Abstract
The rapid development of biosensing technologies together with the advent of deep learning has marked an era in healthcare and biomedical research where widespread devices like smartphones, smartwatches, and health-specific technologies have the potential to facilitate remote and accessible diagnosis, monitoring, and adaptive therapy in a naturalistic environment. This systematic review focuses on the impact of combining multiple biosensing techniques with deep learning algorithms and the application of these models to healthcare. We explore the key areas that researchers and engineers must consider when developing a deep learning model for biosensing: the data modality, the model architecture, and the real-world use case for the model. We also discuss key ongoing challenges and potential future directions for research in this field. We aim to provide useful insights for researchers who seek to use intelligent biosensing to advance precision healthcare.
1. Introduction
Wearable non-invasive biosensors, when combined with machine learning, can enable remote monitoring, diagnosis, and therapy for a wide range of health conditions. Wearable devices can record substantial amounts of unlabeled data from biosensors such as Electrodermal Activity (EDA), Electrocardiography (ECG), and Electroencephalography (EEG). Deep learning [], a family of techniques well-suited for analysis of large data streams, has recently been used for making predictions with these data.
In this systematic review, we explore the synergy between non-invasive biosensors and machine learning, with a focus on deep learning in the field of healthcare and biomedical research. While our review is focused on non-invasive biosensors in particular, the insights and methods that we discuss can also be applied to other types of biosensors.
Rather than conducting one large systematic review, we organize this paper as a narrative review coupled with a series of targeted mini-reviews. Our goal is not to provide an exhaustive list of papers in the field of non-invasive intelligent biosensing for healthcare, as this research field is quite massive. Instead, we aim to provide demonstrative examples of the types of studies, innovations, and trends that have emerged in recent years.
Our semi-systematic review is structured as follows. We begin with an overview of common biosensors used in healthcare. We proceed with a review of essential deep learning architectures that are commonly applied to biosensing. We next describe specific application areas of biosensing in digital health, particularly remote patient monitoring, digital diagnosis, and adaptive digital therapy. In each of these sections, we perform a separate ‘tiny’ systematic review.
2. Common Deep Learning Architectures in Biosensing
Deep learning is improving the field of biosensing by enabling the analysis of the large, complex, and longitudinal data generated by biosensors. Machine learning models can enable personalization of remote treatment strategies (Figure 1). Deep learning models often achieve better performance than classical methods on biosensing-related tasks, as biosensors tend to have high sampling frequencies that result in the generation of large datasets.
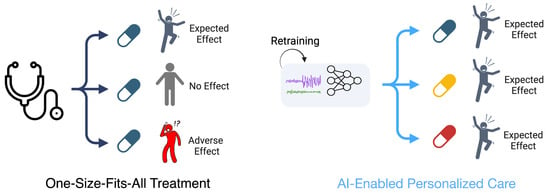
Figure 1.
Comparison between traditional one-size-fits-all treatment strategies and personalized treatment strategies using machine learning. On the left, a single medication is provided to three different people, leading to three different patient outcomes. On the right, individualized patient data are used to personalize a treatment plan for each patient, leading to an optimized care approach with separate medications being prescribed to each patient.
In this section, we review the most common deep learning architectures used with biosensor data. Training and running deep learning models requires a significant amount of computational resources. Due to advancements in model compression, these models may now be integrated into microprocessors and run on smart phones or smart watches, allowing for real-time predictions. The goal of model compression is to create a model with significantly fewer parameters than the original model while maintaining discriminative performance. Many modern microprocessors contain hardware accelerations that are specific to deep learning, enabling close-to-real-time operations.
2.1. Convolutional Neural Networks (CNNs)
Around a decade ago, CNNs revolutionized the processing of images and signal data, including biosignals [,,]. The CNN architecture is inspired by the natural visual perception mechanism of living organisms, performing well on data with a grid-like topology [] such as images (2-dimensional grids of pixels) and time series or audio signals (1-dimensional grids of data). CNNs extract meaningful patterns and features from noisy and complex biological data. 1-dimensional CNNs (1D CNN) in particular are a valuable tool for biosensing.
2.2. Long Short-Term Memory Networks (LSTMs)
LSTM networks are a subclass of the recurrent neural network (RNN) family of architectures. LSTMs aim to solve the vanishing gradient problem that saliently exists for unmodified RNNs []. LSTMs have the ability to learn the dependencies throughout a sequence, making them excel at processing biosensor data, which are inherently sequential in nature [,].
2.3. Autoencoders
Autoencoders are used for dimensionality reduction and feature representation learning [,,]. Autoencoders are made up of two fundamental components: an encoder and a decoder. The encoder compresses the data into a low-dimensional representation, while the decoder then reconstructs the data from this compressed form. Autoencoders can serve as a feature extraction tool for new biosignal data streams, enabling applications such as anomaly detection in the monitoring of symptoms or the diagnosis of diseases.
2.4. Transformers
Transformers have the potential to perform well on sequential data due to the use of the self-attention mechanism. This makes them suitable for health monitoring applications in the analysis of the temporal evolution of the physiological signals. The self-attention mechanism enables parallel processing of data during both training and inference.
2.5. Model Selection Considerations
Selecting an appropriate deep learning model for a healthcare application is a decision that requires careful consideration of several key factors [,,]. The primary deciding factor is the type of data, as different architectures excel at processing differing types of data modalities; certain models are better equipped to capture intricate temporal dependencies, spatial correlations, or anomalies. Specifically, LSTMs are useful for continuous monitoring because of their ability to process and learn from sequential data, making them well-suited for analyzing physiological time-series data. Autoencoders excel in the efficient identification of anomalies and patterns in complex datasets due to their ability to encode data into a compact yet meaningful representation space. Transformers offer advantages in handling large sequences of data due to their self-attention mechanism, enabling the detection of subtle and complex patterns when trained using large datasets. One-dimensional CNNs have also demonstrated utility in making predictions from time-series data. CNNs possess the ability to effectively capture local and global patterns in sequential data.
2.6. Commercial Use of Deep Learning Models for Biosensing
Many products on the market use deep learning for biosensing. For instance, popular products like Fitbit and Apple provide features for tracking heart rate and activity levels using techniques that incorporate deep learning into the process [,]. SpeechVive leverages deep learning algorithms to analyze speech patterns, a voice-based biosignal, to offer aid to individuals living with Parkinson’s disease. The Samsung Watch uses an autoencoder and LSTM for a smart alarm system during sleep []. The Samsung Smartwatch uses a CNN for early detection and burden estimation of atrial fibrillation []. The Oura ring tracks cardiovascular activity using deep learning []. Oura users can follow their autonomic nervous system responses to their daily behavior based on nightly changes in heart rate. Many other wearable device companies, while not publicly releasing the details of their proprietary algorithms, most likely use deep learning.
3. Sensor Modalities and Corresponding Health Applications
Here, we describe common sensor modalities used for biosensing and the corresponding health applications that are common to each modality (Figure 2). We note that the field of biosensing is so vast that we do not provide a comprehensive overview of every biosensor that has ever been used for predictive modeling. Instead, we select 3 common yet complementary modalities as demonstrative examples.
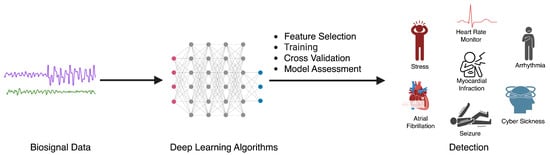
Figure 2.
The process of utilizing deep learning algorithms for the detection of various health conditions using biosignal data, where physiological signals are fed as input to a deep learning model that makes a prediction of a health event of interest.
3.1. Deep Learning with EEG
Prior work has successfully used EEG signals to classify emotions [], detect seizures [], and measure sleep []. EEG is a widely used biosignal for the study of cognitive functions, including perception, memory, focus, language, and emotional processing.
Using Web of Science and PubMed, our search query focused on articles from 2020 to 2023 that addressed EEG-based classification and detection, with titles containing classification or detection and abstracts mentioning CNN and EEG. This approach was also used for LSTMs, Autoencoders, and Transformers. We excluded review articles, pilot studies, and duplicates from our search. There was some overlap between the Web of Science and PubMed results. Table 1 summarizes these papers.

Table 1.
Overview of deep learning algorithms used in different EEG-based studies. In some studies, more than one dataset is used. They are separated by a “/”.
3.2. Deep Learning with EDA
EDA is a common indicator in affective computing due to its sensitivity to physiological changes. However, the challenge of individual variability in EDA responses necessitates advanced analytical approaches to effectively interpret and utilize these signals. EDA is sensitive to skin hydration levels [], making EDA a pivotal tool for capturing physiological arousal and emotional responses [].
Our search in the Web of Science and PubMed focused on deep learning models using EDA, and we searched for papers published between 2020 and 2023. We also applied the research area filter to be Computer Science and Medical Informatics. There was some overlap between the Web of Science and PubMed search results. We further filtered papers based on their relevance to intelligent biosensing. Table 2 summarizes the selected papers.

Table 2.
Overview of deep learning algorithms used in different EDA-based studies. In some studies, more than one dataset is used. They are separated by a “/”.
3.3. Deep Learning with ECG
Cardiovascular disease is a leading cause of death globally. Factors such as stress and psychological distress have been linked to an increased risk of cardiovascular disease, especially in younger people []. ECGs are sensors measuring the electrical activity of the heart. ECGs are used for the diagnosis of heart problems, such as arrhythmias and heart attacks []. However, ECGs produce a large amount of data that are hard to scrutinize manually []. Deep learning algorithms can be trained to identify patterns in ECG data that are associated with different heart conditions []. This can help doctors to diagnose heart problems more efficiently and scalably.
Using Web of Science and PubMed, we performed a targeted search for recent (2020–2023) research using deep learning to analyze ECG data for classification and detection tasks. Specifically querying CNN, LSTM, Autoencoder, and Transformer architectures, we screened for classification or detection in titles and model references in abstracts. After assessing their relevance to computer science and cardiology, we identified a final set of papers. There was some overlap between the Web of Science and PubMed search results. Table 3 provides a summary of the identified papers.

Table 3.
Overview of deep learning algorithms used in different ECG-based studies. In some studies, more than one dataset is used. They are separated by a “/”.
3.4. Consideration for Selecting Biosensors for a Digital Health Application
Each type of biosensor, mentioned above, is often useful for a small range of health conditions. EEG is used for measuring electrical activity of the brain, making it particularly suitable for neurology and cognitive science research. EDA, on the other hand, is less complicated to measure from an end-user perspective than EEG and can be used for applications such as psychological research, stress monitoring, and affective computing. ECG, measuring the electrical activity of the heart, is used in cardiology. We notably did not review multimodal models. However, it is worth noting that it is likely that combining multiple biosensors into a single predictive model might enable the detection of previously unexplored health conditions and events that are infeasible to predict using a single modality alone.
4. Digital Health Applications using Biosensors
Machine learning-based biosensing can be applied to several areas in healthcare. Here, we summarize recent literature in three common applications of biosensing: remote patient monitoring, digital diagnostics, and adaptive digital interventions. We distinguish between remote patient monitoring and digital diagnosis in the following manner: a diagnosis is made of a disease that the patient may or may not have, while remote monitoring focuses on symptoms that can periodically occur for the patient regardless of whether the patient has a disease diagnosis.
4.1. Remote Patient Monitoring
Biosensing technology, when combined with machine learning, opens up new prospects for improving remote patient care through continuous physiological data analysis outside of the clinic. Continuous physiological data analysis provides opportunities to significantly improve patient care by providing the clinician with more detailed status reports about each patient [].
Key technical challenges for remote patient monitoring include efficiently processing and transmitting complex physiological data to optimize energy use and sustainability [,], accurately interpreting multifaceted biosensing signals [], and creating adaptive systems that provide robust monitoring across a diverse array of patients [,]. Additional human-centered challenges involve the need to respond quickly enough to develop supportive technologies amidst the detection of an adverse health event [].
We performed a search on Web of Science using the search terms remote patient monitoring, telehealth, wireless health monitoring, and machine learning along with the use of biosensors mentioned in the abstracts. Table 4 displays the application and overview of these remote patient monitoring papers, focusing on the application rather than the technique or evaluation.

Table 4.
Summary of remote patient monitoring applications at the intersection of biosensing and machine learning.
4.2. Digital Diagnostics
There are multiple key challenges that must be addressed before translation of biosensor- based digital diagnostics into clinical settings. First, the heterogeneity of data can complicate biosensing-based diagnostics. Intrinsic variability common in biosensors [,] and the necessity for sensor stability plus consistent performance throughout a wide range of environmental conditions create a significant challenge for machine learning modeling. Second, the models must be robust enough to handle and interpret the vast, complex datasets that biosensors generate without overlooking nuances that can occur at relatively small time-scales. Finally, biosensors used for nuanced and possibly even subjective diagnoses require high analytical sensitivity and specificity to interpret signals that differ subtly between people [].
We performed a search for digital diagnostics in the Web of Science for papers with a title matching biosensor, healthcare, and ML coupled with the keywords classification or detection, focusing on the last 5 years. We further filtered papers that were not relevant to diagnostics. We list the identified papers in Table 5, focusing on the application rather than the technique or evaluation.

Table 5.
Summary of digital diagnostics using biosensing and machine learning.
4.3. Adaptive Digital Interventions
Digital health interventions powered by machine learning have the potential to improve health outcomes through therapeutics that are delivered in a just-in-time manner []. However, these interventions pose critical challenges, such as data privacy. Though these technologies hold great promise for providing the basis for real-time intervention for a wide spectrum of health conditions, their use must be fine-tuned and validated so that proper, reliable responses may be used in actual practice [,,].
We performed a search on Web of Science for health interventions, using the terms Randomized Controlled Trials, wearable sensor, and biosensor, along with machine learning in the abstract. We excluded review articles and protocols. Table 6 outlines the clinical endpoint, the application effectiveness metrics, and a summary of the study, offering insights into the impact and scope of digital health interventions using intelligent biosensing.

Table 6.
Summary of digital health intervention studies, including clinical endpoints, metrics indicating the effectiveness of the intervention, and a summary of the findings from the study.
4.4. Considerations for Integrating Wearable Sensors in Digital Health Systems
The integration of wearable sensors in digital health systems requires careful consideration of human factors such as interoperability, interpretability, bias, privacy, and user compliance.
Interpretability is necessary to enable clinicians to understand the reasoning behind a models’ decision making []. However, deep learning models are inherently not interpretable, requiring black-box solutions that only provide partial explainability.
Biases in machine learning models can be harmful to biomedical research because incorrect predictions made by digital health technologies may disproportionately affect some demographic groups. Bias may exist at several phases of the model development cycle, including data collection, model optimization, and model calibration [].
There is always a risk of privacy violations when dealing with sensitive health data for patient monitoring or disease diagnosis [,]. A possible solution is federated learning, where a separate model is trained on each individual’s device. This solution integrates well with the idea of model personalization.
Finally, addressing wearability and other user-centered design issues is needed to improve patient compliance. It is well documented that users often do not properly use their wearables. Several machine learning models have been considered for addressing these adherence issues [,,].
5. Challenges and Opportunities
The integration of deep learning with biosensor data presents several challenges and corresponding opportunities for advancing healthcare. We discuss these here.
5.1. Challenges
There are several technical challenges that can hinder the development and deployment of intelligent biosensing. We discuss three key challenges: (1) variability across human subjects, (2) noise, and (3) the necessity of complex and highly parameterized models to make sense of biosignal data streams.
Variability across human subjects is a major challenge for model generalization [,]. This can compromise the ability to generalize a model to new subjects [,,], leading to inconsistent signals being recorded between people. Individual physiological characteristics can have a major impact on the patterns that are present within the data, reducing the accuracy of models when applied to new participants.
Another key challenge is the inherent noisiness of real-world biosignal data. This noise can arise both from the underlying biosensor data as well as from irregular labeling practices. The subjectivity and difficulty of forecasting outcomes, such as stress levels, may add to this noise. These issues have been shown to negatively impact model performance [,,].
Model complexity presents yet another core challenge, leading to difficulties in the interpretation of results [,]. Deep learning models are powerful but can be too complex to understand easily. This ‘black box’ nature of deep learning models can render it difficult for doctors to trust and use these models.
5.2. Opportunities
The challenges described above can be solved by a number of promising techniques that have recently emerged in the literature. We describe two key research opportunities for the field: (1) personalization using self-supervised learning and (2) multimodal explainable artificial intelligence.
There is an opportunity for the research community to innovate in the personalization of deep learning models using biosensor data. Personalizing models to account for variability among subjects has the potential to increase model performance, especially when coupled with multimodal learning approaches [,,,,,,]. Such personalization can be enhanced with self-supervised learning, where a model is pre-trained on the massive unlabeled data streams that are generated when a user passively wears a device with one or more biosensors.
There is also an opportunity to combine innovations in multimodal deep learning with explainable artificial intelligence. Multimodal explainable machine learning approaches designed for time-series data have the potential to improve the clinician and patient’s understanding of automated or semi-automated decision making processes.
6. Conclusions
We have reviewed common deep learning architectures, sensor modalities, and healthcare applications in the field of intelligent biosensing. The fusion of biosensing and artificial intelligence can lead to improved precision healthcare via scalable and accessible remote monitoring platforms, digital diagnostics, and adaptive just-in-time digital interventions.
Funding
This research was funded by National Institute of General Medical Sciences (NIGMS) grant number U54GM138062 and Medical Research Award fund of the Hawaii Community Foundation grant number MedRes_2023_00002689.
Acknowledgments
We used ChatGPT to edit the grammar of our manuscript and to re-phrase sentences that were originally worded unclearly. However, all contents in this manuscript are original ideas and analyses conducted by the authors.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Dong, S.; Wang, P.; Abbas, K. A survey on deep learning and its applications. Comput. Sci. Rev. 2021, 40, 100379. [Google Scholar] [CrossRef]
- Zhou, M.; Tian, C.; Cao, R.; Wang, B.; Niu, Y.; Hu, T.; Guo, H.; Xiang, J. Epileptic seizure detection based on EEG signals and CNN. Front. Neuroinform. 2018, 12, 95. [Google Scholar] [CrossRef]
- Mao, W.; Fathurrahman, H.; Lee, Y.; Chang, T. EEG dataset classification using CNN method. J. Phys. Conf. Ser. 2020, 1456, 012017. [Google Scholar] [CrossRef]
- Liu, Q.; Cai, J.; Fan, S.Z.; Abbod, M.F.; Shieh, J.S.; Kung, Y.; Lin, L. Spectrum analysis of EEG signals using CNN to model patient’s consciousness level based on anesthesiologists’ experience. IEEE Access 2019, 7, 53731–53742. [Google Scholar] [CrossRef]
- Khanday, N.Y.; Sofi, S.A. Taxonomy, state-of-the-art, challenges and applications of visual understanding: A review. Comput. Sci. Rev. 2021, 40, 100374. [Google Scholar] [CrossRef]
- Venugopalan, S.; Xu, H.; Donahue, J.; Rohrbach, M.; Mooney, R.; Saenko, K. Translating videos to natural language using deep recurrent neural networks. arXiv 2014, arXiv:1412.4729. [Google Scholar]
- Saltepe, B.; Bozkurt, E.U.; Güngen, M.A.; Çiçek, A.E.; Şeker, U.Ö.Ş. Genetic circuits combined with machine learning provides fast responding living sensors. Biosens. Bioelectron. 2021, 178, 113028. [Google Scholar] [CrossRef] [PubMed]
- Esmaeili, F.; Cassie, E.; Nguyen, H.P.T.; Plank, N.O.; Unsworth, C.P.; Wang, A. Predicting analyte concentrations from electrochemical aptasensor signals using LSTM recurrent networks. Bioengineering 2022, 9, 529. [Google Scholar] [CrossRef] [PubMed]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef]
- Vincent, P.; Larochelle, H.; Lajoie, I.; Bengio, Y.; Manzagol, P.A.; Bottou, L. Stacked denoising autoencoders: Learning useful representations in a deep network with a local denoising criterion. J. Mach. Learn. Res. 2010, 11, 3371–3408. [Google Scholar]
- Ng, A. Sparse autoencoder. CS294A Lect. Notes 2011, 72, 1–19. [Google Scholar]
- Saadatnejad, S.; Oveisi, M.; Hashemi, M. LSTM-based ECG classification for continuous monitoring on personal wearable devices. IEEE J. Biomed. Health Inform. 2019, 24, 515–523. [Google Scholar] [CrossRef]
- Sun, Y.; Jin, W.; Si, X.; Zhang, X.; Cao, J.; Wang, L.; Yin, S.; Ming, D. Continuous Seizure Detection Based on Transformer and Long-Term iEEG. IEEE J. Biomed. Health Inform. 2022, 26, 5418–5427. [Google Scholar] [CrossRef]
- Yildirim, O.; San Tan, R.; Acharya, U.R. An efficient compression of ECG signals using deep convolutional autoencoders. Cogn. Syst. Res. 2018, 52, 198–211. [Google Scholar] [CrossRef]
- Chien, H.Y.S.; Goh, H.; Sandino, C.M.; Cheng, J.Y. MAEEG: Masked Auto-encoder for EEG Representation Learning. In Proceedings of the NeurIPS Workshop, New Orleans, LA, USA, 28 November 2022. [Google Scholar]
- Nazaret, A.; Tonekaboni, S.; Darnell, G.; Ren, S.; Sapiro, G.; Miller, A.C. Modeling Heart Rate Response to Exercise with Wearable Data. In Proceedings of the NeurIPS, New Orleans, LA, USA, 28 November 2022. [Google Scholar]
- Slyusarenko, K.; Fedorin, I. Smart alarm based on sleep stages prediction. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 4286–4289. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, L.; Nathan, V.; Kuang, J.; Kim, J.; Gao, J.A.; Olgin, J. Towards early detection and burden estimation of atrial fibrillation in an ambulatory free-living environment. In Proceedings of the ACM on Interactive, Mobile, Wearable and Ubiquitous Technologies; ACM: New York, NY, USA, 2021; Volume 5, pp. 1–19. [Google Scholar]
- Kinnunen, H.; Rantanen, A.; Kenttä, T.; Koskimäki, H. Feasible assessment of recovery and cardiovascular health: Accuracy of nocturnal HR and HRV assessed via ring PPG in comparison to medical grade ECG. Physiol. Meas. 2020, 41, 04NT01. [Google Scholar] [CrossRef]
- Song, T.; Zheng, W.; Song, P.; Cui, Z. EEG Emotion Recognition Using Dynamical Graph Convolutional Neural Networks. IEEE Trans. Affect. Comput. 2020, 11, 532–541. [Google Scholar] [CrossRef]
- Park, C.; Choi, G.; Kim, J.; Kim, S.; Kim, T.J.; Min, K.; Jung, K.Y.; Chong, J. Epileptic seizure detection for multi-channel EEG with deep convolutional neural network. In Proceedings of the 2018 International Conference on Electronics, Information, and Communication (ICEIC), Honolulu, HI, USA, 24–27 January 2018; pp. 1–5. [Google Scholar]
- Tsinalis, O.; Matthews, P.M.; Guo, Y.; Zafeiriou, S. Automatic sleep stage scoring with single-channel EEG using convolutional neural networks. arXiv 2016, arXiv:1610.01683. [Google Scholar]
- Roy, A.M. An efficient multi-scale CNN model with intrinsic feature integration for motor imagery EEG subject classification in brain-machine interfaces. Biomed. Signal Process. Control 2022, 74, 103496. [Google Scholar] [CrossRef]
- Leeb, R.; Brunner, C.; Müller-Putz, G. Available online: http://www.bbci.de/competition/iv/ (accessed on 6 March 2021).
- Raghu, S.; Sriraam, N.; Temel, Y.; Rao, S.V.; Kubben, P.L. EEG based multi-class seizure type classification using convolutional neural network and transfer learning. Neural Netw. 2020, 124, 202–212. [Google Scholar] [CrossRef]
- Obeid, I.; Picone, J. The temple university hospital EEG data corpus. Front. Neurosci. 2016, 10, 196. [Google Scholar] [CrossRef]
- Fouladi, S.; Safaei, A.A.; Mammone, N.; Ghaderi, F.; Ebadi, M. Efficient deep neural networks for classification of Alzheimer’s disease and mild cognitive impairment from scalp EEG recordings. Cogn. Comput. 2022, 14, 1247–1268. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, Z.; Xiong, R.; Liao, X.; Tan, X. A Method for Classification and Evaluation of Pilot’s Mental States Based on CNN. Comput. Syst. Sci. Eng. 2023, 46, 1999–2020. [Google Scholar] [CrossRef]
- Zheng, W.L.; Lu, B.L. Investigating Critical Frequency Bands and Channels for EEG-Based Emotion Recognition with Deep Neural Networks. IEEE Trans. Auton. Ment. Dev. 2015, 7, 162–175. [Google Scholar] [CrossRef]
- Parajuli, M.; Amara, A.W.; Shaban, M. Deep-learning detection of mild cognitive impairment from sleep electroencephalography for patients with Parkinson’s disease. PLoS ONE 2023, 18, e0286506. [Google Scholar] [CrossRef] [PubMed]
- Göker, H. Detection of alzheimer’s disease from electroencephalography (EEG) signals using multitaper and ensemble learning methods. Uludağ Üniversitesi Mühendislik Fakültesi Dergisi 2023, 28, 141–152. [Google Scholar] [CrossRef]
- Li, Y.; Shen, Y.; Fan, X.; Huang, X.; Yu, H.; Zhao, G.; Ma, W. A novel EEG-based major depressive disorder detection framework with two-stage feature selection. BMC Med. Inform. Decis. Mak. 2022, 22, 209. [Google Scholar] [CrossRef]
- Masad, I.S.; Alqudah, A.; Qazan, S. Automatic classification of sleep stages using EEG signals and convolutional neural networks. PLoS ONE 2024, 19, e0297582. [Google Scholar] [CrossRef]
- Khalighi, S.; Sousa, T.; Santos, J.M.; Nunes, U. ISRUC-Sleep: A comprehensive public dataset for sleep researchers. Comput. Methods Programs Biomed. 2016, 124, 180–192. [Google Scholar] [CrossRef]
- Abbasi, S.F.; Abbasi, Q.H.; Saeed, F.; Alghamdi, N.S. A convolutional neural network-based decision support system for neonatal quiet sleep detection. Math. Biosci. Eng. 2023, 20, 17018–17036. [Google Scholar] [CrossRef]
- Tao, T.; Gao, Y.; Jia, Y.; Chen, R.; Li, P.; Xu, G. A Multi-Channel Ensemble Method for Error-Related Potential Classification Using 2D EEG Images. Sensors 2023, 23, 2863. [Google Scholar] [CrossRef]
- Wei, L.; Ventura, S.; Ryan, M.A.; Mathieson, S.; Boylan, G.B.; Lowery, M.; Mooney, C. Deep-spindle: An automated sleep spindle detection system for analysis of infant sleep spindles. Comput. Biol. Med. 2022, 150, 106096. [Google Scholar] [CrossRef] [PubMed]
- Barnes, L.D.; Lee, K.; Kempa-Liehr, A.W.; Hallum, L.E. Detection of sleep apnea from single-channel electroencephalogram (EEG) using an explainable convolutional neural network (CNN). PLoS ONE 2022, 17, e0272167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.Q.; Cui, L.; Mueller, R.; Tao, S.; Kim, M.; Rueschman, M.; Mariani, S.; Mobley, D.; Redline, S. The National Sleep Research Resource: Towards a sleep data commons. J. Am. Med. Inform. Assoc. 2018, 25, 1351–1358. [Google Scholar] [CrossRef] [PubMed]
- Shaban, M.; Amara, A.W. Resting-state electroencephalography based deep-learning for the detection of Parkinson’s disease. PLoS ONE 2022, 17, e0263159. [Google Scholar] [CrossRef] [PubMed]
- Rockhill, A.P.; Jackson, N.; George, J.; Aron, A.; Swann, N.C. UC San Diego Resting State EEG Data from Patients with Parkinson’s Disease. 2020. Available online: https://openneuro.org/datasets/ds002778/versions/1.0.5 (accessed on 1 January 2020).
- Ghosh, R.; Phadikar, S.; Deb, N.; Sinha, N.; Das, P.; Ghaderpour, E. Automatic Eyeblink and Muscular Artifact Detection and Removal From EEG Signals Using k-Nearest Neighbor Classifier and Long Short-Term Memory Networks. IEEE Sens. J. 2023, 23, 5422–5436. [Google Scholar] [CrossRef]
- Ghosh, R.; Deb, N.; Sengupta, K.; Phukan, A.; Choudhury, N.; Kashyap, S.; Phadikar, S.; Saha, R.; Das, P.; Sinha, N.; et al. SAM 40: Dataset of 40 subject EEG recordings to monitor the induced-stress while performing Stroop color-word test, arithmetic task, and mirror image recognition task. Data Brief 2022, 40, 107772. [Google Scholar] [CrossRef] [PubMed]
- Michielli, N.; Acharya, U.R.; Molinari, F. Cascaded LSTM recurrent neural network for automated sleep stage classification using single-channel EEG signals. Comput. Biol. Med. 2019, 106, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef]
- Soleymani, M.; Asghari-Esfeden, S.; Fu, Y.; Pantic, M. Analysis of EEG Signals and Facial Expressions for Continuous Emotion Detection. IEEE Trans. Affect. Comput. 2016, 7, 17–28. [Google Scholar] [CrossRef]
- Soleymani, M.; Lichtenauer, J.; Pun, T.; Pantic, M. A Multimodal Database for Affect Recognition and Implicit Tagging. IEEE Trans. Affect. Comput. 2012, 3, 42–55. [Google Scholar] [CrossRef]
- Alessandrini, M.; Biagetti, G.; Crippa, P.; Falaschetti, L.; Luzzi, S.; Turchetti, C. EEG-Based Neurodegenerative Disease Classification using LSTM Neural Networks. In Proceedings of the 2023 IEEE Statistical Signal Processing Workshop (SSP), Hanoi, Vietnam, 2–5 July 2023; pp. 428–432. [Google Scholar]
- Zhang, S.; Wang, J.; Pei, L.; Liu, K.; Gao, Y.; Fang, H.; Zhang, R.; Zhao, L.; Sun, S.; Wu, J.; et al. Interpretable CNN for ischemic stroke subtype classification with active model adaptation. BMC Med. Inform. Decis. Mak. 2022, 22, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Alvi, A.M.; Siuly, S.; Wang, H. A long short-term memory based framework for early detection of mild cognitive impairment from EEG signals. IEEE Trans. Emerg. Top. Comput. Intell. 2022, 7, 375–388. [Google Scholar] [CrossRef]
- Kashefpoor, M.; Rabbani, H.; Barekatain, M. Automatic diagnosis of mild cognitive impairment using electroencephalogram spectral features. J. Med. Signals Sens. 2016, 6, 25–32. [Google Scholar] [PubMed]
- Khan, S.U.; Jan, S.U.; Koo, I. Robust Epileptic Seizure Detection Using Long Short-Term Memory and Feature Fusion of Compressed Time–Frequency EEG Images. Sensors 2023, 23, 9572. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, A.; Siddiqui, F.; Alam, M.A.; Idrees, S.M. Tri-model classifiers for EEG based mental task classification: Hybrid optimization assisted framework. BMC Bioinform. 2023, 24, 406. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Tang, L.; Pan, J. AGL-Net: An efficient neural network for EEG-based driver fatigue detection. J. Integr. Neurosci. 2023, 22, 146. [Google Scholar] [CrossRef] [PubMed]
- Najafi, T.; Jaafar, R.; Remli, R.; Wan Zaidi, W.A. A classification model of EEG signals based on rnn-lstm for diagnosing focal and generalized epilepsy. Sensors 2022, 22, 7269. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, F.; Mostafa, S.S.; Freitas, D.; Morgado-Dias, F.; Ravelo-García, A.G. Multiple Time Series Fusion based on LSTM: An application to CAP A phase classification using EEG. Int. J. Environ. Res. Public Health 2022, 19, 10892. [Google Scholar] [CrossRef] [PubMed]
- Phutela, N.; Relan, D.; Gabrani, G.; Kumaraguru, P.; Samuel, M. Stress classification using brain signals based on LSTM network. Comput. Intell. Neurosci. 2022, 2022, 7607592. [Google Scholar] [CrossRef]
- Muhammad, G.; Hossain, M.S.; Kumar, N. EEG-Based Pathology Detection for Home Health Monitoring. IEEE J. Sel. Areas Commun. 2021, 39, 603–610. [Google Scholar] [CrossRef]
- Amin, S.U.; Alsulaiman, M.; Muhammad, G.; Mekhtiche, M.A.; Hossain, M.S. Deep Learning for EEG motor imagery classification based on multi-layer CNNs feature fusion. Future Gener. Comput. Syst. 2019, 101, 542–554. [Google Scholar] [CrossRef]
- Brunner, C.; Leeb, R.; Müller-Putz, G.; Schlögl, A.; Pfurtscheller, G. BCI Competition 2008–Graz Data Set A; Institute for Knowledge Discovery (Laboratory of Brain-Computer Interfaces), Graz University of Technology: Graz, Austria, 2008; Volume 16, pp. 1–6. [Google Scholar]
- Schirrmeister, R.T.; Springenberg, J.T.; Fiederer, L.D.J.; Glasstetter, M.; Eggensperger, K.; Tangermann, M.; Hutter, F.; Burgard, W.; Ball, T. Deep learning with convolutional neural networks for EEG decoding and visualization. Hum. Brain Mapp. 2017, 38, 5391–5420. [Google Scholar] [CrossRef] [PubMed]
- Ieracitano, C.; Mammone, N.; Hussain, A.; Morabito, F.C. A novel multi-modal machine learning based approach for automatic classification of EEG recordings in dementia. Neural Netw. 2020, 123, 176–190. [Google Scholar] [CrossRef] [PubMed]
- Andrzejak, R.G.; Lehnertz, K.; Mormann, F.; Rieke, C.; David, P.; Elger, C.E. Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: Dependence on recording region and brain state. Phys. Rev. E 2001, 64, 061907. [Google Scholar] [CrossRef] [PubMed]
- Waqar, H.; Xiang, J.; Zhou, M.; Hu, T.; Ahmed, B.; Shapor, S.H.; Iqbal, M.S.; Raheel, M. Towards classifying epileptic seizures using entropy variants. In Proceedings of the 2019 IEEE Fifth International Conference on Big Data Computing Service and Applications (BigDataService), Newark, CA, USA, 4–9 April 2019; pp. 296–300. [Google Scholar]
- Bark, B.; Nam, B.; Kim, I.Y. SelANet: Decision-assisting selective sleep apnea detection based on confidence score. BMC Med. Inform. Decis. Mak. 2023, 23, 190. [Google Scholar] [CrossRef] [PubMed]
- Ghassemi, M.M.; Moody, B.E.; Lehman, L.W.H.; Song, C.; Li, Q.; Sun, H.; Mark, R.G.; Westover, M.B.; Clifford, G.D. You snooze, you win: The physionet/computing in cardiology challenge 2018. In Proceedings of the 2018 Computing in Cardiology Conference (CinC), Maastricht, The Netherlands, 23–26 September 2018; Volume 45, pp. 1–4. [Google Scholar]
- Srinivasan, S.; Dayalane, S.; Mathivanan, S.k.; Rajadurai, H.; Jayagopal, P.; Dalu, G.T. Detection and classification of adult epilepsy using hybrid deep learning approach. Sci. Rep. 2023, 13, 17574. [Google Scholar] [CrossRef] [PubMed]
- Miltiadous, A.; Gionanidis, E.; Tzimourta, K.D.; Giannakeas, N.; Tzallas, A.T. DICE-Net: A Novel Convolution-Transformer Architecture for Alzheimer Detection in EEG Signals. IEEE Access 2023, 11, 71840–71858. [Google Scholar] [CrossRef]
- Miltiadous, A.; Tzimourta, K.D.; Afrantou, T.; Ioannidis, P.; Grigoriadis, N.; Tsalikakis, D.G.; Angelidis, P.; Tsipouras, M.G.; Glavas, E.; Giannakeas, N.; et al. A Dataset of Scalp EEG Recordings of Alzheimer’s Disease, Frontotemporal Dementia and Healthy Subjects from Routine EEG. Data 2023, 8, 95. [Google Scholar] [CrossRef]
- Rukhsar, S.; Tiwari, A.K. Lightweight convolution transformer for cross-patient seizure detection in multi-channel EEG signals. Comput. Methods Programs Biomed. 2023, 242, 107856. [Google Scholar] [CrossRef]
- Shoeb, A.; Edwards, H.; Connolly, J.; Bourgeois, B.; Treves, S.T.; Guttag, J. Patient-specific seizure onset detection. Epilepsy Behav. 2004, 5, 483–498. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, Y.; Pan, Y.; Xu, P.; Guan, C. A transformer-based deep neural network model for SSVEP classification. Neural Netw. 2023, 164, 521–534. [Google Scholar] [CrossRef]
- Nakanishi, M.; Wang, Y.; Wang, Y.T.; Jung, T.P. A comparison study of canonical correlation analysis based methods for detecting steady-state visual evoked potentials. PLoS ONE 2015, 10, e0140703. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, X.; Gao, X.; Gao, S. A benchmark dataset for SSVEP-based brain–computer interfaces. IEEE Trans. Neural Syst. Rehabil. Eng. 2016, 25, 1746–1752. [Google Scholar] [CrossRef]
- Lih, O.S.; Jahmunah, V.; Palmer, E.E.; Barua, P.D.; Dogan, S.; Tuncer, T.; Garcia, S.; Molinari, F.; Acharya, U.R. EpilepsyNet: Novel automated detection of epilepsy using transformer model with EEG signals from 121 patient population. Comput. Biol. Med. 2023, 164, 107312. [Google Scholar] [CrossRef] [PubMed]
- Tasci, I.; Tasci, B.; Barua, P.D.; Dogan, S.; Tuncer, T.; Palmer, E.E.; Fujita, H.; Acharya, U.R. Epilepsy detection in 121 patient populations using hypercube pattern from EEG signals. Inf. Fusion 2023, 96, 252–268. [Google Scholar] [CrossRef]
- Huang, X.; Shirahama, K.; Irshad, M.T.; Nisar, M.A.; Piet, A.; Grzegorzek, M. Sleep stage classification in children using self-attention and Gaussian noise data augmentation. Sensors 2023, 23, 3446. [Google Scholar] [CrossRef]
- Huang, X.; Shirahama, K.; Li, F.; Grzegorzek, M. Sleep stage classification for child patients using DeConvolutional Neural Network. Artif. Intell. Med. 2020, 110, 101981. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Liu, G.; Dong, X.; Li, C.; Li, H.; Cui, H.; Zhou, W. Automatic Seizure Detection Based on Stockwell Transform and Transformer. Sensors 2023, 24, 77. [Google Scholar] [CrossRef] [PubMed]
- Giannakakis, G.; Grigoriadis, D.; Giannakaki, K.; Simantiraki, O.; Roniotis, A.; Tsiknakis, M. Review on psychological stress detection using biosignals. IEEE Trans. Affect. Comput. 2019, 13, 440–460. [Google Scholar] [CrossRef]
- Healey, J.A.; Picard, R.W. Detecting stress during real-world driving tasks using physiological sensors. IEEE Trans. Intell. Transp. Syst. 2005, 6, 156–166. [Google Scholar] [CrossRef]
- Ganapathy, N.; Veeranki, Y.R.; Swaminathan, R. Convolutional neural network based emotion classification using electrodermal activity signals and time-frequency features. Expert Syst. Appl. 2020, 159, 113571. [Google Scholar] [CrossRef]
- Koelstra, S.; Muhl, C.; Soleymani, M.; Lee, J.S.; Yazdani, A.; Ebrahimi, T.; Pun, T.; Nijholt, A.; Patras, I. Deap: A database for emotion analysis; using physiological signals. IEEE Trans. Affect. Comput. 2011, 3, 18–31. [Google Scholar] [CrossRef]
- Pinzon-Arenas, J.O.; Kong, Y.; Chon, K.H.; Posada-Quintero, H.F. Design and Evaluation of Deep Learning Models for Continuous Acute Pain Detection Based on Phasic Electrodermal Activity. IEEE J. Biomed. Health Inform. 2023, 27, 4250–4260. [Google Scholar] [CrossRef]
- Posada-Quintero, H.F.; Kong, Y.; Nguyen, K.; Tran, C.; Beardslee, L.; Chen, L.; Guo, T.; Cong, X.; Feng, B.; Chon, K.H. Using electrodermal activity to validate multilevel pain stimulation in healthy volunteers evoked by thermal grills. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2020, 319, R366–R375. [Google Scholar] [CrossRef] [PubMed]
- Islam, T.; Washington, P. Individualized Stress Mobile Sensing Using Self-Supervised Pre-Training. Appl. Sci. 2023, 13, 12035. [Google Scholar] [CrossRef]
- Schmidt, P.; Reiss, A.; Duerichen, R.; Marberger, C.; Van Laerhoven, K. Introducing wesad, a multimodal dataset for wearable stress and affect detection. In Proceedings of the 20th ACM International Conference on Multimodal Interaction, Boulder, CO, USA, 16–20 October 2018; pp. 400–408. [Google Scholar]
- Eom, S.; Eom, S.; Washington, P. SIM-CNN: Self-Supervised Individualized Multimodal Learning for Stress Prediction on Nurses Using Biosignals. In Workshop on Machine Learning for Multimodal Healthcare Data, Proceedings of the First International Workshop, ML4MHD 2023, Honolulu, HI, USA, 29 July 2023; Springer: Cham, Switzerland, 2023; pp. 155–171. [Google Scholar]
- Hosseini, S.; Gottumukkala, R.; Katragadda, S.; Bhupatiraju, R.T.; Ashkar, Z.; Borst, C.W.; Cochran, K. A multimodal sensor dataset for continuous stress detection of nurses in a hospital. Sci. Data 2022, 9, 255. [Google Scholar] [CrossRef]
- Gouverneur, P.J.; Li, F.; M. Szikszay, T.; M. Adamczyk, W.; Luedtke, K.; Grzegorzek, M. Classification of Heat-Induced Pain Using Physiological Signals. In Information Technology in Biomedicine; Springer: Berlin/Heidelberg, Germany, 2021; pp. 239–251. [Google Scholar]
- Qu, C.; Che, X.; Ma, S.; Zhu, S. Bio-physiological-signals-based vr cybersickness detection. CCF Trans. Pervasive Comput. Interact. 2022, 4, 268–284. [Google Scholar] [CrossRef]
- Kim, J.; Kim, W.; Oh, H.; Lee, S.; Lee, S. A deep cybersickness predictor based on brain signal analysis for virtual reality contents. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 10580–10589. [Google Scholar]
- Pouromran, F.; Lin, Y.; Kamarthi, S. Personalized Deep Bi-LSTM RNN based model for pain intensity classification using EDA signal. Sensors 2022, 22, 8087. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Xiao, Y.; Wang, L.; Zhu, W.; Schreiber, K.L. Experimental exploration of objective human pain assessment using multimodal sensing signals. Front. Neurosci. 2022, 16, 831627. [Google Scholar] [CrossRef]
- Liaqat, S.; Dashtipour, K.; Rizwan, A.; Usman, M.; Shah, S.A.; Arshad, K.; Assaleh, K.; Ramzan, N. Personalized wearable electrodermal sensing-based human skin hydration level detection for sports, health and wellbeing. Sci. Rep. 2022, 12, 3715. [Google Scholar] [CrossRef]
- Da Silva, H.P.; Guerreiro, J.; Lourenço, A.; Fred, A.; Martins, R. BITalino: A novel hardware framework for physiological computing. In Proceedings of the International Conference on Physiological Computing Systems, Lisbon, Portugal, 7–9 January 2014; SciTePress: Setúbal, Portugal, 2014; Volume 2, pp. 246–253. [Google Scholar]
- Kuttala, R.; Subramanian, R.; Oruganti, V.R.M. Hierarchical Autoencoder Frequency Features for Stress Detection. IEEE Access 2023, 11, 103232–103241. [Google Scholar] [CrossRef]
- Abadi, M.K.; Subramanian, R.; Kia, S.M.; Avesani, P.; Patras, I.; Sebe, N. DECAF: MEG-based multimodal database for decoding affective physiological responses. IEEE Trans. Affect. Comput. 2015, 6, 209–222. [Google Scholar] [CrossRef]
- Markova, V.; Ganchev, T.; Kalinkov, K. Clas: A database for cognitive load, affect and stress recognition. In Proceedings of the 2019 International Conference on Biomedical Innovations and Applications (BIA), Varna, Bulgaria, 8–9 November 2019; pp. 1–4. [Google Scholar]
- Beh, W.K.; Wu, Y.H. MAUS: A dataset for mental workload assessmenton N-back task using wearable sensor. arXiv 2021, arXiv:2111.02561. [Google Scholar]
- Albuquerque, I.; Tiwari, A.; Parent, M.; Cassani, R.; Gagnon, J.F.; Lafond, D.; Tremblay, S.; Falk, T.H. Wauc: A multi-modal database for mental workload assessment under physical activity. Front. Neurosci. 2020, 14, 549524. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; El Atrache, R.; Tang, J.; Jackson, M.; Makarucha, A.; Cantley, S.; Sheehan, T.; Vieluf, S.; Zhang, B.; Rogers, J.L.; et al. Artificial intelligence-enhanced epileptic seizure detection by wearables. Epilepsia 2023, 64, 3213–3226. [Google Scholar] [CrossRef]
- Li, Y.; Li, K.; Chen, J.; Wang, S.; Lu, H.; Wen, D. Pilot Stress Detection Through Physiological Signals Using a Transformer-Based Deep Learning Model. IEEE Sens. J. 2023, 23, 11774–11784. [Google Scholar] [CrossRef]
- Wang, H.; Naghavi, M.; Allen, C.; Barber, R.; Bhutta, Z.; Carter, A. A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1459–1544. [Google Scholar] [CrossRef]
- Lilly, L.S. Pathophysiology of Heart Disease: A Collaborative Project of Medical Students and Faculty; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2012. [Google Scholar]
- George, S.; Rodriguez, I.; Ipe, D.; Sager, P.T.; Gussak, I.; Vajdic, B. Computerized extraction of electrocardiograms from continuous 12-lead holter recordings reduces measurement variability in a thorough QT study. J. Clin. Pharmacol. 2012, 52, 1891–1900. [Google Scholar] [CrossRef]
- Benali, R.; Bereksi Reguig, F.; Hadj Slimane, Z. Automatic classification of heartbeats using wavelet neural network. J. Med. Syst. 2012, 36, 883–892. [Google Scholar] [CrossRef]
- Huang, J.; Chen, B.; Yao, B.; He, W. ECG Arrhythmia Classification Using STFT-Based Spectrogram and Convolutional Neural Network. IEEE Access 2019, 7, 92871–92880. [Google Scholar] [CrossRef]
- Moody, G.B.; Mark, R.G. The impact of the MIT-BIH arrhythmia database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef]
- Yao, Q.; Wang, R.; Fan, X.; Liu, J.; Li, Y. Multi-class Arrhythmia detection from 12-lead varied-length ECG using Attention-based Time-Incremental Convolutional Neural Network. Inf. Fusion 2020, 53, 174–182. [Google Scholar] [CrossRef]
- Jahmunah, V.; Ng, E.; Tan, R.S.; Oh, S.L.; Acharya, U.R. Explainable detection of myocardial infarction using deep learning models with Grad-CAM technique on ECG signals. Comput. Biol. Med. 2022, 146, 105550. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Fujita, H.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adam, M. Application of deep convolutional neural network for automated detection of myocardial infarction using ECG signals. Inf. Sci. 2017, 415, 190–198. [Google Scholar] [CrossRef]
- Andersen, R.S.; Peimankar, A.; Puthusserypady, S. A deep learning approach for real-time detection of atrial fibrillation. Expert Syst. Appl. 2019, 115, 465–473. [Google Scholar] [CrossRef]
- Colloca, R.; Johnson, A.E.; Mainardi, L.; Clifford, G.D. A Support Vector Machine approach for reliable detection of atrial fibrillation events. In Proceedings of the Computing in Cardiology 2013, Zaragoza, Spain, 22–25 September 2013; pp. 1047–1050. [Google Scholar]
- Darmawahyuni, A.; Nurmaini, S.; Rachmatullah, M.N.; Tutuko, B.; Sapitri, A.I.; Firdaus, F.; Fansyuri, A.; Predyansyah, A. Deep learning-based electrocardiogram rhythm and beat features for heart abnormality classification. PeerJ Comput. Sci. 2022, 8, e825. [Google Scholar] [CrossRef] [PubMed]
- Jahan, M.S.; Mansourvar, M.; Puthusserypady, S.; Wiil, U.K.; Peimankar, A. Short-term atrial fibrillation detection using electrocardiograms: A comparison of machine learning approaches. Int. J. Med. Inform. 2022, 163, 104790. [Google Scholar] [CrossRef] [PubMed]
- Moody, G. A new method for detecting atrial fibrillation using RR intervals. Proc. Comput. Cardiol. 1983, 10, 227–230. [Google Scholar]
- Ehrlich, F.; Bender, J.; Malberg, H.; Goldammer, M. Automatic Sleep Arousal Detection Using Heart Rate From a Single-Lead Electrocardiogram. In Proceedings of the 2022 Computing in Cardiology (CinC), Tampere, Finland, 4–7 September 2022; Volume 498, pp. 1–4. [Google Scholar]
- Quan, S.F.; Howard, B.V.; Iber, C.; Kiley, J.P.; Nieto, F.J.; O’Connor, G.T.; Rapoport, D.M.; Redline, S.; Robbins, J.; Samet, J.M.; et al. The sleep heart health study: Design, rationale, and methods. Sleep 1997, 20, 1077–1085. [Google Scholar]
- Liu, Y.; Liu, J.; Qin, C.; Jin, Y.; Li, Z.; Zhao, L.; Liu, C. A deep learning-based acute coronary syndrome-related disease classification method: A cohort study for network interpretability and transfer learning. Appl. Intell. 2023, 53, 25562–25580. [Google Scholar] [CrossRef]
- Topalidis, P.I.; Baron, S.; Heib, D.P.; Eigl, E.S.; Hinterberger, A.; Schabus, M. From Pulses to Sleep Stages: Towards Optimized Sleep Classification Using Heart-Rate Variability. Sensors 2023, 23, 9077. [Google Scholar] [CrossRef] [PubMed]
- Loh, H.W.; Ooi, C.P.; Oh, S.L.; Barua, P.D.; Tan, Y.R.; Molinari, F.; March, S.; Acharya, U.R.; Fung, D.S.S. Deep neural network technique for automated detection of ADHD and CD using ECG signal. Comput. Methods Programs Biomed. 2023, 241, 107775. [Google Scholar] [CrossRef] [PubMed]
- Donati, M.; Olivelli, M.; Giovannini, R.; Fanucci, L. ECG-Based Stress Detection and Productivity Factors Monitoring: The Real-Time Production Factory System. Sensors 2023, 23, 5502. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.P.; Freed, B.C.; Walter, D.P.; Perry, J.C.; Barakat, A.F.; Elashery, A.R.A.; Shah, K.S.; Kutty, S.; McGillion, M.; Ng, F.S.; et al. Convolution neural network algorithm for shockable arrhythmia classification within a digitally connected automated external defibrillator. J. Am. Heart Assoc. 2023, 12, e026974. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Lee, G.; Kim, S.H. An ECG stitching scheme for driver arrhythmia classification based on deep learning. Sensors 2023, 23, 3257. [Google Scholar] [CrossRef] [PubMed]
- Yoon, T.; Kang, D. Bimodal CNN for cardiovascular disease classification by co-training ECG grayscale images and scalograms. Sci. Rep. 2023, 13, 2937. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Mallik, A.; Kumar, A.; Del Ser, J.; Yang, G. Fuzz-ClustNet: Coupled fuzzy clustering and deep neural networks for Arrhythmia detection from ECG signals. Comput. Biol. Med. 2023, 153, 106511. [Google Scholar] [CrossRef] [PubMed]
- Bousseljot, R.; Kreiseler, D.; Schnabel, A. Nutzung der EKG-Signaldatenbank CARDIODAT der PTB über das Internet; De Gruyter: Berlin, Germany, 1995. [Google Scholar]
- Farag, M.M. A tiny matched filter-based cnn for inter-patient ecg classification and arrhythmia detection at the edge. Sensors 2023, 23, 1365. [Google Scholar] [CrossRef]
- Pokaprakarn, T.; Kitzmiller, R.R.; Moorman, R.; Lake, D.E.; Krishnamurthy, A.K.; Kosorok, M.R. Sequence to Sequence ECG Cardiac Rhythm Classification Using Convolutional Recurrent Neural Networks. IEEE J. Biomed. Health Inform. 2022, 26, 572–580. [Google Scholar] [CrossRef]
- Moss, T.J.; Lake, D.E.; Moorman, J.R. Local dynamics of heart rate: Detection and prognostic implications. Physiol. Meas. 2014, 35, 1929. [Google Scholar] [CrossRef]
- Yeh, C.Y.; Chang, H.Y.; Hu, J.Y.; Lin, C.C. Contribution of different subbands of ECG in sleep apnea detection evaluated using filter bank decomposition and a convolutional neural network. Sensors 2022, 22, 510. [Google Scholar] [CrossRef] [PubMed]
- Penzel, T.; Moody, G.B.; Mark, R.G.; Goldberger, A.L.; Peter, J.H. The apnea-ECG database. In Proceedings of the Computers in Cardiology 2000. Vol. 27 (Cat. 00CH37163), Cambridge, MA, USA, 24–27 September 2000; pp. 255–258. [Google Scholar]
- Yildirim, O.; Baloglu, U.B.; Tan, R.S.; Ciaccio, E.J.; Acharya, U.R. A new approach for arrhythmia classification using deep coded features and LSTM networks. Comput. Methods Programs Biomed. 2019, 176, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, O. A novel wavelet sequence based on deep bidirectional LSTM network model for ECG signal classification. Comput. Biol. Med. 2018, 96, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.J.; Tang, F.Q.; Cai, H.P.; Qian, Y.F. Heart failure classification using deep learning to extract spatiotemporal features from ECG. BMC Med. Inform. Decis. Mak. 2024, 24, 17. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.E.; Pollard, T.J.; Shen, L.; Lehman, L.w.H.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Anthony Celi, L.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Ahmed, J. Cloud-based healthcare framework for real-time anomaly detection and classification of 1-D ECG signals. PLoS ONE 2022, 17, e0279305. [Google Scholar] [CrossRef]
- De Marco, F.; Ferrucci, F.; Risi, M.; Tortora, G. Classification of QRS complexes to detect Premature Ventricular Contraction using machine learning techniques. PLoS ONE 2022, 17, e0268555. [Google Scholar] [CrossRef]
- Kumar, D.; Peimankar, A.; Sharma, K.; Domínguez, H.; Puthusserypady, S.; Bardram, J.E. Deepaware: A hybrid deep learning and context-aware heuristics-based model for atrial fibrillation detection. Comput. Methods Programs Biomed. 2022, 221, 106899. [Google Scholar] [CrossRef]
- Xia, Y.; Zhang, H.; Xu, L.; Gao, Z.; Zhang, H.; Liu, H.; Li, S. An Automatic Cardiac Arrhythmia Classification System with Wearable Electrocardiogram. IEEE Access 2018, 6, 16529–16538. [Google Scholar] [CrossRef]
- Belkadi, M.A.; Daamouche, A.; Melgani, F. A deep neural network approach to QRS detection using autoencoders. Expert Syst. Appl. 2021, 184, 115528. [Google Scholar] [CrossRef]
- Sugimoto, K.; Kon, Y.; Lee, S.; Okada, Y. Detection and localization of myocardial infarction based on a convolutional autoencoder. Knowl.-Based Syst. 2019, 178, 123–131. [Google Scholar] [CrossRef]
- National Institute of General Medical Sciences and National Institute of Biomedical Imaging and Bioengineering. PhysioBank. 2018. Available online: https://physionet.org/physiobank/ (accessed on 1 June 2018).
- Siouda, R.; Nemissi, M.; Seridi, H. ECG beat classification using neural classifier based on deep autoencoder and decomposition techniques. Prog. Artif. Intell. 2021, 10, 333–347. [Google Scholar] [CrossRef]
- Thill, M.; Konen, W.; Wang, H.; Back, T. Temporal convolutional autoencoder for unsupervised anomaly detection in time series. Appl. Soft Comput. 2021, 112, 107751. [Google Scholar] [CrossRef]
- Arslan, N.N.; Ozdemir, D.; Temurtas, H. ECG heartbeats classification with dilated convolutional autoencoder. Signal Image Video Process. 2024, 18, 417–426. [Google Scholar] [CrossRef]
- Moody, G.B.; Mark, R.G.; Goldberger, A.L. PhysioNet: A web-based resource for the study of physiologic signals. IEEE Eng. Med. Biol. Mag. 2001, 20, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Sharma, A. Attention-based convolutional denoising autoencoder for two-lead ECG denoising and arrhythmia classification. IEEE Trans. Instrum. Meas. 2022, 71, 4007710. [Google Scholar] [CrossRef]
- Silva, R.; Fred, A.; Plácido da Silva, H. Morphological autoencoders for beat-by-beat atrial fibrillation detection using single-lead ecg. Sensors 2023, 23, 2854. [Google Scholar] [CrossRef] [PubMed]
- Clifford, G.D.; Liu, C.; Moody, B.; Li-wei, H.L.; Silva, I.; Li, Q.; Johnson, A.; Mark, R.G. AF classification from a short single lead ECG recording: The PhysioNet/computing in cardiology challenge 2017. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017; pp. 1–4. [Google Scholar]
- Tutuko, B.; Darmawahyuni, A.; Nurmaini, S.; Tondas, A.E.; Naufal Rachmatullah, M.; Teguh, S.B.P.; Firdaus, F.; Sapitri, A.I.; Passarella, R. DAE-ConvBiLSTM: End-to-end learning single-lead electrocardiogram signal for heart abnormalities detection. PLoS ONE 2022, 17, e0277932. [Google Scholar] [CrossRef] [PubMed]
- Laguna, P.; Mark, R.G.; Goldberg, A.; Moody, G.B. A database for evaluation of algorithms for measurement of QT and other waveform intervals in the ECG. In Proceedings of the Computers in cardiology 1997, Lund, Sweden, 7–10 September 1997; pp. 673–676. [Google Scholar]
- Che, C.; Zhang, P.; Zhu, M.; Qu, Y.; Jin, B. Constrained transformer network for ECG signal processing and arrhythmia classification. BMC Med. Inform. Decis. Mak. 2021, 21, 184. [Google Scholar] [CrossRef]
- Liu, F.; Liu, C.; Zhao, L.; Zhang, X.; Wu, X.; Xu, X.; Liu, Y.; Ma, C.; Wei, S.; He, Z.; et al. An open access database for evaluating the algorithms of electrocardiogram rhythm and morphology abnormality detection. J. Med. Imaging Health Inform. 2018, 8, 1368–1373. [Google Scholar] [CrossRef]
- Meng, L.; Tan, W.; Ma, J.; Wang, R.; Yin, X.; Zhang, Y. Enhancing dynamic ECG heartbeat classification with lightweight transformer model. Artif. Intell. Med. 2022, 124, 102236. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Liu, C.; Gao, H.; Wang, X.; Zhao, L.; Shen, Q.; Ng, E.; Li, J. An open-access long-term wearable ECG database for premature ventricular contractions and supraventricular premature beat detection. J. Med. Imaging Health Inform. 2020, 10, 2663–2667. [Google Scholar] [CrossRef]
- Hu, R.; Chen, J.; Zhou, L. A transformer-based deep neural network for arrhythmia detection using continuous ECG signals. Comput. Biol. Med. 2022, 144, 105325. [Google Scholar] [CrossRef] [PubMed]
- Akan, T.; Alp, S.; Bhuiyan, M.A.N. ECGformer: Leveraging transformer for ECG heartbeat arrhythmia classification. arXiv 2024, arXiv:2401.05434. [Google Scholar]
- Zou, C.; Djajapermana, M.; Martens, E.; Müller, A.; Rückert, D.; Müller, P.; Steger, A.; Becker, M.; Wolfgang, U. DWT-CNNTRN: A Convolutional Transformer for ECG Classification with Discrete Wavelet Transform. In Proceedings of the 2023 45th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Sydney, Australia, 24–27 July 2023; pp. 1–6. [Google Scholar]
- Alshareef, M.S.; Alturki, B.; Jaber, M. A transformer-based model for effective and exportable IoMT-based stress detection. In Proceedings of the GLOBECOM 2022-2022 IEEE Global Communications Conference, Rio de Janeiro, Brazil, 4–8 December 2022; pp. 1158–1163. [Google Scholar] [CrossRef]
- Lu, P.; Creagh, A.P.; Lu, H.Y.; Hai, H.B.; Consortium, V.; Thwaites, L.; Clifton, D.A. 2D-WinSpatt-Net: A Dual Spatial Self-Attention Vision Transformer Boosts Classification of Tetanus Severity for Patients Wearing ECG Sensors in Low-and Middle-Income Countries. Sensors 2023, 23, 7705. [Google Scholar] [CrossRef] [PubMed]
- Van, H.M.T.; Van Hao, N.; Quoc, K.P.N.; Hai, H.B.; Yen, L.M.; Nhat, P.T.H.; Duong, H.T.H.; Thuy, D.B.; Zhu, T.; Greeff, H.; et al. Vital sign monitoring using wearable devices in a Vietnamese intensive care unit. BMJ Innov. 2021, 7. [Google Scholar] [CrossRef]
- Liu, T.; Si, Y.; Yang, W.; Huang, J.; Yu, Y.; Zhang, G.; Zhou, R. Inter-patient congestive heart failure detection using ECG-convolution-vision transformer network. Sensors 2022, 22, 3283. [Google Scholar] [CrossRef]
- Baim, D.S.; Colucci, W.S.; Monrad, E.S.; Smith, H.S.; Wright, R.F.; Lanoue, A.; Gauthier, D.F.; Ransil, B.J.; Grossman, W.; Braunwald, E. Survival of patients with severe congestive heart failure treated with oral milrinone. J. Am. Coll. Cardiol. 1986, 7, 661–670. [Google Scholar] [CrossRef]
- Chi, W.N.; Reamer, C.; Gordon, R.; Sarswat, N.; Gupta, C.; White VanGompel, E.; Dayiantis, J.; Morton-Jost, M.; Ravichandran, U.; Larimer, K.; et al. Continuous Remote Patient Monitoring: Evaluation of the Heart Failure Cascade Soft Launch. Appl. Clin. Inform. 2021, 12, 1161–1173. [Google Scholar] [CrossRef]
- Idrees, A.K.; Idrees, S.K.; Ali-Yahiya, T.; Couturier, R. Multibiosensor Data Sampling and Transmission Reduction With Decision-Making for Remote Patient Monitoring in IoMT Networks. IEEE Sens. J. 2023, 23, 15140–15152. [Google Scholar] [CrossRef]
- Idrees, A.K.; Abd Alhussein, D.; Harb, H. Energy-efficient multisensor adaptive sampling and aggregation for patient monitoring in edge computing based IoHT networks. J. Ambient. Intell. Smart Environ. 2023, 15, 235–253. [Google Scholar] [CrossRef]
- Alam, M.G.R.; Haw, R.; Kim, S.S.; Azad, M.A.K.; Abedin, S.F.; Hong, C.S. EM-Psychiatry: An Ambient Intelligent System for Psychiatric Emergency. IEEE Trans. Ind. Inform. 2016, 12, 2321–2330. [Google Scholar] [CrossRef]
- Ganti, V.G.; Gazi, A.H.; An, S.; Srivatsa, V.A.; Nevius, B.N.; Nichols, C.J.; Carek, A.M.; Fares, M.; Abdulkarim, M.; Hussain, T.; et al. Wearable Seismocardiography-Based Assessment of Stroke Volume in Congenital Heart Disease. J. Am. Heart Assoc. 2022, 11, e026067. [Google Scholar] [CrossRef] [PubMed]
- Shaik, T.; Tao, X.; Higgins, N.; Li, L.; Gururajan, R.; Zhou, X.; Acharya, U.R. Remote patient monitoring using artificial intelligence: Current state, applications, and challenges. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2023, 13, e1485. [Google Scholar] [CrossRef]
- Larimer, K.; Wegerich, S.; Splan, J.; Chestek, D.; Prendergast, H.; Vanden Hoek, T. Personalized Analytics and a Wearable Biosensor Platform for Early Detection of COVID-19 Decompensation (DeCODe): Protocol for the Development of the COVID-19 Decompensation Index. JMIR Res. Protoc. 2021, 10, e27271. [Google Scholar] [CrossRef] [PubMed]
- Annis, T.; Pleasants, S.; Hultman, G.; Lindemann, E.; Thompson, J.A.; Billecke, S.; Badlani, S.; Melton, G.B. Rapid implementation of a COVID-19 remote patient monitoring program. J. Am. Med. Inform. Assoc. 2020, 27, 1326–1330. [Google Scholar] [CrossRef] [PubMed]
- Catarinucci, L.; de Donno, D.; Mainetti, L.; Palano, L.; Patrono, L.; Stefanizzi, M.L.; Tarricone, L. An IoT-Aware Architecture for Smart Healthcare Systems. IEEE Internet Things J. 2015, 2, 515–526. [Google Scholar] [CrossRef]
- Alizadeh, M.; Shaker, G.; Martins De Almeida, J.C.; Morita, P.P.; Safavi-Naeini, S. Remote Monitoring of Human Vital Signs Using mm-Wave FMCW Radar. IEEE Access 2019, 7, 54958–54968. [Google Scholar] [CrossRef]
- Kenaan, A.; Li, K.; Barth, I.; Johnson, S.; Song, J.; Krauss, T.F. Guided mode resonance sensor for the parallel detection of multiple protein biomarkers in human urine with high sensitivity. Biosens. Bioelectron. 2020, 153, 112047. [Google Scholar] [CrossRef]
- Ben Halima, H.; Bellagambi, F.G.; Hangouet, M.; Alcacer, A.; Pfeiffer, N.; Heuberger, A.; Zine, N.; Bausells, J.; Elaissari, A.; Errachid, A. A novel electrochemical strategy for NT-proBNP detection using IMFET for monitoring heart failure by saliva analysis. Talanta 2023, 251, 123759. [Google Scholar] [CrossRef]
- Amin, M.; Abdullah, B.M.M.; Wylie, S.R.R.; Rowley-Neale, S.J.J.; Banks, C.E.E.; Whitehead, K.A.A. The Voltammetric Detection of Cadaverine Using a Diamine Oxidase and Multi-Walled Carbon Nanotube Functionalised Electrochemical Biosensor. Nanomaterials 2023, 13, 36. [Google Scholar] [CrossRef] [PubMed]
- Goswami, P.P.; Deshpande, T.; Rotake, D.R.; Singh, S.G. Near perfect classification of cardiac biomarker Troponin-I in human serum assisted by SnS2-CNT composite, explainable ML, and operating-voltage-selection-algorithm. Biosens. Bioelectron. 2023, 220, 114915. [Google Scholar] [CrossRef] [PubMed]
- So, S.; Khalaf, A.; Yi, X.; Herring, C.; Zhang, Y.; Simon, M.A.; Akcakaya, M.; Lee, S.; Yun, M. Induced bioresistance via BNP detection for machine learning-based risk assessment. Biosens. Bioelectron. 2021, 175, 112903. [Google Scholar] [CrossRef] [PubMed]
- Samman, N.; El-Boubbou, K.; Al-Muhalhil, K.; Ali, R.; Alaskar, A.; Alharbi, N.K.; Nehdi, A. MICaFVi: A Novel Magnetic Immuno-Capture Flow Virometry Nano-Based Diagnostic Tool for Detection of Coronaviruses. Biosensors 2023, 13, 553. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Towers, D.; Myers, S.; Galvin, C.; Kireev, D.; Ellington, A.D.; Akinwande, D. Graphene Field Effect Biosensor for Concurrent and Specific Detection of SARS-CoV-2 and Influenza. ACS Nano 2023, 17, 18629–18640. [Google Scholar] [CrossRef] [PubMed]
- Le Brun, G.; Hauwaert, M.; Leprince, A.; Glinel, K.; Mahillon, J.; Raskin, J.P. Electrical Characterization of Cellulose-Based Membranes towards Pathogen Detection in Water. Biosensors 2021, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, X.; Guo, J.; Zhang, Y.; Guo, J.; Wu, X.; Wang, B.; Ma, X. Simultaneous Detection of Inflammatory Biomarkers by SERS Nanotag-Based Lateral Flow Assay with Portable Cloud Raman Spectrometer. Nanomaterials 2021, 11, 1496. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hu, Y.; Jiang, N.; Yetisen, A.K. Wearable artificial intelligence biosensor networks. Biosens. Bioelectron. 2023, 219, 114825. [Google Scholar] [CrossRef]
- Kim, S.Y.; Lee, J.C.; Seo, G.; Woo, J.H.; Lee, M.; Nam, J.; Sim, J.Y.; Kim, H.R.; Park, E.C.; Park, S. Computational Method-Based Optimization of Carbon Nanotube Thin-Film Immunosensor for Rapid Detection of SARS-CoV-2 Virus. Small Sci. 2022, 2, 2100111. [Google Scholar] [CrossRef]
- Krokidis, M.G.; Dimitrakopoulos, G.N.; Vrahatis, A.G.; Tzouvelekis, C.; Drakoulis, D.; Papavassileiou, F.; Exarchos, T.P.; Vlamos, P. A Sensor-Based Perspective in Early-Stage Parkinson’s Disease: Current State and the Need for Machine Learning Processes. Sensors 2022, 22, 409. [Google Scholar] [CrossRef]
- Soundararajan, R.; Prabu, A.V.; Routray, S.; Malla, P.P.; Ray, A.K.; Palai, G.; Faragallah, O.S.; Baz, M.; Abualnaja, M.M.; Eid, M.M.A.; et al. Deeply Trained Real-Time Body Sensor Networks for Analyzing the Symptoms of Parkinson’s Disease. IEEE Access 2022, 10, 63403–63421. [Google Scholar] [CrossRef]
- Wang, J.B.; Cadmus-Bertram, L.A.; Natarajan, L.; White, M.M.; Madanat, H.; Nichols, J.F.; Ayala, G.X.; Pierce, J.P. Wearable Sensor/Device (Fitbit One) and SMS Text-Messaging Prompts to Increase Physical Activity in Overweight and Obese Adults: A Randomized Controlled Trial. Telemed. E-Health 2015, 21, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Baca-Motes, K.; Edwards, A.M.; Waalen, J.; Edmonds, S.; Mehta, R.R.; Ariniello, L.; Ebner, G.S.; Talantov, D.; Fastenau, J.M.; Carter, C.T.; et al. Digital recruitment and enrollment in a remote nationwide trial of screening for undiagnosed atrial fibrillation: Lessons from the randomized, controlled mSToPS trial. Contemp. Clin. Trials Commun. 2019, 14, 100318. [Google Scholar] [CrossRef] [PubMed]
- Elzinga, W.O.; Prins, S.; Borghans, L.G.J.M.; Gal, P.; Vargas, G.A.; Groeneveld, G.J.; Doll, R.J. Detection of Clenbuterol-Induced Changes in Heart Rate Using At-Home Recorded Smartwatch Data: Randomized Controlled Trial. JMIR Form. Res. 2021, 5, e31890. [Google Scholar] [CrossRef] [PubMed]
- Fascio, E.; Vitale, J.A.; Sirtori, P.; Peretti, G.; Banfi, G.; Mangiavini, L. Early Virtual-Reality-Based Home Rehabilitation after Total Hip Arthroplasty: A Randomized Controlled Trial. J. Clin. Med. 2022, 11, 1766. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, N.; Eisenkraft, A.; Arguello, C.J.; Yang, G.J.; Sand, E.; Ishay, A.B.; Merin, R.; Fons, M.; Littman, R.; Nachman, D.; et al. Exploring Early Pre-Symptomatic Detection of Influenza Using Continuous Monitoring of Advanced Physiological Parameters during a Randomized Controlled Trial. J. Clin. Med. 2021, 10, 5202. [Google Scholar] [CrossRef] [PubMed]
- Gatsios, D.; Antonini, A.; Gentile, G.; Marcante, A.; Pellicano, C.; Macchiusi, L.; Assogna, F.; Spalletta, G.; Gage, H.; Touray, M.; et al. Feasibility and Utility of mHealth for the Remote Monitoring of Parkinson Disease: Ancillary Study of the PD_manager Randomized Controlled Trial. JMIR Mhealth Uhealth 2020, 8, e16414. [Google Scholar] [CrossRef] [PubMed]
- Browne, S.H.; Umlauf, A.; Tucker, A.J.; Low, J.; Moser, K.; Garcia, J.G.; Peloquin, C.A.; Blaschke, T.; Vaida, F.; Benson, C.A. Wirelessly observed therapy compared to directly observed therapy to confirm and support tuberculosis treatment adherence: A randomized controlled trial. PLoS Med. 2019, 16, e1002891. [Google Scholar] [CrossRef]
- Zhang, J.; Kailkhura, B.; Han, T.Y.J. Leveraging uncertainty from deep learning for trustworthy material discovery workflows. ACS Omega 2021, 6, 12711–12721. [Google Scholar] [CrossRef]
- Radin, J.M.; Quer, G.; Jalili, M.; Hamideh, D.; Steinhubl, S.R. The hopes and hazards of using personal health technologies in the diagnosis and prognosis of infections. Lancet Digit. Health 2021, 3, e455–e461. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, J.; Liu, T.; Luo, Y.; Loh, X.J.; Chen, X. Machine learning-reinforced noninvasive biosensors for healthcare. Adv. Healthc. Mater. 2021, 10, 2100734. [Google Scholar] [CrossRef] [PubMed]
- Krittanawong, C.; Rogers, A.J.; Johnson, K.W.; Wang, Z.; Turakhia, M.P.; Halperin, J.L.; Narayan, S.M. Integration of novel monitoring devices with machine learning technology for scalable cardiovascular management. Nat. Rev. Cardiol. 2021, 18, 75–91. [Google Scholar] [CrossRef] [PubMed]
- Hezarjaribi, N.; Fallahzadeh, R.; Ghasemzadeh, H. A machine learning approach for medication adherence monitoring using body-worn sensors. In Proceedings of the 2016 Design, Automation & Test in Europe Conference & Exhibition (DATE), Dresden, Germany, 14–18 March 2016; pp. 842–845. [Google Scholar]
- Jourdan, T.; Debs, N.; Frindel, C. The contribution of machine learning in the validation of commercial wearable sensors for gait monitoring in patients: A systematic review. Sensors 2021, 21, 4808. [Google Scholar] [CrossRef] [PubMed]
- Tucker, C.S.; Behoora, I.; Nembhard, H.B.; Lewis, M.; Sterling, N.W.; Huang, X. Machine learning classification of medication adherence in patients with movement disorders using non-wearable sensors. Comput. Biol. Med. 2015, 66, 120–134. [Google Scholar] [CrossRef]
- Fuest, J.; Tacke, M.; Ullman, L.; Washington, P. Individualized, self-supervised deep learning for blood glucose prediction. medRxiv 2023, 2023-08. [Google Scholar]
- Kargarandehkordi, A.; Washington, P. Personalized Prediction of Stress-Induced Blood Pressure Spikes in Real Time from FitBit Data using Artificial Intelligence: A Research Protocol. medRxiv 2023, 2023-12. [Google Scholar]
- Kargarandehkordi, A.; Washington, P. Computer Vision Estimation of Stress and Anxiety Using a Gamified Mobile-based Ecological Momentary Assessment and Deep Learning: Research Protocol. medRxiv 2023, 2023-04. [Google Scholar]
- Li, J.; Washington, P. A Comparison of Personalized and Generalized Approaches to Emotion Recognition Using Consumer Wearable Devices: Machine Learning Study. arXiv 2023, arXiv:2308.14245. [Google Scholar] [CrossRef]
- Washington, P. Personalized Machine Learning using Passive Sensing and Ecological Momentary Assessments for Meth Users in Hawaii: A Research Protocol. medRxiv 2023, 2023-08. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).