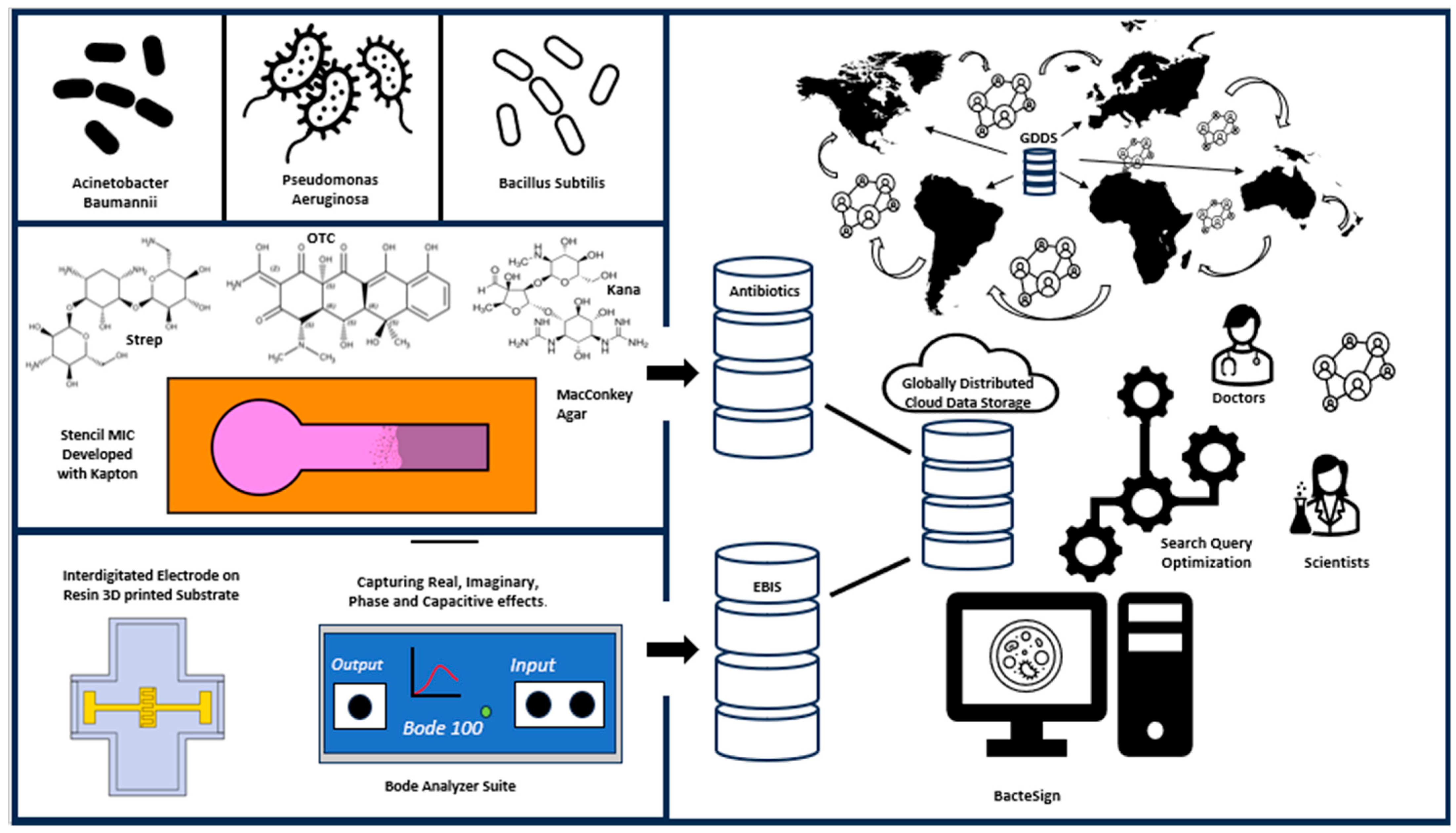
BacteSign: Building a Findable, Accessible, Interoperable, and Reusable (FAIR) Database for Universal Bacterial Identification
Abstract
1. Introduction
2. Materials and Methods
2.1. Methods of Imaging
2.2. Stencil Mask Fabrication for Minimum Inhibitory Concentration Tests
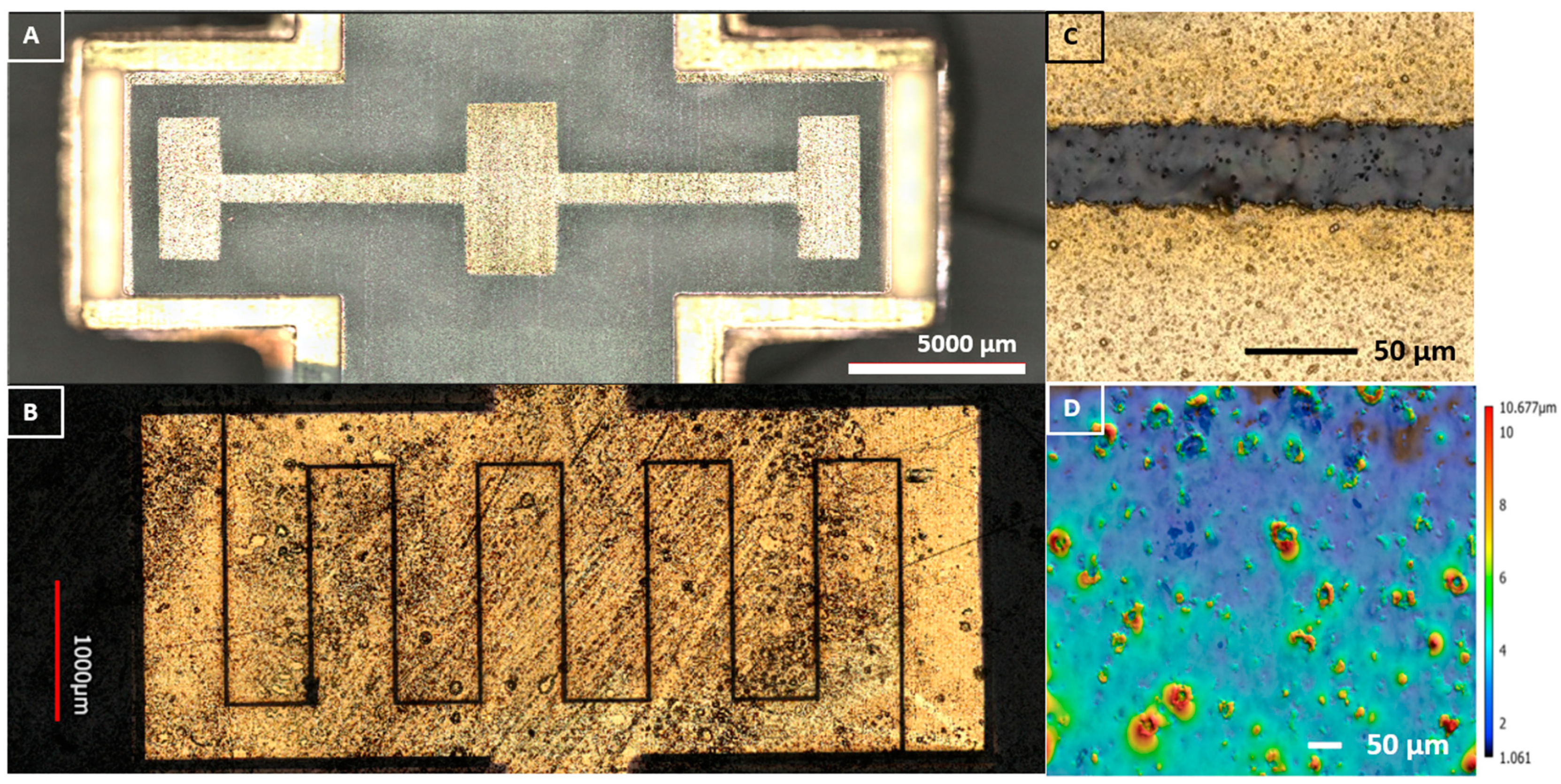
2.3. Laser Interdigitated Electrode (IDE) Substrate on µbiochamber Development
2.4. Bacterial Interdigitated Electrode µbiochamber Assembly
2.5. Bacteria Strain Handling, Growth Conditions, and Media Culture for EBIS
2.6. Impedance Measurements
2.7. Database Building
3. Results
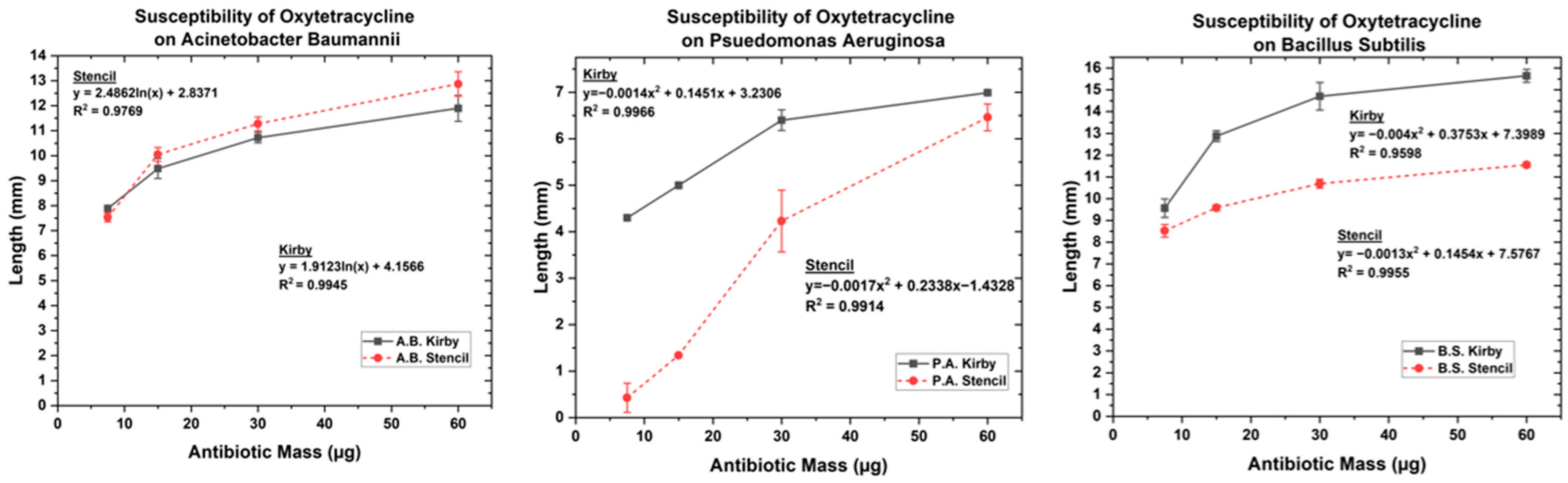
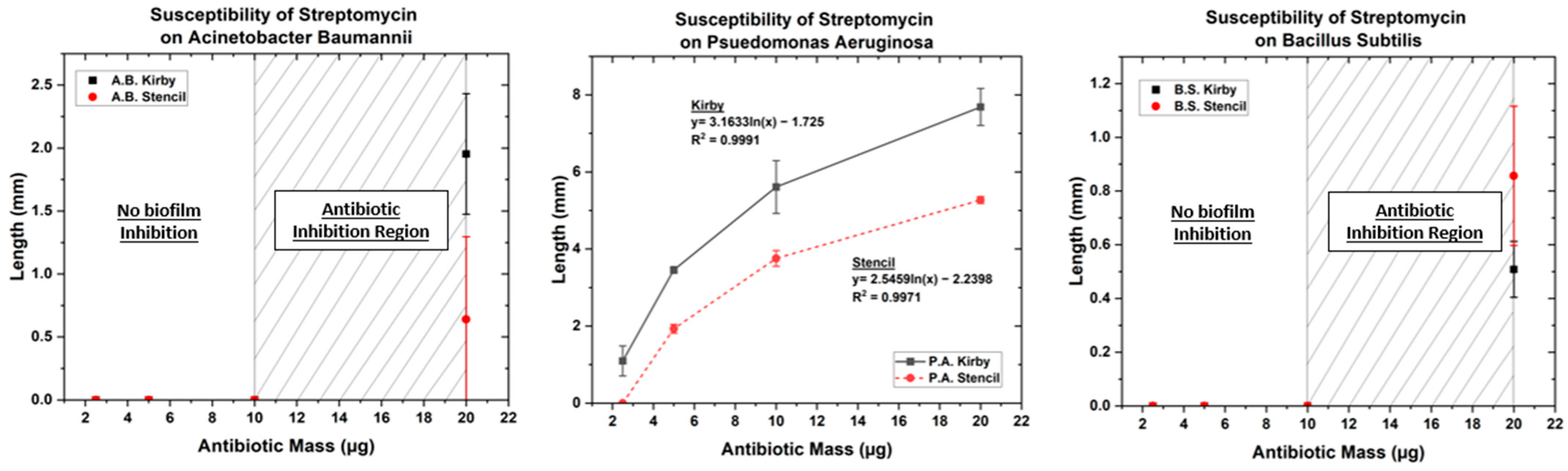
3.1. Optimized Kirby Bauer Stencil Mask
3.2. Electrochemical Biological Impedance Spectroscopy (EBIS)
Characterization of Interdigitated Electrode
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gupta, A.; Landis, R.F.; Rotello, V.M. Nanoparticle-based antimicrobials: Surface functionality is critical. F1000Research 2016, 5, 364. [Google Scholar] [CrossRef] [PubMed]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Healthcare associated infection: Surgical site infections. In ECDC Annual Epidemiological Report for 2018–2020; ECDC: Stockhom, Sweden, 2023. [Google Scholar]
- O’Brien, W.J.; Gupta, K.; Itani, K.M.F. Association of Postoperative Infection with Risk of Long-term Infection and Mortality. JAMA Surg. 2020, 155, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Dobson, G.P. Trauma of major surgery: A global problem that is not going away. Int. J. Surg. 2020, 81, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, S.D.; Knuuti, J.; Saraste, A.; Anker, S.; Bøtker, H.E.; Hert, S.D.; Ford, I.; Gonzalez-Juanatey, J.R.; Gorenek, B.; Heyndrickx, G.R.; et al. 2014 ESC/ESA Guidelines on non-cardiac surgery: Cardiovascular assessment and management: The Joint Task Force on non-cardiac surgery: Cardiovascular assessment and management of the European Society of Cardiology (ESC) and the European Society of Anaesthesiology (ESA). Eur. Heart J. 2014, 35, 2383–2431. [Google Scholar]
- Oliveira, J.; Reygaert, W.C. Gram-Negative Bacteria. StatPearls. 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK538213/ (accessed on 12 November 2023).
- Nikaido, H. Multidrug resistance in bacteria. Annu. Rev. Biochem. 2009, 78, 119–146. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.; Acevedo, J.; Wiest, R.; Gustot, T.; Amoros, A.; Deulofeu, C.; Reverter, E.; Martínez, J.; Saliba, F.; Jalan, R.; et al. Bacterial and fungal infections in acute-on-chronic liver failure: Prevalence, characteristics and impact on prognosis. Gut 2017, 67, 1870–1880. [Google Scholar] [CrossRef] [PubMed]
- Howard, A.; O’Donoghue, M.; Feeney, A.; Sleator, R.D. Acinetobacter baumannii: An emerging opportunistic pathogen . Virulence 2012, 3, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Jiménez, E.; del Campo, R.; Toledano, V.; Vallejo-Cremades, M.T.; Muñoz, A.; Largo, C.; Arnalich, F.; García-Rio, F.; Cubillos-Zapata, C.; López-Collazo, E. Biofilm vs. planktonic bacterial mode of growth: Which do human macrophages prefer? Biochem. Biophys. Res. Commun. 2013, 441, 947–952. [Google Scholar] [CrossRef]
- Melander, R.J.; Melander, C. Innovative strategies for combating biofilm-based infections. In Biofilm-Based Healthcare-Associated Infections; Springer: Cham, Switzerland, 2014; Volume 2, pp. 69–91. [Google Scholar] [CrossRef]
- Lemon, K.P.; Earl, A.M.; Vlamakis, H.C.; Aguilar, C.; Kolter, R. Biofilm development with an emphasis on Bacillus subtilis. Curr. Top. Microbiol. Immunol. 2008, 322, 1–16. [Google Scholar] [CrossRef]
- Römling, U.; Balsalobre, C. Biofilm infections, their resilience to therapy and Innovative Treatment Strategies. J. Intern. Med. 2012, 272, 541–561. [Google Scholar] [CrossRef] [PubMed]
- Lamireau, T.; Martin, S.; Lallier, M.; Marcotte, J.E.; Alvarez, F. Liver transplantation for cirrhosis in cystic fibrosis. Can. J. Gastroenterol. 2006, 20, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Hoyle, B.D.; Costerton, J.W. Bacterial resistance to antibiotics: The role of biofilms. In Progress in Drug Research; Birkhäuser: Basel, Switzerland, 1991; pp. 91–105. [Google Scholar] [CrossRef]
- Mah, T.-F.; Pitts, B.; Pellock, B.; Walker, G.C.; Stewart, P.S.; O’Toole, G.A. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 2003, 426, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Sebeny, P.J.; Riddle, M.S.; Petersen, K. Acinetobacter baumannii skin and soft-tissue infection associated with war trauma. Clin. Infect. Dis. 2008, 47, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Higgins, P.G.; Poirel, L.; Lehmann, M.; Nordmann, P.; Seifert, H. OXA-143, a novel carbapenem-hydrolyzing class D beta-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009, 53, 5035–5038. [Google Scholar] [CrossRef] [PubMed]
- Kanno, E.; Toriyabe, S.; Zhang, L.; Imai, Y.; Tachi, M. Biofilm Formation on rat skin wounds by Pseudomonas aeruginosa carrying the green fluorescent protein gene. Exp. Dermatol. 2010, 19, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.E.; Caiazza, N.C.; O’Toole, G.A. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas Aeruginosa PAO1. J. Bacteriol. 2003, 185, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Branda, S.S.; González-Pastor, J.E.; Ben-Yehuda, S.; Losick, R.; Kolter, R. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2001, 98, 11621–11626. [Google Scholar] [CrossRef] [PubMed]
- Branda, S.S.; Chu, F.; Kearns, D.B.; Losick, R.; Kolter, R. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 2005, 59, 1229–1238. [Google Scholar] [CrossRef]
- World Health Organization. Disease Outbreak News. Available online: https://www.who.int/emergencies/disease-outbreak-news (accessed on 17 November 2023).
- Centers for Disease Control and Prevention. CDC Current Outbreak List. Centers for Disease Control and 693 Prevention. 2023. Available online: https://www.cdc.gov/outbreaks/index.html (accessed on 17 November 2023).
- Huttenhower, C.; Finn, R.D.; McHardy, A.C. Challenges and opportunities in sharing microbiome data and analyses. Nat. Microbiol. 2023, 8, 1960–1970. [Google Scholar] [CrossRef]
- Cochrane, G.; Karsch-Mizrachi, I.; Takagi, T.; International Nucleotide Sequence Database, Collaboration. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2016, 44, D48–D50. [Google Scholar] [CrossRef]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020, 48, D440–D444. [Google Scholar] [CrossRef] [PubMed]
- Vizcaíno, J.A.; Deutsch, E.W.; Wang, R.; Csordas, A.; Reisinger, F.; Ríos, D.; Dianes, J.A.; Sun, Z.; Farrah, T.; Bandeira, N.; et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014, 32, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, I.C.; Allan, J.-M.; Burel, D.; Creager, A.; Falconi, H.; Hochheiser, J.; Johnston, J.; Mellen, P.K.; Sorger Swedlow, J.R. The Open Microscopy Environment (OME) Data Model and XML File: Open Tools for Informatics and Quantitative Analysis in Biological Imaging. Genome Biol. 2005, 6, R47. [Google Scholar] [CrossRef] [PubMed]
- Hartley, M.; Kleywegt, G.J.; Patwardhan, A.; Sarkans, U.; Swedlow, J.R.; Brazma, A. The BioImage Archive—Building a Home for Life—Sciences Microscopy Data. J. Mol. Biol. 2022, 434, 167505. [Google Scholar] [CrossRef] [PubMed]
- Baker, R.E.; Mahmud, A.S.; Miller, I.F.; Rajeev, M.; Rasambainarivo, F.; Rice, B.L.; Takahashi, S.; Tatem, A.J.; Wagner, C.E.; Wang, L.-F.; et al. Infectious disease in an ERA of global change. Nat. Rev. Microbiol. 2021, 20, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Lau, H.J.; Lim, C.H.; Foo, S.C.; Tan, H.S. The role of Artificial Intelligence in the battle against antimicrobial-resistant bacteria. Curr. Genet. 2021, 67, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Rong, R.; Jiang, S.; Xu, L.; Xiao, G.; Xie, Y.; Liu, D.J.; Li, Q.; Zhan, X. MB-GAN: Microbiome Simulation via Generative Adversarial Network. GigaScience 2021, 10, giab005. [Google Scholar] [CrossRef] [PubMed]
- Golshan, A.; Myers, J.; Watson, A. The Synthetic Data Generation Platform for Developers. Gretel AI. Available online: https://gretel.ai/ (accessed on 15 December 2023).
- Platzer, M.; Kalcher, K.; Boubela, R. Synthetic Data Generation with the Highest Accuracy. Mostly AI. Available online: https://mostly.ai/ (accessed on 15 December 2023).
- Kotwal, S.; Rani, P.; Arif, T.; Manhas, J.; Sharma, S. Automated bacterial classifications using machine learning based computational techniques: Architectures, challenges and open research issues. Arch. Comput. Methods Eng. 2021, 29, 2469–2490. [Google Scholar] [CrossRef]
- Akova, F.; Dundar, M.; Davisson, V.J.; Hirleman, E.D.; Bhunia, A.K.; Robinson, J.P.; Rajwa, B. A machine-learning approach to detecting unknown bacterial serovars. Stat. Anal. Data Min. ASA Data Sci. J. 2010, 3, 289–301. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed]
- Stirling, D. DNA extraction from fungi, yeast, and bacteria. Methods Mol. Biol. 2003, 226, 53–54. [Google Scholar] [CrossRef]
- Priyadarshi, N.; Singhal, N.K. Quartz Crystal Microbalance (qcm)-based nanosensors for the detection of pathogenic bacteria. In Nanosensors for Point-of-Care Diagnostics of Pathogenic Bacteria; Springer: Singapore, 2023; pp. 143–167. [Google Scholar] [CrossRef]
- Thawany, P.; Tiwari, U.K.; Deep, A. Surface plasmon resonance (spr)-based nanosensors for the detection of pathogenic bacteria. In Nanosensors for Point-of-Care Diagnostics of Pathogenic Bacteria; Springer: Singapore, 2023; pp. 41–57. [Google Scholar] [CrossRef]
- Giana, H.E.; Silveira, L., Jr.; Zângaro, R.A.; Pacheco, M.T. Rapid identification of bacterial species by fluorescence spectroscopy and classification through Principal Components Analysis. J. Fluoresc. 2003, 13, 489–493. [Google Scholar] [CrossRef]
- Smith, A.C.; Hussey, M.A. Gram Stain Protocols. American Society for Microbiology. 2005. Available online: https://asm.org/getattachment/5c95a063-326b-4b2f-98ce-001de9a5ece3/gram-stain-protocol-2886.pdf (accessed on 2 December 2023).
- Lähnemann, D.; Köster, J.; Szczurek, E.; McCarthy, D.J.; Hicks, S.C.; Robinson, M.D.; Vallejos, C.A.; Campbell, K.R.; Beerenwinkel, N.; Mahfouz, A.; et al. Eleven grand challenges in single-cell data science. Genome Biol. 2020, 21, 31. [Google Scholar] [CrossRef] [PubMed]
- Mohd Maidin, N.N.; Buyong, M.R.; ARahim, R.; Mohamed, M.A. Dielectrophoresis applications in biomedical field and future perspectives in biomedical technology. Electrophoresis 2021, 42, 2033–2059. [Google Scholar] [CrossRef] [PubMed]
- Percival, S.L.; McCarty, S.M.; Lipsky, B. Biofilms and wounds: An overview of the evidence. Adv. Wound Care 2015, 4, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Song, H.; Ahn, H.; Kim, T.; Jung, J.; Cho, S.K.; Shin, D.-M.; Choi, J.-r.; Hwang, Y.-H.; Kim, K. A Review of Advanced Impedance Biosensors with Microfluidic Chips for Single-Cell Analysis. Biosensors 2021, 11, 412. [Google Scholar] [CrossRef] [PubMed]
- Ivnitski, D.; Abdel-Hamid, I.; Atanasov, P.; Wilkins, E. Biosensors for detection of pathogenic bacteria. Biosens. Bioelectron. 1999, 14, 599–624. [Google Scholar] [CrossRef]
- Gnaim, R.; Golberg, A.; Sheviryov, J.; Rubinsky, B.; González, C.A. Detection and differentiation of bacteria by electrical bioimpedance spectroscopy. BioTechniques 2020, 69, 384–394. [Google Scholar] [CrossRef]
- Lee, W.; Kwon, D.; Choi, W.; Jung, G.Y.; Au, A.K.; Folch, A.; Jeon, S. 3D-Printed Microfluidic Device for the Detection of Pathogenic Bacteria Using Size-based Separation in Helical Channel with Trapezoid Cross-Section. Sci. Rep. 2015, 5, 7717. [Google Scholar] [CrossRef]
- Yeh, P.-C.; Chen, J.; Karakurt, I.; Lin, L. 3D printed bio-sensing chip for the determination of bacteria antibiotic-resistant profile. In Proceedings of the 20th International Conference on Solid-State Sensors, Actuators and Microsystems Eurosensors XXXIII, Berlin, Germany, 23–27 June 2019. [Google Scholar] [CrossRef]
- Bancalari, E.; Bernini, V.; Bottari, B.; Neviani, E.; Gatti, M. Application of impedance microbiology for evaluating potential acidifying performances of starter lactic acid bacteria to employ in milk transformation. Front. Microbiol. 2016, 7, 213420. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, M.; Lagier, J.C.; Raoult, D.; Khelaifia, S. Bacterial culture through selective and non-selective conditions: The evolution of culture media in clinical microbiology. New Microbes New Infect. 2019, 34, 100622. [Google Scholar] [CrossRef] [PubMed]
- Elazhary, M.A.; Saheb, S.A.; Roy, R.S.; Lagacé, A. A simple procedure for the preliminary identification of aerobic gram negative intestinal bacteria with special reference to the Enterobacteriaceae. Can. J. Comp. Med. 1973, 37, 43–46. [Google Scholar]
- Childs, A.; Pereira, J.; Didier, C.M.; Baksh, A.; Johnson, I.; Castro, J.M.; Davidson, E.; Santra, S.; Rajaraman, S. Plotter Cut Stencil Masks for the Deposition of Organic and Inorganic Materials and a New Rapid, Cost Effective Technique for Antimicrobial Evaluations. Micromachines 2023, 14, 14. [Google Scholar] [CrossRef] [PubMed]
- Kundu, A.; Ausaf, T.; Rajasekaran, P.; Rajaraman, S. Multimodal Microfluidic Biosensor with Interdigitated Electrodes (IDE) And Microelectrode Array (MEA) For Bacterial Detection and Identification. In Proceedings of the 2019 20th International Conference on Solid-State Sensors, Actuators and Microsystems & Eurosensors XXXIII (TRANSDUCERS & EUROSENSORS XXXIII), Berlin, Germany, 23–27 June 2019; pp. 1199–1202. [Google Scholar] [CrossRef]
- Azim, N.; Orrico, J.F.; Appavoo, D.; Zhai, L.; Rajaraman, S. Polydopamine surface functionalization of 3D printed resin material for enhanced polystyrene adhesion towards insulation layers for 3D microelectrode arrays (3D MEAS). RSC Adv. 2022, 12, 25605–25616. [Google Scholar] [CrossRef]
- Ding, Y.H.; Floren, M.; Tan, W. Mussel-inspired polydopamine for bio-surface functionalization. Biosurface Biotribology 2016, 2, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Bonev, B.; Hooper, J.; Parisot, J. Principles of assessing bacterial susceptibility to antibiotics using the agar diffusion method. J. Antimicrob. Chemother. 2008, 61, 1295–1301. [Google Scholar] [CrossRef]
- Kosztołowicz, T.; Metzler, R. Diffusion of antibiotics through a biofilm in the presence of diffusion and absorption barriers. Phys. Rev. E 2020, 102, 032408. [Google Scholar] [CrossRef] [PubMed]
- Hart, C.; Rajaraman, S. Low-Power, Multimodal Laser Micromachining of Materials for Applications in sub-5 µm Shadow Masks and sub-10 µm Interdigitated Electrodes (IDEs) Fabrication. Micromachines 2020, 11, 178. [Google Scholar] [CrossRef]
- Champigneux, P.; Delia, M.-L.; Bergel, A. Impact of electrode micro- and nano-scale topography on the formation and performance of microbial electrodes. Biosens. Bioelectron. 2018, 118, 231–246. [Google Scholar] [CrossRef]
- Green, E.R.; Mecsas, J. Bacterial secretion systems: An overview. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Gomez, J.B.; Waters, C.M. Switching on cyclic di-GMP heterogeneity in Pseudomonas aeruginosa biofilms. Nat. Microbiol. 2023, 8, 1380–1381. [Google Scholar] [CrossRef] [PubMed]
- Chabowski, K.; Junka, A.F.; Szymczyk, P.; Piasecki, T.; Sierakowski, A.; Maczynska, B.; Nitsch, K. The application of impedance microsensors for real-time analysis of Pseudomonas aeruginosa biofilm formation. Pol. J. Microbiol. 2015, 64, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Silley, P.; Forsythe, S. Impedance microbiology—A rapid change for microbiologists. J. Appl. Bacteriol. 1996, 80, 233–243. [Google Scholar] [CrossRef]
- Pickens, L.B.; Tang, Y. Oxytetracycline biosynthesis. J. Biol. Chem. 2010, 285, 27509–27515. [Google Scholar] [CrossRef] [PubMed]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed]
- Schnappinger, D.; Hillen, W. Tetracyclines: Antibiotic action, uptake, and resistance mechanisms. Arch. Microbiol. 1996, 165, 359–369. [Google Scholar] [CrossRef] [PubMed]
- John, T.; Thomas, T.; Abel, B.; Wood, B.R.; Chalmers, D.K.; Martin, L.L. How kanamycin a interacts with bacterial and mammalian mimetic membranes. Biochim. Biophys. Acta (BBA) Biomembr. 2017, 1859, 2242–2252. [Google Scholar] [CrossRef] [PubMed]
- Kaloyanides, G.J. Drug-phospholipid interactions: Role in aminoglycoside nephrotoxicity. Ren. Fail. 1992, 14, 351–357. [Google Scholar] [CrossRef]
- Wang, X.; Lan, B.; Fei, H.; Wang, S.; Zhu, G. Heavy metal-induced co-selection for antibiotic resistance in terrestrial subsurface soils. J. Hazard. Mater. 2021, 411, 124848. [Google Scholar] [CrossRef]
- Habimana, O.; Steenkeste, K.; Fontaine-Aupart, M.P.; Bellon-Fontaine, M.N.; Kulakauskas, S.; Briandet, R. Diffusion of nanoparticles in biofilms is altered by bacterial cell wall hydrophobicity. Appl. Environ. Microbiol. 2011, 77, 367–368. [Google Scholar] [CrossRef]
- Choi, C.H.; Lee, E.Y.; Lee, Y.C.; Park, T.I.; Kim, H.J.; Hyun, S.H.; Kim, S.A.; Lee, S.-K.; Lee, J.C. Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell. Microbiol. 2005, 7, 1127–1138. [Google Scholar] [CrossRef] [PubMed]
- Seth, A.K.; Geringer, M.R.; Galiano, R.D.; Leung, K.P.; Mustoe, T.A.; Hong, S.J. Quantitative comparison and analysis of species-specific wound biofilm virulence using an in vivo, rabbit-ear model. J. Am. Coll. Surg. 2012, 215, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Kearns, D.B. The structure and regulation of flagella in Bacillus subtilis. Annu. Rev. Genet. 2014, 48, 319–340. [Google Scholar] [CrossRef] [PubMed]
- Moradali, M.F.; Ghods, S.; Rehm, B.H. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.M.; Zuber, P. Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu. Rev. Microbiol. 1998, 52, 165–190. [Google Scholar] [CrossRef] [PubMed]
- Furst, A.L.; Francis, M.B. Impedance-based detection of bacteria. Chem. Rev. 2018, 119, 700–726. [Google Scholar] [CrossRef] [PubMed]
- Bot, C.T.; Prodan, C. Quantifying the membrane potential during E. coli growth stages. Biophys. Chem. 2010, 146, 133–137. [Google Scholar] [CrossRef]
- Noble, P.A.; Dziuba, M.; Harrison, D.J.; Albritton, W.L. Factors influencing capacitance-based monitoring of Microbial Growth. J. Microbiol. Methods 1999, 37, 51–64. [Google Scholar] [CrossRef]
- Dziuba, M.; Noble, P.A.; Albritton, W.L. A study of the nutritional requirements of a selected haemophilus ducreyi strain by impedance and conventional methods. Curr. Microbiol. 1993, 27, 109–113. [Google Scholar] [CrossRef]
- Noble, P.A. Hypothetical model for monitoring microbial growth by using capacitance measurements—A minireview. J. Microbiol. Methods 1999, 37, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.C.; Jason, A.C.; Hobbs, G.; Gibson, D.M.; Christie, R.H. Electronic measurement of bacterial growth. J. Phys. E Sci. Instrum. 1978, 11, 560–568. [Google Scholar] [CrossRef]
- Paidhungat, M.; Setlow, P. Role of ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 2000, 182, 2513–2519. [Google Scholar] [CrossRef] [PubMed]
- Paidhungat, M.; Setlow, P. Spore germination and outgrowth. In Bacillus Subtilis and Its Closest Relatives; ASM Press: Washington, DC, USA, 2014; pp. 537–548. [Google Scholar] [CrossRef]
- Gould, P.; Marquis, R.E. Germination. In The Bacterial Spore; Could, G.W., Hurs, A., Eds.; Academic Press: London, UK, 1969; pp. 397–444. [Google Scholar]
- Paidhungat, M.; Ragkousi, K.; Setlow, P. Genetic requirements for induction of germination of spores of Bacillus subtilis by Ca2+-dipicolinate. J. Bacteriol. 2001, 183, 4886–4893. [Google Scholar] [CrossRef] [PubMed]
- Paidhungat, M.; Setlow, B.; Daniels, W.B.; Hoover, D.; Papafragkou, E.; Setlow, P. Mechanisms of induction of germination of bacillus subtilis spores by high pressure. Appl. Environ. Microbiol. 2002, 68, 3172–3175. [Google Scholar] [CrossRef]
- González-Pastor, J.E. Cannibalism: A social behavior in sporulating Bacillus subtilis. FEMS Microbiol. Rev. 2011, 35, 415–424. [Google Scholar] [CrossRef]
- Rada, B.; Leto, T.L. Pyocyanin effects on respiratory epithelium: Relevance in Pseudomonas aeruginosa airway infections. Trends Microbiol. 2013, 21, 73–81. [Google Scholar] [CrossRef]
- Darland, G. Principal component analysis of infraspecific bacteria. Appl. Microbiol. 1975, 30, 282–289. [Google Scholar] [CrossRef]



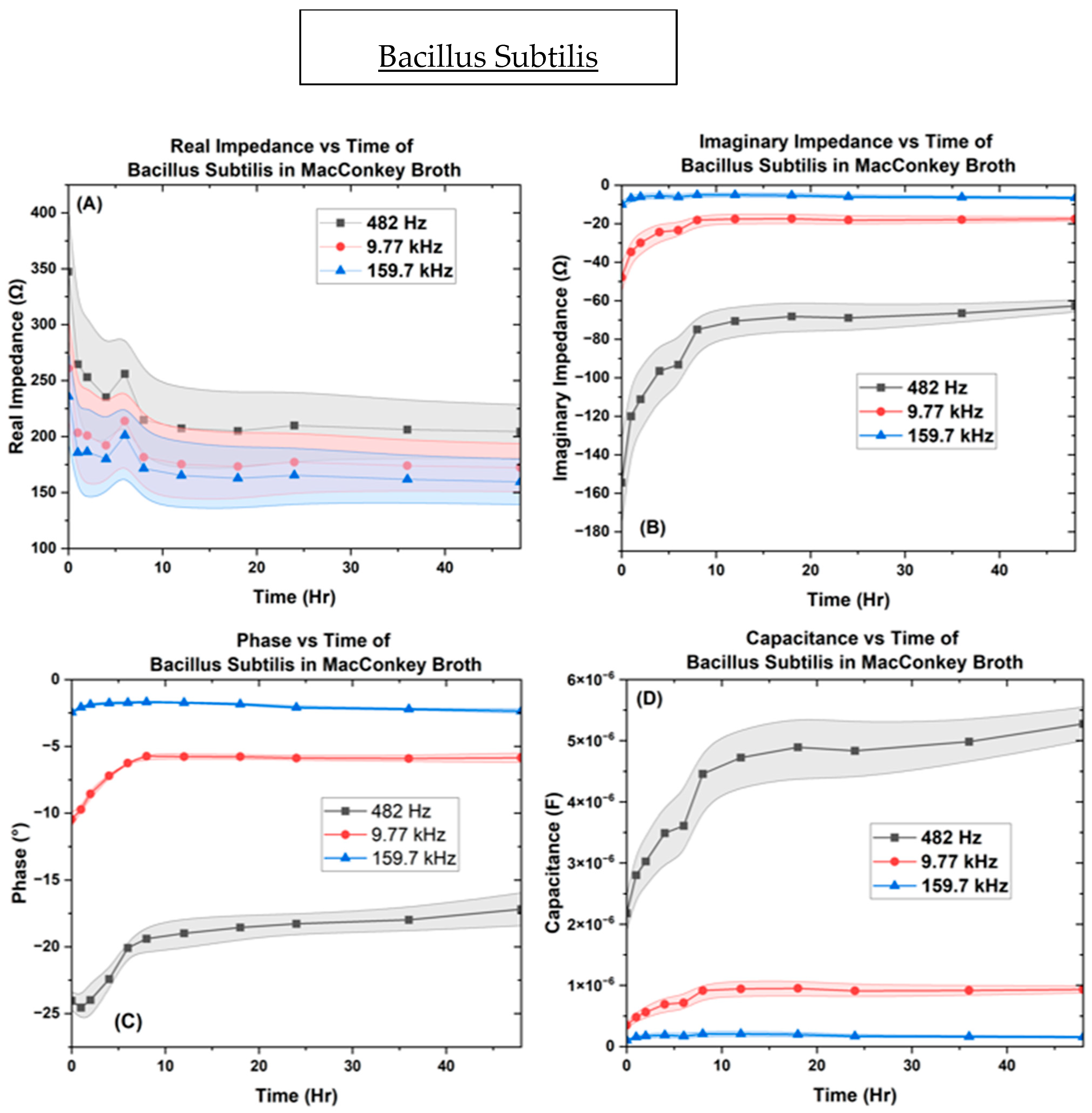
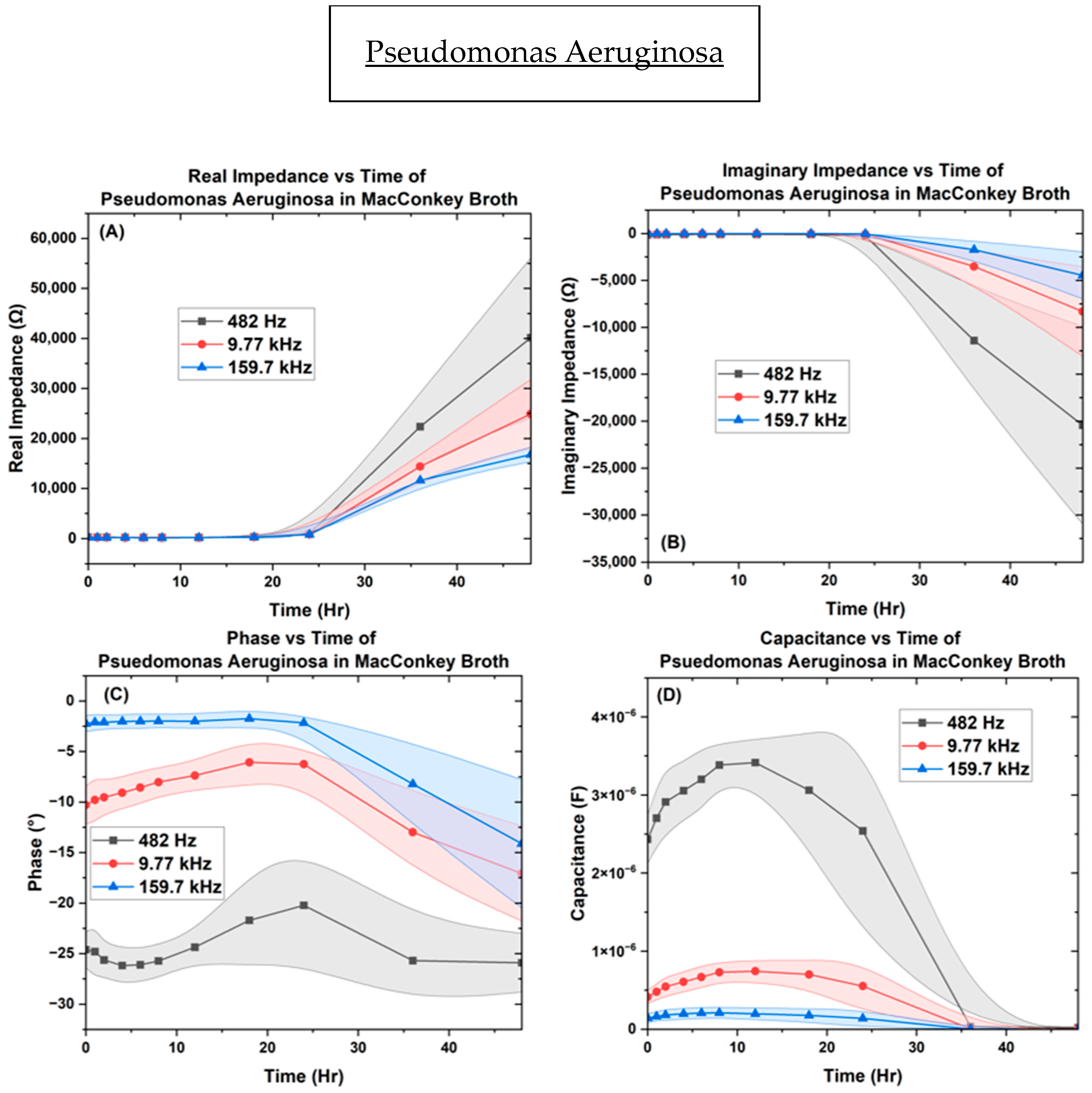
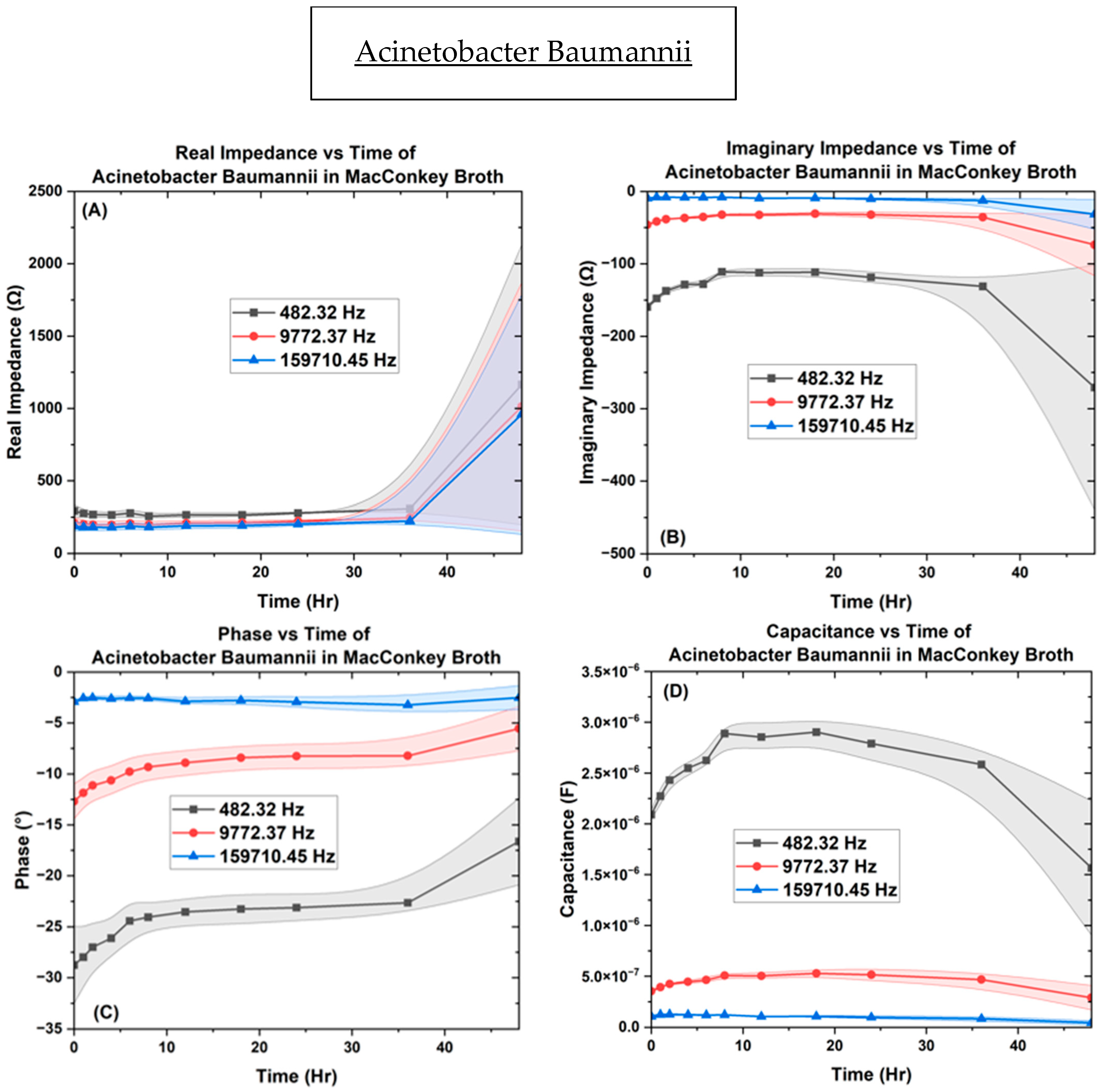
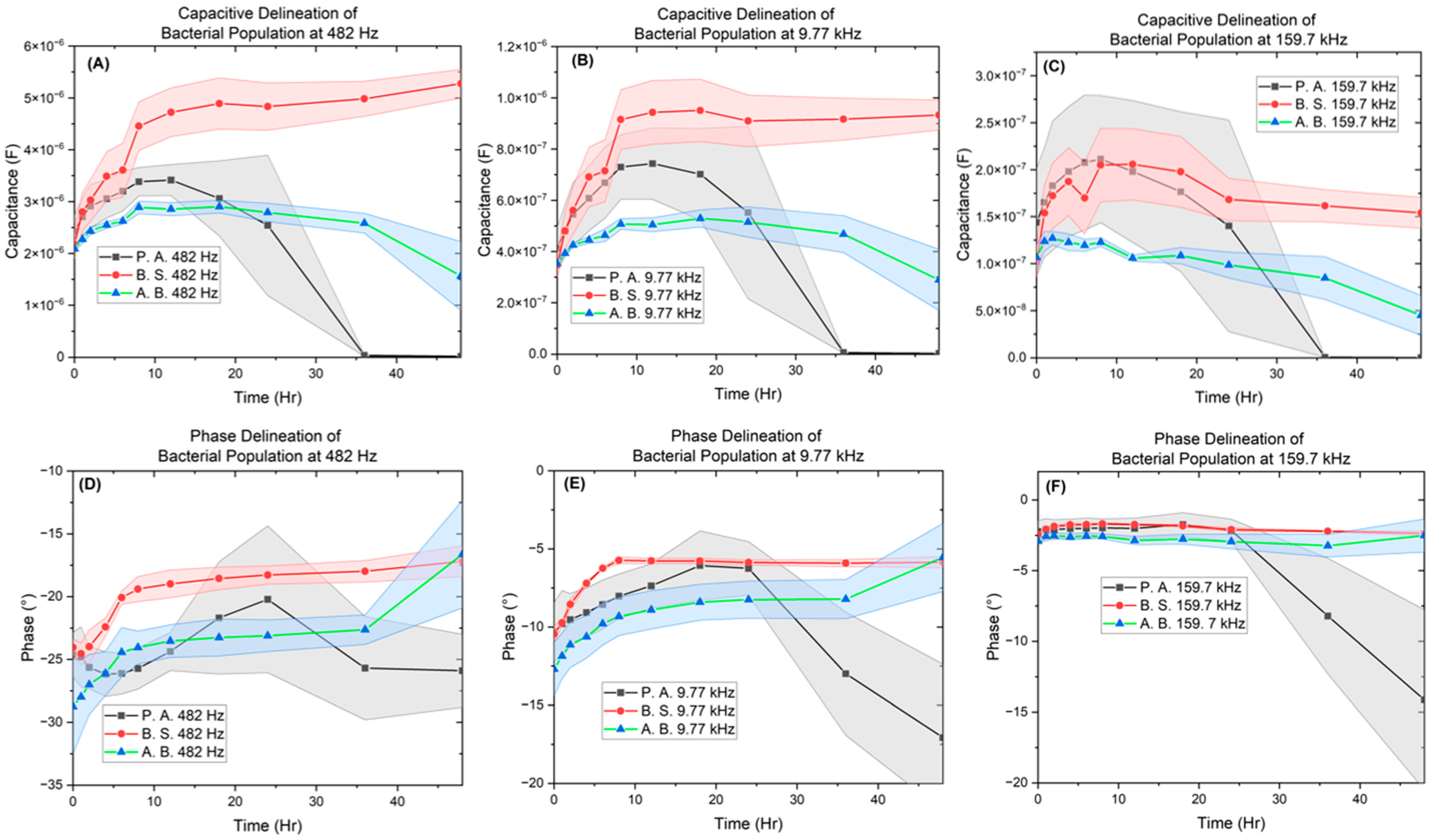

| Bacteria | OD600 |
|---|---|
| A. baumannii | 0.1 |
| P. aeruginosa | 0.4 |
| B. subtilis | 1.0 |
| Antibiotic | Antibiotic Mass (µg) | A.B Kirby (mm) | A. B. Stencil (mm) | B. S. Kirby (mm) | B.S Stencil (mm) | P.A. Kirby (mm) | P.A Stencil (mm) |
|---|---|---|---|---|---|---|---|
| Oxytetracycline | 7.5 | 7.89 | 7.53 | 9.56 | 8.52 | 4.29 | 0.42 |
| 15 | 9.485 | 10.05 | 12.87 | 9.58 | 4.99 | 1.34 | |
| 30 | 10.72 | 11.27 | 14.71 | 10.69 | 6.39 | 4.23 | |
| 60 | 11.89 | 12.87 | 15.64 | 11.55 | 6.99 | 6.46 | |
| Kanamycin | 7.5 | 4.42 | 4.25 | 3.8 | 4.15 | 0 | 0 |
| 15 | 6.75 | 6.42 | 5.57 | 5.39 | 1.048 | 0.42 | |
| 30 | 7.81 | 7.89 | 7.17 | 6.61 | 2.53 | 1.78 | |
| 60 | 9.21 | 9.21 | 8.65 | 7.66 | 3.56 | 3.27 | |
| Streptomycin | 2.5 | 0 | 0 | 0 | 0 | 1.09 | 0 |
| 5 | 0 | 0 | 0 | 0 | 3.45 | 1.93 | |
| 10 | 0 | 0 | 0 | 0 | 5.61 | 3.75 | |
| 20 | 1.95 | 0.64 | 0.51 | 0.86 | 7.68 | 5.27 | |
| Penicillin G. | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 12 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 24 | 0 | 0 | 5.13 * | 0.36 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Childs, A.; Chand, D.; Pereira, J.; Santra, S.; Rajaraman, S. BacteSign: Building a Findable, Accessible, Interoperable, and Reusable (FAIR) Database for Universal Bacterial Identification. Biosensors 2024, 14, 176. https://doi.org/10.3390/bios14040176
Childs A, Chand D, Pereira J, Santra S, Rajaraman S. BacteSign: Building a Findable, Accessible, Interoperable, and Reusable (FAIR) Database for Universal Bacterial Identification. Biosensors. 2024; 14(4):176. https://doi.org/10.3390/bios14040176
Chicago/Turabian StyleChilds, Andre, David Chand, Jorge Pereira, Swadeshmukul Santra, and Swaminathan Rajaraman. 2024. "BacteSign: Building a Findable, Accessible, Interoperable, and Reusable (FAIR) Database for Universal Bacterial Identification" Biosensors 14, no. 4: 176. https://doi.org/10.3390/bios14040176
APA StyleChilds, A., Chand, D., Pereira, J., Santra, S., & Rajaraman, S. (2024). BacteSign: Building a Findable, Accessible, Interoperable, and Reusable (FAIR) Database for Universal Bacterial Identification. Biosensors, 14(4), 176. https://doi.org/10.3390/bios14040176





