lee, G.H.; Ligler, F.S.; Diekman, B.O.; Polacheck, W.J. Microfluidics for the study of mechanotransduction. J. Phys. D Appl. Phys. 2020, 53, 224004. [Google Scholar] [CrossRef] [PubMed]
- van der Meer, A.D.; Poot, A.A.; Duits, M.H.G.; Feijen, J.; Vermes, I. Microfluidic Technology in Vascular Research. Biomed Res. Int. 2009, 2009, 823148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, M.; Tseng, T.-M.; Schlichtmann, U. Open-source interactive design platform for 3D-printed microfluidic devices. Comms. Eng. 2024, 3, 71. [Google Scholar] [CrossRef]
- Yu, F.; Hunziker, W.; Choudhury, D. Engineering Microfluidic Organoid-on-a-Chip Platforms. Micromachines 2019, 10, 165. [Google Scholar] [CrossRef] [PubMed]
- Zanotelli, M.R.; Goldblatt, Z.E.; Miller, J.P.; Bordeleau, F.; Li, J.; VanderBurgh, J.A.; Reinhart-King, C.A. Regulation of ATP utilization during metastatic cell migration by collagen architecture. Mol. Biol. Cell 2018, 29, 1–9. [Google Scholar] [CrossRef]
- Desai, S.P.; Bhatia, S.N.; Toner, M.; Irimia, D. Mitochondrial localization and the persistent migration of epithelial cancer cells. Biophys. J. 2013, 104, 2077–2088. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Pang, S.W. Detecting Nanotopography Induced Changes in Cell Migration Directions Using Oxygen Sensors. Biosensors 2024, 14, 389. https://doi.org/10.3390/bios14080389
Wang M, Pang SW. Detecting Nanotopography Induced Changes in Cell Migration Directions Using Oxygen Sensors. Biosensors. 2024; 14(8):389. https://doi.org/10.3390/bios14080389
Chicago/Turabian StyleWang, Muting, and Stella W. Pang. 2024. "Detecting Nanotopography Induced Changes in Cell Migration Directions Using Oxygen Sensors" Biosensors 14, no. 8: 389. https://doi.org/10.3390/bios14080389
APA StyleWang, M., & Pang, S. W. (2024). Detecting Nanotopography Induced Changes in Cell Migration Directions Using Oxygen Sensors. Biosensors, 14(8), 389. https://doi.org/10.3390/bios14080389





