Aptamer-Based Targeting of Cancer: A Powerful Tool for Diagnostic and Therapeutic Aims
Abstract
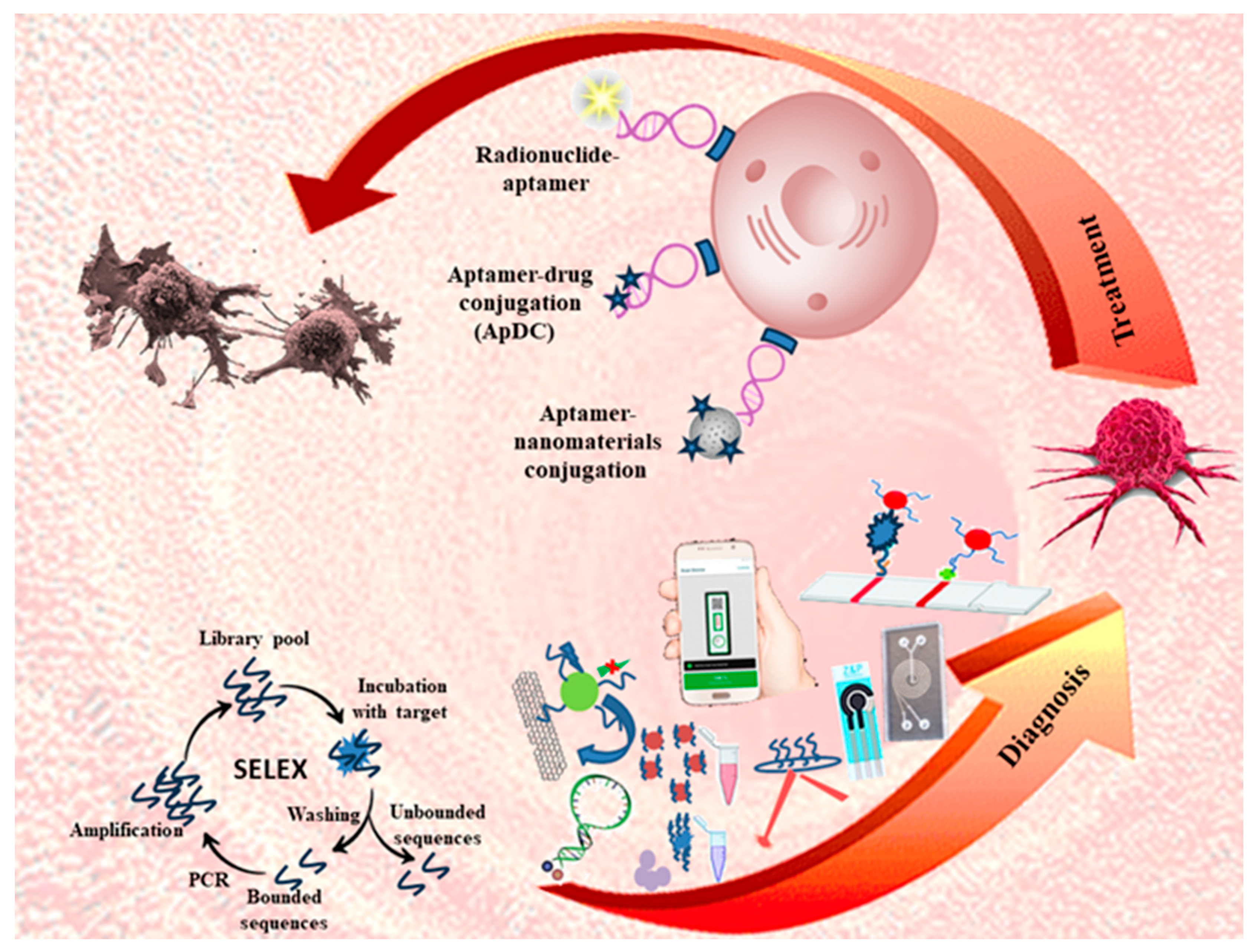
1. Introduction
2. Aptamer-Based Targeted Delivery Systems
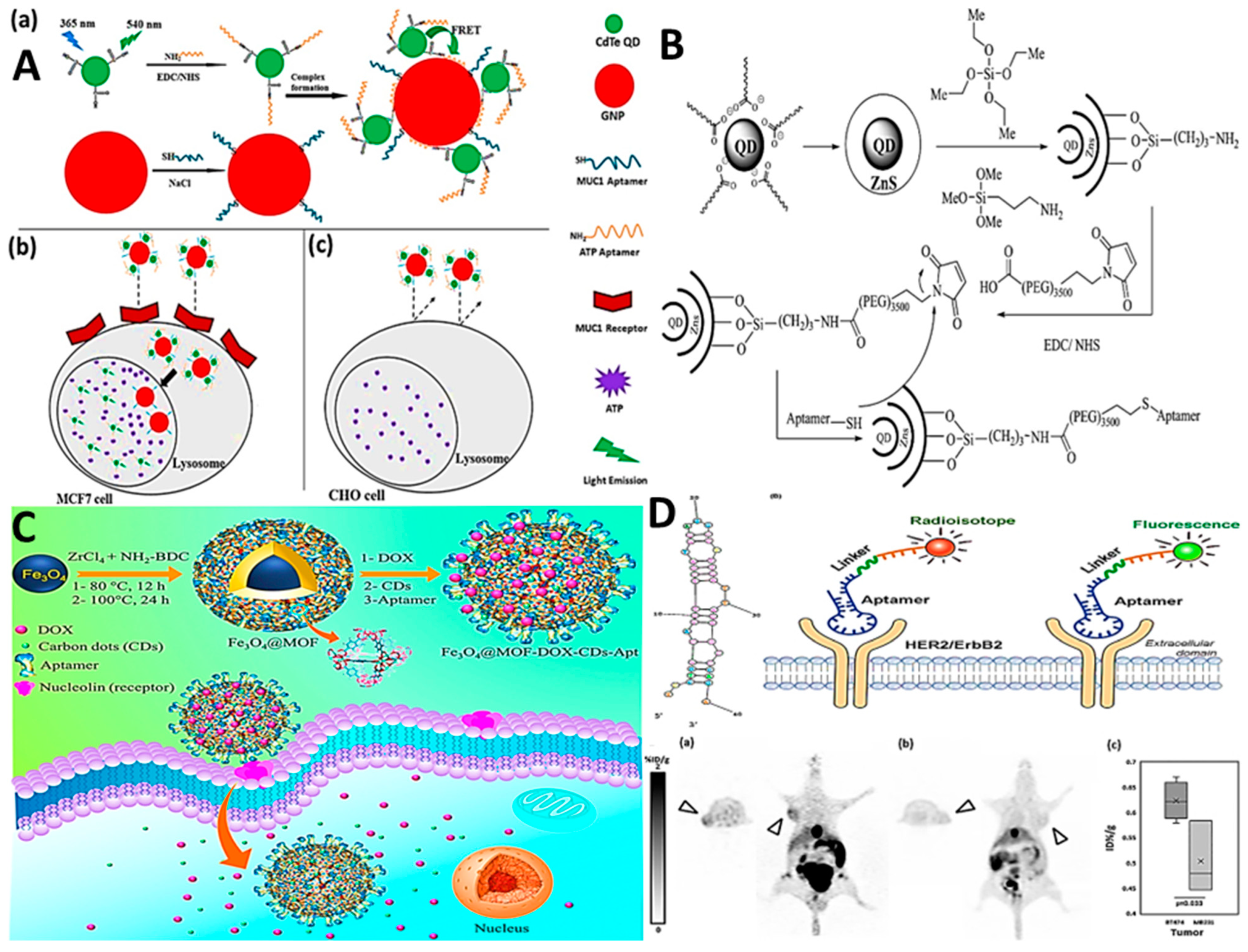
2.1. Aptamer–Nanomaterial Conjugation
2.2. Critical Note
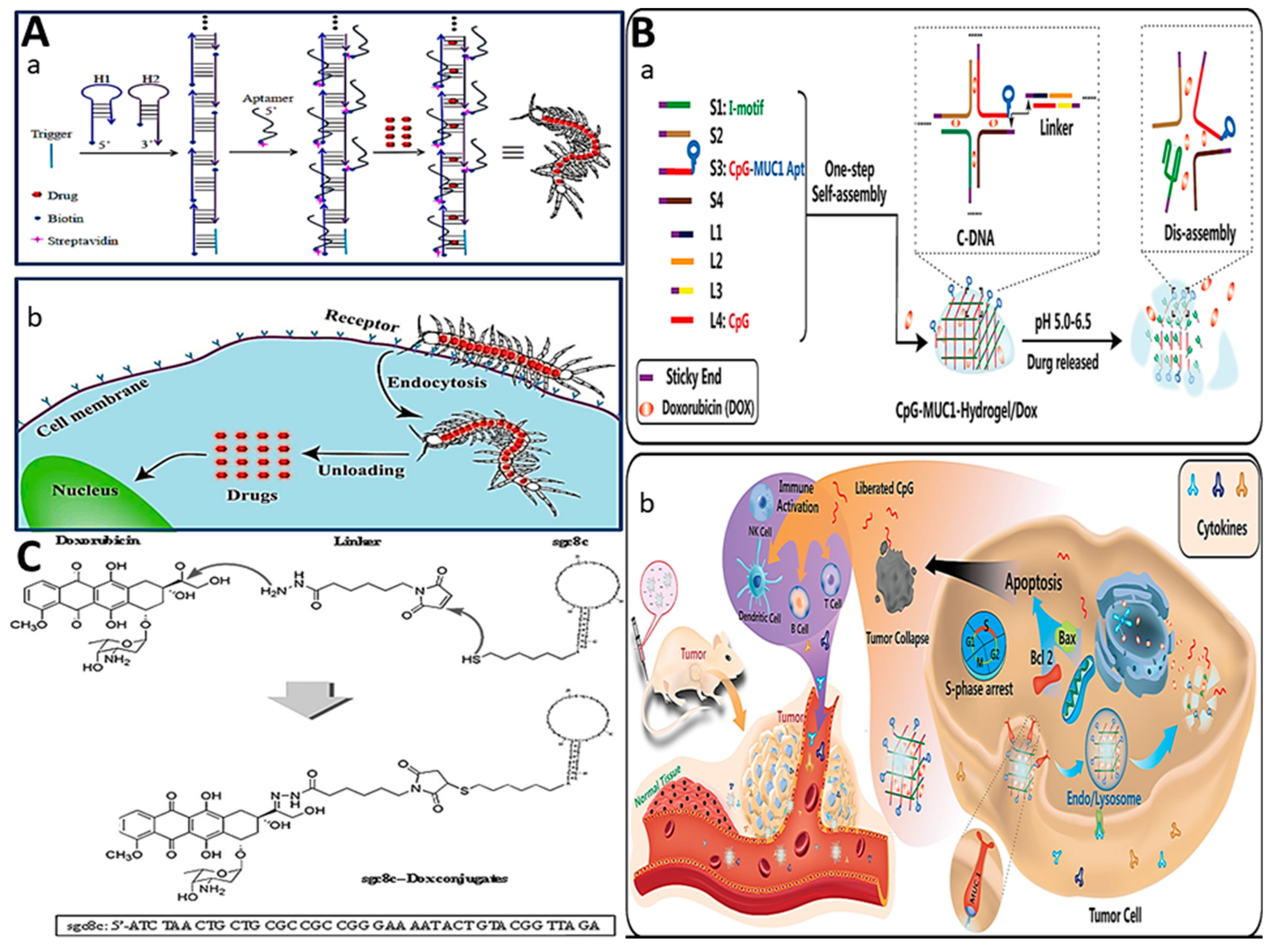
2.3. Aptamer–Drug Conjugation (ApDC)
2.3.1. Physical Conjugation (Intercalation)
2.3.2. Covalent Conjugation
2.3.3. Critical Note
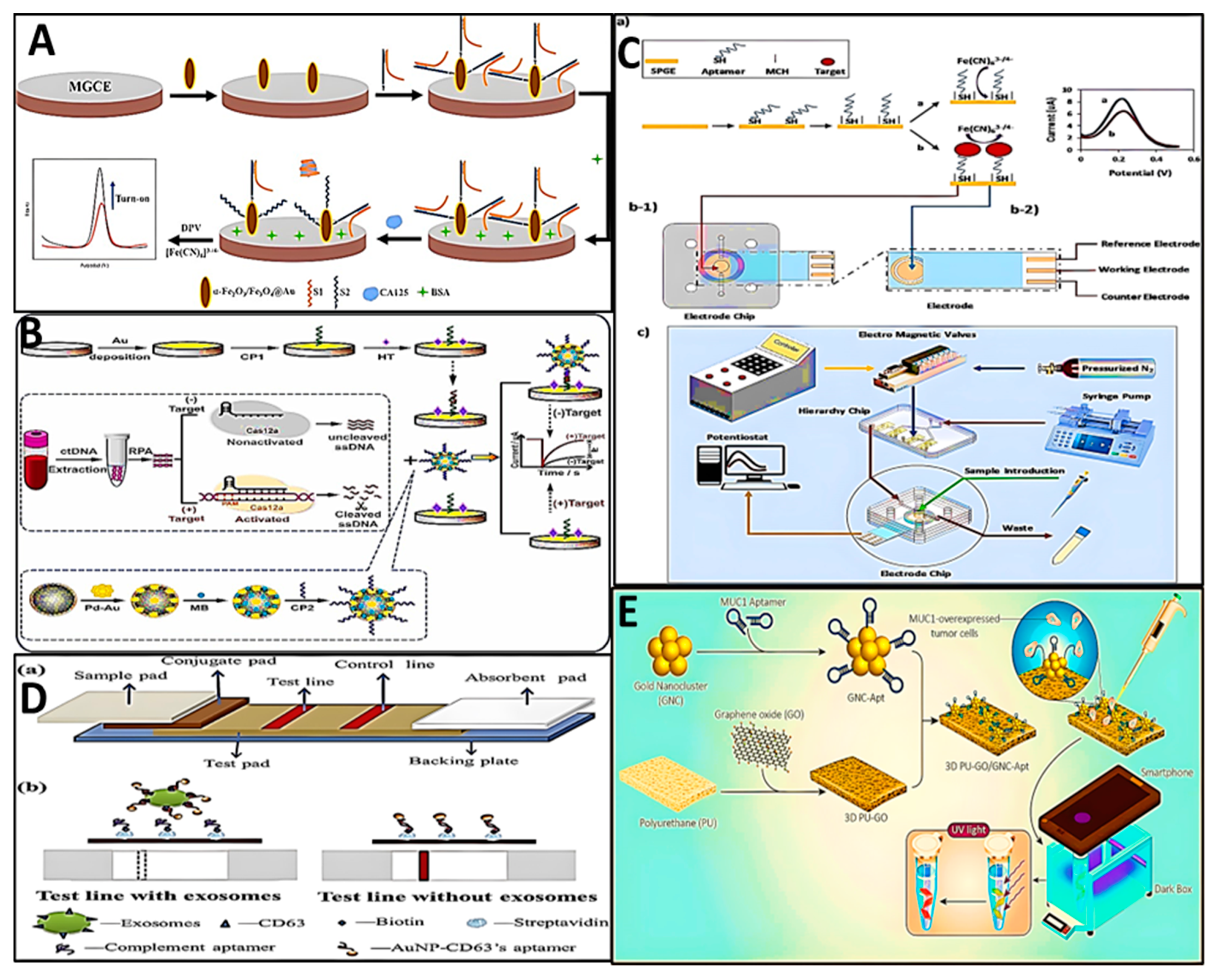
3. Biosensors
4. Critical Note
5. Conclusions and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, P.-L.; Wang, Z.-K.; Chen, Q.-Y.; Du, X.; Gao, J. Biocompatible G-Quadruplex/BODIPY assembly for cancer cell imaging and the attenuation of mitochondria. Bioorganic Med. Chem. Lett. 2019, 29, 1943–1947. [Google Scholar] [CrossRef]
- Kong, H.Y.; Byun, J. Nucleic acid aptamers: New methods for selection, stabilization, and application in biomedical science. Biomol. Ther. 2013, 21, 423. [Google Scholar] [CrossRef] [PubMed]
- Bavi, R.; Hang, Z.; Banerjee, P.; Aquib, M.; Jadhao, M.; Rane, N.; Bavi, S.; Bhosale, R.; Kodam, K.; Jeon, B.-H. Doxorubicin-conjugated innovative 16-mer DNA aptamer-based Annexin A1 targeted anti-cancer drug delivery. Mol. Ther.-Nucleic Acids 2020, 21, 1074–1086. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Wu, X.; Luo, L.; Liu, J.; Yang, L.; Wang, F. Fluorescent Ag clusters conjugated with anterior gradient-2 antigen aptamer for specific detection of cancer cells. Talanta 2019, 197, 86–91. [Google Scholar] [CrossRef]
- You, X.; Gopinath, S.C.; Lakshmipriya, T.; Li, D. High-affinity detection of alpha-synuclein by aptamer-gold conjugates on an amine-modified dielectric surface. J. Anal. Methods Chem. 2019, 2019, 6526850. [Google Scholar] [CrossRef]
- Yang, Q.; Deng, Z.; Wang, D.; He, J.; Zhang, D.; Tan, Y.; Peng, T.; Wang, X.-Q.; Tan, W. Conjugating aptamer and mitomycin C with reductant-responsive linker leading to synergistically enhanced anticancer effect. J. Am. Chem. Soc. 2020, 142, 2532–2540. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent advances in aptamer discovery and applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.W.; Shima, D.T.; Calias, P.; Cunningham Jr, E.T.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Iqbal, M.J.; Javed, Z.; Herrera-Bravo, J.; Sadia, H.; Anum, F.; Raza, S.; Tahir, A.; Shahwani, M.N.; Sharifi-Rad, J.; Calina, D. Biosensing chips for cancer diagnosis and treatment: A new wave towards clinical innovation. Cancer Cell Int. 2022, 22, 354. [Google Scholar] [CrossRef]
- Stein, C.A.; Castanotto, D. FDA-approved oligonucleotide therapies in 2017. Mol. Ther. 2017, 25, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Tsuchikama, K.; An, Z. Antibody-drug conjugates: Recent advances in conjugation and linker chemistries. Protein Cell 2018, 9, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Mohammadinejad, A.; Taghdisi, S.M.; Es’ haghi, Z.; Abnous, K.; Mohajeri, S.A. Targeted imaging of breast cancer cells using two different kinds of aptamers-functionalized nanoparticles. Eur. J. Pharm. Sci. 2019, 134, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Salmasi, Z.; Hashemi, M.; Mosaffa, F.; Abnous, K.; Ramezani, M. Single-walled carbon nanotubes functionalized with aptamer and piperazine–polyethylenimine derivative for targeted siRNA delivery into breast cancer cells. Int. J. Pharm. 2015, 485, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Akbarzadeh, M.; Babaei, M.; Abnous, K.; Taghdisi, S.M.; Peivandi, M.T.; Ramezani, M.; Alibolandi, M. Hybrid silica-coated Gd-Zn-Cu-In-S/ZnS bimodal quantum dots as an epithelial cell adhesion molecule targeted drug delivery and imaging system. Int. J. Pharm. 2019, 570, 118645. [Google Scholar] [CrossRef]
- Yang, Y.; Zhao, W.; Tan, W.; Lai, Z.; Fang, D.; Jiang, L.; Zuo, C.; Yang, N.; Lai, Y. An efficient cell-targeting drug delivery system based on aptamer-modified mesoporous silica nanoparticles. Nanoscale Res. Lett. 2019, 14, 390. [Google Scholar]
- Li, X.; Yu, Y.; Ji, Q.; Qiu, L. Targeted delivery of anticancer drugs by aptamer AS1411 mediated Pluronic F127/cyclodextrin-linked polymer composite micelles. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 175–184. [Google Scholar]
- Li, X.; Wu, X.; Yang, H.; Li, L.; Ye, Z.; Rao, Y. A nuclear targeted Dox-aptamer loaded liposome delivery platform for the circumvention of drug resistance in breast cancer. Biomed. Pharmacother. 2019, 117, 109072. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, P.; Ma, J.; Li, D.; Yang, H.; Chen, W.; Jiang, Y. Enhancement of radiosensitization by silver nanoparticles functionalized with polyethylene glycol and aptamer As1411 for glioma irradiation therapy. Int. J. Nanomed. 2019, 14, 9483–9496. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Liu, B.; Chen, X.; Lin, H.; Peng, Y.; Li, Y.; Zheng, H.; Xu, Y.; Ou, X.; Yan, S. Aptamer-assisted superparamagnetic iron oxide nanoparticles as multifunctional drug delivery platform for chemo-photodynamic combination therapy. J. Mater. Sci. Mater. Med. 2019, 30, 76. [Google Scholar] [CrossRef] [PubMed]
- Alijani, H.; Noori, A.; Faridi, N.; Bathaie, S.Z.; Mousavi, M.F. Aptamer-functionalized Fe3O4@ MOF nanocarrier for targeted drug delivery and fluorescence imaging of the triple-negative MDA-MB-231 breast cancer cells. J. Solid State Chem. 2020, 292, 121680. [Google Scholar] [CrossRef]
- Lin, H.-C.; Li, W.-T.; Madanayake, T.W.; Tao, C.; Niu, Q.; Yan, S.-Q.; Gao, B.-A.; Ping, Z. Aptamer-guided upconversion nanoplatform for targeted drug delivery and near-infrared light-triggered photodynamic therapy. J. Biomater. Appl. 2020, 34, 875–888. [Google Scholar] [CrossRef]
- Chang, M.; Yang, C.-S.; Huang, D.-M. Aptamer-conjugated DNA icosahedral nanoparticles as a carrier of doxorubicin for cancer therapy. ACS Nano 2011, 5, 6156–6163. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wei, T.; Zhao, J.; Huang, Y.; Deng, H.; Kumar, A.; Wang, C.; Liang, Z.; Ma, X.; Liang, X.-J. Multifunctional aptamer-based nanoparticles for targeted drug delivery to circumvent cancer resistance. Biomaterials 2016, 91, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Huang, K.-W.; Reebye, V.; Spalding, D.; Przytycka, T.M.; Wang, Y.; Swiderski, P.; Li, L.; Armstrong, B.; Reccia, I. Aptamer-drug conjugates of active metabolites of nucleoside analogs and cytotoxic agents inhibit pancreatic tumor cell growth. Mol. Ther.-Nucleic Acids 2017, 6, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.F.; Shangguan, D.; Liu, H.; Phillips, J.A.; Zhang, X.; Chen, Y.; Tan, W. Molecular assembly of an aptamer–drug conjugate for targeted drug delivery to tumor cells. ChemBioChem 2009, 10, 862–868. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, X.; He, L.; Wang, K.; Wang, Q.; Huang, J.; Liu, J.; Wu, B.; Xu, C. Self-assembled DNA nanocentipede as multivalent drug carrier for targeted delivery. ACS Appl. Mater. Interfaces 2016, 8, 25733–25740. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Zhao, Z.; Wang, Y.; Zou, J.; Lin, Q.; Duan, Y. One-step self-assembly of multifunctional DNA nanohydrogels: An enhanced and harmless strategy for guiding combined antitumor therapy. ACS Appl. Mater. Interfaces 2019, 11, 46479–46489. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Rahimizadeh, K.; Veedu, R.N. Development of a novel DNA oligonucleotide targeting low-density lipoprotein receptor. Mol. Ther.-Nucleic Acids 2020, 19, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Lu, J.; Liu, J.; Liang, C.; Wang, M.; Wang, L.; Li, D.; Yao, H.; Zhang, Q.; Wen, J. A water-soluble nucleolin aptamer-paclitaxel conjugate for tumor-specific targeting in ovarian cancer. Nat. Commun. 2017, 8, 1390. [Google Scholar] [CrossRef]
- Jeong, H.Y.; Kim, H.; Lee, M.; Hong, J.; Lee, J.H.; Kim, J.; Choi, M.J.; Park, Y.S.; Kim, S.-C. Development of HER2-specific aptamer-drug conjugate for breast cancer therapy. Int. J. Mol. Sci. 2020, 21, 9764. [Google Scholar] [CrossRef]
- Thiel, K.W.; Hernandez, L.I.; Dassie, J.P.; Thiel, W.H.; Liu, X.; Stockdale, K.R.; Rothman, A.M.; Hernandez, F.J.; McNamara, J.O.; Giangrande, P.H. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res. 2012, 40, 6319–6337. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Park, J.Y.; Lee, T.S.; Song, I.H.; Cho, Y.L.; Chae, J.R.; Kang, H.; Lim, J.H.; Lee, J.H.; Kang, W.J. PET imaging of HER2 expression with an 18F-fluoride labeled aptamer. PLoS ONE 2019, 14, e0211047. [Google Scholar] [CrossRef] [PubMed]
- Alshaer, W.; Hillaireau, H.; Fattal, E. Aptamer-guided nanomedicines for anticancer drug delivery. Adv. Drug Deliv. Rev. 2018, 134, 122–137. [Google Scholar] [CrossRef] [PubMed]
- Serrano, C.M.; Freeman, R.; Godbe, J.; Lewis, J.A.; Stupp, S.I. DNA-peptide amphiphile nanofibers enhance aptamer function. ACS Appl. Bio Mater. 2019, 2, 2955–2963. [Google Scholar] [CrossRef]
- Vazquez-Gonzalez, M.; Willner, I. Aptamer-functionalized micro-and nanocarriers for controlled release. ACS Appl. Mater. Interfaces 2021, 13, 9520–9541. [Google Scholar] [CrossRef]
- Bai, J.; Luo, Y.; Wang, X.; Li, S.; Luo, M.; Yin, M.; Zuo, Y.; Li, G.; Yao, J.; Yang, H. A protein-independent fluorescent RNA aptamer reporter system for plant genetic engineering. Nat. Commun. 2020, 11, 3847. [Google Scholar] [CrossRef] [PubMed]
- Nooranian, S.; Mohammadinejad, A.; Mohajeri, T.; Aleyaghoob, G.; Kazemi Oskuee, R. Biosensors based on aptamer-conjugated gold nanoparticles: A review. Biotechnol. Appl. Biochem. 2022, 69, 1517–1534. [Google Scholar] [CrossRef]
- Debnath, S.K.; Srivastava, R. Drug delivery with carbon-based nanomaterials as versatile nanocarriers: Progress and prospects. Front. Nanotechnol. 2021, 3, 644564. [Google Scholar] [CrossRef]
- Nair, A.; Haponiuk, J.T.; Thomas, S.; Gopi, S. Natural carbon-based quantum dots and their applications in drug delivery: A review. Biomed. Pharmacother. 2020, 132, 110834. [Google Scholar] [CrossRef]
- Mohammadinejad, A.; Es’ haghi, Z.; Abnous, K.; Mohajeri, S.A. Tandem determination of mitoxantrone and ribonucleic acid using mercaptosuccinic acid-capped CdTe quantum dots. J. Lumin. 2017, 190, 254–260. [Google Scholar] [CrossRef]
- Croissant, J.G.; Butler, K.S.; Zink, J.I.; Brinker, C.J. Synthetic amorphous silica nanoparticles: Toxicity, biomedical and environmental implications. Nat. Rev. Mater. 2020, 5, 886–909. [Google Scholar] [CrossRef]
- Ghosh, B.; Biswas, S. Polymeric micelles in cancer therapy: State of the art. J. Control. Release 2021, 332, 127–147. [Google Scholar] [CrossRef] [PubMed]
- Malam, Y.; Loizidou, M.; Seifalian, A.M. Liposomes and nanoparticles: Nanosized vehicles for drug delivery in cancer. Trends Pharmacol. Sci. 2009, 30, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Vangijzegem, T.; Stanicki, D.; Laurent, S. Magnetic iron oxide nanoparticles for drug delivery: Applications and characteristics. Expert Opin. Drug Deliv. 2019, 16, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Deng, K.; Hou, Z.; Li, X.; Li, C.; Zhang, Y.; Deng, X.; Cheng, Z.; Lin, J. Aptamer-mediated up-conversion core/MOF shell nanocomposites for targeted drug delivery and cell imaging. Sci. Rep. 2015, 5, 7851. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamid, H.N. Nanoparticle assisted laser desorption/ionization mass spectrometry for small molecule analytes. Microchim. Acta 2018, 185, 200. [Google Scholar] [CrossRef] [PubMed]
- Soontornworajit, B.; Zhou, J.; Shaw, M.T.; Fan, T.-H.; Wang, Y. Hydrogel functionalization with DNA aptamers for sustained PDGF-BB release. Chem. Commun. 2010, 46, 1857–1859. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-Y.; Chen, L.-Q.; Sun, W.; Du, H.-H.; Dong, S.; Ahmed, A.M.Q.; Cao, D.; Cui, J.-H.; Zhang, Y.; Cao, Q.-R. Collagenase IV and clusterin-modified polycaprolactone-polyethylene glycol nanoparticles for penetrating dense tumor tissues. Theranostics 2021, 11, 906. [Google Scholar] [CrossRef] [PubMed]
- Patil, Y.P.; Jadhav, S. Novel methods for liposome preparation. Chem. Phys. Lipids 2014, 177, 8–18. [Google Scholar] [CrossRef]
- Gao, J.; Gu, H.; Xu, B. Multifunctional magnetic nanoparticles: Design, synthesis, and biomedical applications. Acc. Chem. Res. 2009, 42, 1097–1107. [Google Scholar] [CrossRef]
- Azzouz, A.; Goud, K.Y.; Raza, N.; Ballesteros, E.; Lee, S.-E.; Hong, J.; Deep, A.; Kim, K.-H. Nanomaterial-based electrochemical sensors for the detection of neurochemicals in biological matrices. TrAC Trends Anal. Chem. 2019, 110, 15–34. [Google Scholar]
- Wen, M.; Li, G.; Liu, H.; Chen, J.; An, T.; Yamashita, H. Metal–organic framework-based nanomaterials for adsorption and photocatalytic degradation of gaseous pollutants: Recent progress and challenges. Environ. Sci. Nano 2019, 6, 1006–1025. [Google Scholar] [CrossRef]
- Mehtab, T.; Yasin, G.; Arif, M.; Shakeel, M.; Korai, R.M.; Nadeem, M.; Muhammad, N.; Lu, X. Metal-organic frameworks for energy storage devices: Batteries and supercapacitors. J. Energy Storage 2019, 21, 632–646. [Google Scholar] [CrossRef]
- Yan, Y.; Li, C.; Wu, Y.; Gao, J.; Zhang, Q. From isolated Ti-oxo clusters to infinite Ti-oxo chains and sheets: Recent advances in photoactive Ti-based MOFs. J. Mater. Chem. A 2020, 8, 15245–15270. [Google Scholar] [CrossRef]
- Wang, S.; Chen, Y.; Wang, S.; Li, P.; Mirkin, C.A.; Farha, O.K. DNA-functionalized metal–organic framework nanoparticles for intracellular delivery of proteins. J. Am. Chem. Soc. 2019, 141, 2215–2219. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, Q.; Feng, W.; Sun, Y.; Li, F. Upconversion luminescent materials: Advances and applications. Chem. Rev. 2015, 115, 395–465. [Google Scholar] [CrossRef]
- Mohammadinejad, A.; Abnous, K.; Nameghi, M.A.; Yahyazadeh, R.; Hamrah, S.; Senobari, F.; Mohajeri, S.A. Application of green-synthesized carbon dots for imaging of cancerous cell lines and detection of anthraquinone drugs using silica-coated CdTe quantum dots-based ratiometric fluorescence sensor. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 288, 122200. [Google Scholar] [CrossRef]
- Yang, Q.; Zhao, C.; Zhao, J.; Ye, Y. Synthesis and singlet oxygen activities of near infrared photosensitizers by conjugation with upconversion nanoparticles. Opt. Mater. Express 2017, 7, 913–923. [Google Scholar] [CrossRef]
- Lim, C.-K.; Heo, J.; Shin, S.; Jeong, K.; Seo, Y.H.; Jang, W.-D.; Park, C.R.; Park, S.Y.; Kim, S.; Kwon, I.C. Nanophotosensitizers toward advanced photodynamic therapy of Cancer. Cancer Lett. 2013, 334, 176–187. [Google Scholar] [CrossRef]
- Yano, S.; Hirohara, S.; Obata, M.; Hagiya, Y.; Ogura, S.-i.; Ikeda, A.; Kataoka, H.; Tanaka, M.; Joh, T. Current states and future views in photodynamic therapy. J. Photochem. Photobiol. C Photochem. Rev. 2011, 12, 46–67. [Google Scholar] [CrossRef]
- Del Rosal, B.; Jaque, D. Upconversion nanoparticles for in vivo applications: Limitations and future perspectives. Methods Appl. Fluoresc. 2019, 7, 022001. [Google Scholar] [CrossRef] [PubMed]
- Institute, N.C. Radiation Therapy Side Effects. Available online: https://www.cancer.gov/about-cancer/treatment/types/radiation-therapy/side-effects (accessed on 27 January 2021).
- Yan, A.C.; Levy, M. Aptamer-mediated delivery and cell-targeting aptamers: Room for improvement. Nucleic Acid Ther. 2018, 28, 194–199. [Google Scholar] [CrossRef]
- Chandola, C.; Neerathilingam, M. Aptamers for targeted delivery: Current challenges and future opportunities. In Role of Novel Drug Delivery Vehicles in Nanobiomedicine; BoD–Books on Demand: Norderstedt, Germany, 2019; pp. 1–22. [Google Scholar]
- Mohammadinejad, A.; Oskuee, R.K.; Eivazzadeh-Keihan, R.; Rezayi, M.; Baradaran, B.; Maleki, A.; Hashemzaei, M.; Mokhtarzadeh, A.; de la Guardia, M. Development of biosensors for detection of alpha-fetoprotein: As a major biomarker for hepatocellular carcinoma. TrAC Trends Anal. Chem. 2020, 130, 115961. [Google Scholar] [CrossRef]
- Schneider-Futschik, E.K.; Reyes-Ortega, F. Advantages and disadvantages of using magnetic nanoparticles for the treatment of complicated ocular disorders. Pharmaceutics 2021, 13, 1157. [Google Scholar] [CrossRef] [PubMed]
- Gu, K.; Meng, F. Former research and recent advances of metal-organic frameworks (MOF) for anti-cancer drug delivery. In Journal of Physics: Conference Series; IOP Publishing: Bristol, UK, 2021; p. 012021. [Google Scholar]
- Hu, R.; Zhang, X.; Zhao, Z.; Zhu, G.; Chen, T.; Fu, T.; Tan, W. DNA nanoflowers for multiplexed cellular imaging and traceable targeted drug delivery. Angew. Chem. 2014, 126, 5931–5936. [Google Scholar] [CrossRef]
- Huang, F.; You, M.; Chen, T.; Zhu, G.; Liang, H.; Tan, W. Self-assembled hybrid nanoparticles for targeted co-delivery of two drugs into cancer cells. Chem. Commun. 2014, 50, 3103–3105. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, L.; Wang, J.; Jiang, G.; Cheng, H.; Pei, R. The Study of the Interaction between Doxorubicin and Single-Stranded DNA. ChemistrySelect 2016, 1, 3823–3828. [Google Scholar] [CrossRef]
- Richards, A.D.; Rodger, A. Synthetic metallomolecules as agents for the control of DNA structure. Chem. Soc. Rev. 2007, 36, 471–483. [Google Scholar] [CrossRef]
- Zhu, G.; Chen, X. Aptamer-based targeted therapy. Adv. Drug Deliv. Rev. 2018, 134, 65–78. [Google Scholar] [CrossRef]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef]
- Zhu, G.; Niu, G.; Chen, X. Aptamer–drug conjugates. Bioconjugate Chem. 2015, 26, 2186–2197. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Lifson, M.A.; Inci, F.; Liang, L.-G.; Sheng, Y.-F.; Demirci, U. Advances in addressing technical challenges of point-of-care diagnostics in resource-limited settings. Expert Rev. Mol. Diagn. 2016, 16, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Seo, J.-M.; Shin, K.-J.; Yang, S.-G. Design and clinical developments of aptamer-drug conjugates for targeted cancer therapy. Biomater. Res. 2021, 25, 42. [Google Scholar] [CrossRef] [PubMed]
- Testing.com. Point-of-Care Testing. Available online: https://www.testing.com/articles/point-of-care-testing/ (accessed on 27 January 2021).
- Feng, D.; Ren, M.; Miao, Y.; Liao, Z.; Zhang, T.; Chen, S.; Ye, K.; Zhang, P.; Ma, X.; Ni, J. Dual selective sensor for exosomes in serum using magnetic imprinted polymer isolation sandwiched with aptamer/graphene oxide based FRET fluorescent ignition. Biosens. Bioelectron. 2022, 207, 114112. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Qiao, Y.; Gu, D.; Wu, Z.; Zhao, W.; Li, X.; Yin, Y.; Zhao, W.; Kong, D.; Xi, R. Reliable FRET-ON imaging of telomerase in living cells by a tetrahedral DNA nanoprobe integrated with structure-switchable molecular beacon. Sens. Actuators B Chem. 2020, 312, 127943. [Google Scholar] [CrossRef]
- Hasan, M.R.; Sharma, P.; Pilloton, R.; Khanuja, M.; Narang, J. Colorimetric biosensor for the naked-eye detection of ovarian cancer biomarker PDGF using citrate modified gold nanoparticles. Biosens. Bioelectron. X 2022, 11, 100142. [Google Scholar] [CrossRef]
- Shahbazlou, S.V.; Vandghanooni, S.; Dabirmanesh, B.; Eskandani, M.; Hasannia, S. Biotinylated aptamer-based SPR biosensor for detection of CA125 antigen. Microchem. J. 2023, 194, 109276. [Google Scholar] [CrossRef]
- Ni, Y.; Ouyang, H.; Yu, L.; Ling, C.; Zhu, Z.; He, A.; Liu, R. Label-free electrochemical aptasensor based on magnetic α-Fe2O3/Fe3O4 heterogeneous hollow nanorods for the detection of cancer antigen 125. Bioelectrochemistry 2022, 148, 108255. [Google Scholar] [CrossRef]
- Liu, F.; Peng, J.; Lei, Y.-M.; Liu, R.-S.; Jin, L.; Liang, H.; Liu, H.-F.; Ma, S.-Y.; Zhang, X.-H.; Zhang, Y.-P. Electrochemical detection of ctDNA mutation in non-small cell lung cancer based on CRISPR/Cas12a system. Sens. Actuators B Chem. 2022, 362, 131807. [Google Scholar] [CrossRef]
- Khaksari, S.; Ameri, A.R.; Taghdisi, S.M.; Sabet, M.; Bami, S.M.J.G.; Abnous, K.; Shaegh, S.A.M. A microfluidic electrochemical aptasensor for highly sensitive and selective detection of A549 cells as integrin α6β4-containing cell model via IDA aptamers. Talanta 2023, 252, 123781. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Zhao, Q.; Wang, S.; Zhao, S.; Zhang, S.; Yin, Y.; Dong, Y. Development of a lateral flow aptamer assay strip for facile identification of theranostic exosomes isolated from human lung carcinoma cells. Anal. Biochem. 2020, 594, 113591. [Google Scholar] [CrossRef] [PubMed]
- Sanati, A.; Esmaeili, Y.; Khavani, M.; Bidram, E.; Rahimi, A.; Dabiri, A.; Rafienia, M.; Jolfaie, N.A.; Mofrad, M.R.; Javanmard, S.H. Smartphone-assisted lab-in-a-tube device using gold nanocluster-based aptasensor for detection of MUC1-overexpressed tumor cells. Anal. Chim. Acta 2023, 1252, 341017. [Google Scholar] [CrossRef] [PubMed]
- Moutsiopoulou, A.; Broyles, D.; Dikici, E.; Daunert, S.; Deo, S.K. Molecular aptamer beacons and their applications in sensing, imaging, and diagnostics. Small 2019, 15, 1902248. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhao, J.; Zhang, R.; Han, G.; Zhang, C.; Liu, B.; Zhang, Z.; Han, M.-Y.; Gao, X. Cross-platform cancer cell identification using telomerase-specific spherical nucleic acids. ACS Nano 2018, 12, 3629–3637. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Panchal, A.; Yadav, N.; Narang, J. Analytical techniques for the detection of glycated haemoglobin underlining the sensors. Int. J. Biol. Macromol. 2020, 155, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Hasanzadeh, M.; Shadjou, N.; Eskandani, M.; de la Guardia, M.; Omidinia, E. Electrochemical nano-immunosensing of effective cardiac biomarkers for acute myocardial infarction. TrAC Trends Anal. Chem. 2013, 49, 20–30. [Google Scholar] [CrossRef]
- Zhong, Q.; Ding, H.; Gao, B.; He, Z.; Gu, Z. Advances of microfluidics in biomedical engineering. Adv. Mater. Technol. 2019, 4, 1800663. [Google Scholar] [CrossRef]
- Mohammadinejad, A.; Aleyaghoob, G.; Ertas, Y.N. Nanomaterials in Lateral Flow Assay. In Functionalized Smart Nanomaterials for Point-of-Care Testing; Springer: Berlin/Heidelberg, Germany, 2023; pp. 49–81. [Google Scholar]
- Khandan-Nasab, N.; Askarian, S.; Mohammadinejad, A.; Aghaee-Bakhtiari, S.H.; Mohajeri, T.; Oskuee, R.K. Biosensors, microfluidics systems and lateral flow assays for circulating microRNA detection: A review. Anal. Biochem. 2021, 633, 114406. [Google Scholar] [CrossRef]
- ul ain Zahra, Q.; Mohsan, S.A.H.; Shahzad, F.; Qamar, M.; Qiu, B.; Luo, Z.; Zaidi, S.A. Progress in smartphone-enabled aptasensors. Biosens. Bioelectron. 2022, 215, 114509. [Google Scholar]
- Burnett, J.C.; Rossi, J.J. RNA-based therapeutics: Current progress and future prospects. Chem. Biol. 2012, 19, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Mi, J.; Liu, Y.; Rabbani, Z.N.; Yang, Z.; Urban, J.H.; Sullenger, B.A.; Clary, B.M. In vivo selection of tumor-targeting RNA motifs. Nat. Chem. Biol. 2010, 6, 22–24. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Hu, Y.; Yin, L.; Tang, C.; Yin, C. Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials 2010, 31, 3657–3666. [Google Scholar] [CrossRef] [PubMed]

| Method of Delivery | Sequences of Aptamer (5′ to 3′) | Cell or Animal | Marker | Therapeutic Agent | Ref. |
|---|---|---|---|---|---|
| Nanocomplex of ATP aptamer/QDs and MUC-1 aptamer/AuNPs | ATP-aptamer: NH2-AACCTGGGGGAGTATTGCGGAGGAAGGTMUC1-aptamer: 5′-SH-GAAGTGAAAATGACAGAACACAACA-3′ | MCF-7 | Muc-1, ATP | - | [13] |
| EpCAM aptamer-conjugated SWNT/piperazine–polyethylenimine | EpCAM aptamer: GCG ACU GGU UAC CCG GUC G SiRNA: GGAUGUUCAAGAUCCCAUGCAGCTC | MCF-7 | EpCAM | siRNA for suppressing BCL9l | [14] |
| silica coated-Gd-Zn-Cu-In-S/ZnS QDs/PEG/EpCAM DNA | EpCAM aptamer: CAC TAC AGA GGT TGC GTC TGT CCC ACG TTG TCA TGG GGG GTT GGC CTG | 4T1, MCF-7 | EpCAM | DOX | [15] |
| Sgc8 aptamer-modified silica nanoparticles system | Sgc8 aptamer: ATCTAACTGCCGCCGCGGGAAAATGTACGGTTA G(T)10-COOH | CCRF-CEM human acute T lymphocyte leukemia | protein tyrosine kinase-7 (PTK-7) | DOX | [16] |
| As141 aptamer-conjugated pluronic F127/beta-cyclodextrin-linked poly (ethylene glycol)-b-polylactide block copolymers (β-CD-PELA) | As141 aptamer: TTGGTGGTGGTGGTTGTGGTGGTGGTGG | MCF-7, female BALB/c nude mice | Nucleolin | DOX | [17] |
| Encapsulation of aptamer-DOX in liposome | Aptamer AS1411: GGT GGT GGT GGT TGT GGT GGT GGT GGT T | Human breast tumor MCF-7/Adr cells | nucleolin | DOX | [18] |
| irradiation therapy using AgNPs functionalized with PEG and As141 aptamer | As1411 aptamer: (CH2)6-NH2-GGTGGTGGTGGTTGTGGTGGTG GTGG | C6 glioma, C6 glioma-bearing mice | nucleolin | - | [19] |
| Photodynamic therapy by aptamer-conjugated superparamagnetic iron oxide nanoparticles (SPION) loaded by daunomycin (DNM) and 5, 10, 15, 20-tetra (phenyl-4-N-methyl-4-pyridyl) (TMPyP) | As1411 and DNM aptamer: 5′-NH2-GGG GGG GGT TGT CCC CCC CCT TTT TTG GTG GTG GTG GTT GTG GTG GTG GTG G | C26, A549 | nucleolin | DNM, TMPyP | [20] |
| Fe3O4@ UiO-66-NH2 MOF/DOX/CDs/AS1411 aptamer | As1411 aptamer: GGTGGTGGTGGTTGTGGTGGTG GTGG | MDA-MB-231 | nucleolin | DOX | [21] |
| photosensitizer protoporphyrin IX/ AS1411/NaYF4:Yb, Er nanocluster (UCNP) | As1411 aptamer: NH2–GG TGGTGGTGG TTG TGG TGGTGG TGG | MCF7, Hella | nucleolin | photosensitizer protoporphyrin IX produced ROS | [22] |
| assembling five DNA strands to form five-point-star motif | strand I: ATAGTGAGTCGTATTAATTAACCCTCACTAAAAAGGATCCGGATCCTT strand II: TTTAGTGAGGGTTAATCATACGATTTAGGTGAAAGGATCCGGATCCTT strand III: TCACCTAAATCGTATGGGAGCTCTGCTTATATAAGGATCCGGATCCTT strand IV: ATATAAGCAGAGCTCCTAGAAGGCACAGTCGAAAGGATCCGGATCCTT strand V: TCGACTGTGCCTTCTATAATACGACTCACTATAAGGATCCGGATCCTT | MCF7 | MUC1 | DOX | [23] |
| Assembling two strands containing a DNA aptamer with G-quadruplex and double-stranded DNA | CCCCCCCCCCTGTTGGGGGGGGGGTTTTTTTTTGGTGGTGGTGGTTGTGGTGGTGGTGG | MCF7 | MUC1 | DOX | [24] |
| Conjugation of aptamer P19 to gemcitabine, 5-fluorouracil (5-FU), monomethyl auristatin E (MMAE) and derivative of maytansine 1 (DM1) | GGGAGACAAGAAUAAACGCUCAAUGGCGAAUGCCCGCCUAAUAGGGCGUUAUGACUUGUUGAGUUCGACAGGAGGCUCACAACAGGC | PANC-1 AsPC-1 | cells | Gemcitabine 5-FU MMAE DM1 | [25] |
| Covalently binding sgc8ca aptamer–doxorubicin | ATC TAA CTG CTG CGC CGC CGG GAA AAT ACT GTA CGG TTA GA | Human T-cell ALL (CCRF-CEM), human B-cell Burkitt’s lymphoma (Ramos) | kinase 7 (PTK7) | DOX | [26] |
| self-assembly of biotinylated hairpin DNAs followed by streptavidin–aptamer (Zy1) conjugation | H1: Biotin-CGT CGT GCA GCA GCA GCA GCA GCA ACG GCT TGC TGC TGC TGC TGC TGC H2: Biotin-TGC TGC TGC TGC TGC TGC ACG ACG GCA GCA GCA GCA GCA GCA AGC CGT Trigger: TGC TGC TGC TGC TGC TGC ACG ACG Zy1: ACG CGC GCG CGC ATA GCG CGC TGA GCT GAA GAT CGT ACC GTG AGC GCG T(T)10- streptavidin | SMMC-7721 | cell | DOX | [27] |
| self-assembly of 8 sequences of S1–S4 DNA and linker L1–L4 DNA to form nanohydrogels: -unmethylated cytosine-phosphate-guanine oligonucleotides (CpG ODNs) -DNA nanohydrogels (CpG-MUC1-hydrogel) - I-motif cytosine (C)-rich single-stranded DNA | S1(I-motif): TCAACACTAATCCCCAATCCCAATCCCAATCCCAAACG A S2: TCAACACTAATCCGTTTGGGATTGGGACAAAACGACGAA S3(CpG-MUC1): TCAACACTAATCCCCAATCGTCGTTTTGTCGTTTTGTCGTT-S-S GCAGTTGATCCTTTGGATACCCTGG S4: TCAACACTAATCAAAACGACAAAACGATTGGGATTGGGA L1: GATTAGTGTTGAAACGACA-S-S-AAACG L2: ACAAAA-S-S-CGACGAGCCCTCCCCC L3: GATTAGTGTTGAGGGGGAGGGC L4 (CpG): TCGTCGTTTTGTCGTTTTGTCGTT | MCF-7, A549, HepG-2, Female BALB/c (nu/nu) athymic nude mice (5–6 wk) | MUC1 | DOX | [28] |
| antimiR-21 DNAzyme linked to the aptamer of low-density lipoprotein receptor (LDL-R) | TCA ACA GGC TAG CTA CAA CGA CAG TCT GAT AAG CTA TTTTTA GGA CAG GAC CAC ACC CAG CGC GGT CGG CGG GTG GGC GGG GGG AGA ACG AGG TAG GG | Huh-7, MDA-MB-231 | LDL-R | antimiR-21 DNAzyme | [29] |
| Covalently bonding paclitaxel (PTX) to the nucleolin AS1411 aptamer (NucA) through dipeptide bond | TTGGTGGTGGTGGTTGTGGTGGTGGTGG | SKOV3 (ATCC HTB-77), OVCAR3 (ATCC HTB-161) | Nucleolin and cathepsin | PTX | [30] |
| HER2- aptamer-conjugated to the mertansine (DM1) | AGC CGCGAG GGG AGG GAU AGG GUA GGG CGC GGC U | BT-474, MDA-MB-231 MCF-7, A549, mouse xenografts with BT-474 breast cancer | HER2 | DM1 | [31] |
| conjugation of HER2 aptamer to siRNAs targeting Bcl-2 | GGGAGGACGAUGCGGCGAUGCUUACGUGCACGCGCCAGACGACUCGCCCGAGCUGUCACAGAGGGGCUACUU | N202.1A | HER2 | siRNAs targeting Bcl-2 | [32] |
| 18F-fluoride- HER2 aptamer | TCCTGGCATGTTCGATGGAGGCCTTTGATTACAGCCCAGA | tumor-bearing mice | HER2 | 18F-fluoride- HER2 aptamer | [33] |
| Method | Sequence | Biomarker | Linear Range | LOD | Ref. |
|---|---|---|---|---|---|
| Exosomes on the molecularly imprinted polymer (MIP)-coated Fe3O4 release the aptamer-FAM from GO | CD63: FAM-CACCCCACCTCGCTCCCGTGACACTAATGCTAMUC1: FAM-CAGCCTGCACTCTAACGCAGTTGATCCTTTGGATAGCCTGGGTTAGA | CD63, MUC1 | 1.19 × 10−6–4.76 × 10−5 mol/L | 2.27 × 10−6 mol/L | [79] |
| tetrahedral DNA (L1–L4) hybridized with MAB | L1: ACATTCCTAAGTCTGAAACATTACAGCTTGCTACACGAGAAGA GCCGCCATAGTA L2: TCAACTGCCTGGTGATAAAACGACACTACGTGGGAATCTACTA TGGCGGCTCTTCTTTTTAATCCGTCGAGCAGAGTT L3: TATCACCAGGCAGTTGACAGTGTAGCAAGCTGTAATAGATGCG AGGGTCCAATACTTTTT/iBHQ2dT/TGCACCCTAACCCTAACCCT L4: TTCAGACTTAGGAATGTGCTTCCCACGTAGTGTCGTTTGTATTG GACCCTCGCAT MB: Cy3-AACGATTAGGGTTAGGGTCGTT-Cy5 | telomerase | 50–2000 HeLa cells | 35 HeLa cells | [80] |
| Prevention of AuNP aggregation by aptamer | CCGATCTCTCCCACTCTCTCCAACTCACAGGCTACGGCACGTAGAGCATCACCATGATCCTGTGGGTGTGTTGTTGATGGATCGGATCATCATGGTGAT | platelet-derived growth factor (PDGF) | 0.01–10 μg/mL | 0.01 μg/mL | [81] |
| Gold chip modified with CA125 aptamer through streptavidin–biotin | CTC ACT ATA GGG AGA CAA GAA TAA ACG CTC AA-biotin | Mucin 16 (MUC16) or cancer antigen 125 (CA125) | 10–100 U/mL | 0.01 U/mL | [82] |
| Magnetic glass carbon electrode (MGCE)/α-Fe2O3/Fe3O4/Au/complementary strand/aptamer | Complementary strand: SH-TTTTTTTTTTTTTTTTTTTTCCCTATAGTGAG Aptamer: CTCACTATAGGGAGACAAGAATAAACGCTCAA | cancer antigen 125 (CA125) | 5–125 U/mL | 2.99 U/mL | [83] |
| CRISPR/Cas12a + RPA + electrochemistry (Modified electrode with complementary 1 (CP1) and MB/Fe3O4@COF/PdAu modified with complementary 2 (CP2)) | crRNA template: AAGTACCCAGCAGTTTGGCCCGCCATCTACACTTAGTAGAAATTCCtatagtgagtcgtattag CP1: SH-ACACTTGAAGTGTATTTCCTAAATA CP2: AATTGCAAGTATGTAGAAGTTCACA-SH Synthetic target: ATGTCAAGATCACAGATTTTGGGCTGGCCAAACTGCTGGGTGCG | ctDNA EGFR L858R | 10 aM–100 pM | 3.3 aM | [84] |
| Microfluidic system incorporated with screen-printed gold electrode was modified with integrin α6β4-specific aptamer (IDA) | GCCTGTTGTGAGCCTCCTAACCGTGCGTATTCGTACTGGAACTGATATCGATGTCCCCATGCTTATTCTTGTCTCCC–SH | α6β4 integrin on A549 cells | 50–5 × 105 cells/mL | 14 cells/mL | [85] |
| Lateral flow assay (LFA) with streptavidin (SA)-biotin-CD63 aptamer on T-line | CD63 aptamer: 5′-GTGGGGTGGACGAGGGCACGTGATTACGTA-3′ complement aptamer: CACCCCACCTCGCTCCCGTGACACTAATGCTA -Biotin | CD63 on the non-small cell lung cancer (NSCLC) | - | 6.4 × 109 particles/mL | [86] |
| Smartphone control of emission from gold nanocluster (GNC)-aptamer as emitter and polyurethane (PU) coated with GO as quencher | SH-CCCCCCGATCCTTTGGATA | Mucin 1 (MUC1) | 250–20,000 cells/mL | 221 cells/mL | [87] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammadinejad, A.; Gaman, L.E.; Aleyaghoob, G.; Gaceu, L.; Mohajeri, S.A.; Moga, M.A.; Badea, M. Aptamer-Based Targeting of Cancer: A Powerful Tool for Diagnostic and Therapeutic Aims. Biosensors 2024, 14, 78. https://doi.org/10.3390/bios14020078
Mohammadinejad A, Gaman LE, Aleyaghoob G, Gaceu L, Mohajeri SA, Moga MA, Badea M. Aptamer-Based Targeting of Cancer: A Powerful Tool for Diagnostic and Therapeutic Aims. Biosensors. 2024; 14(2):78. https://doi.org/10.3390/bios14020078
Chicago/Turabian StyleMohammadinejad, Arash, Laura Elena Gaman, Ghazaleh Aleyaghoob, Liviu Gaceu, Seyed Ahmad Mohajeri, Marius Alexandru Moga, and Mihaela Badea. 2024. "Aptamer-Based Targeting of Cancer: A Powerful Tool for Diagnostic and Therapeutic Aims" Biosensors 14, no. 2: 78. https://doi.org/10.3390/bios14020078
APA StyleMohammadinejad, A., Gaman, L. E., Aleyaghoob, G., Gaceu, L., Mohajeri, S. A., Moga, M. A., & Badea, M. (2024). Aptamer-Based Targeting of Cancer: A Powerful Tool for Diagnostic and Therapeutic Aims. Biosensors, 14(2), 78. https://doi.org/10.3390/bios14020078









