Post-Assay Chemical Enhancement for Highly Sensitive Lateral Flow Immunoassays: A Critical Review
Abstract
:1. Introduction
Factors Determining the LOD of LFIA
2. Post-Assay Chemical Enhancement Approaches
- Approaches focused on the modification of physicochemical properties of the nanosized labels. The ultimate goal is increasing the “visibility” of nanoparticles on the membrane. Among these properties are size, shape, and chemical composition affecting the optical properties of nanoparticles [26,27]. Further in this paper, we focus on GNPs as the most widely used nanolabel in LFIA. The intensity of the coloration of GNPs on the membrane is determined by light absorption and scattering, and the impact of each parameter is determined by the size of the particles [23]. Larger GNPs have a higher scattering intensity and molar extinction coefficient, which provides higher coloration for the given number of particles [28,29,30]. Non-spherical GNPs also have a higher extinction coefficient than similarly sized spherical GNPs [31]. Changing the chemical composition of nanoparticles by in situ formation of metal shells (Cu, Ag, Pt) over initial GNPs also increases the molar extinction coefficient and facilitates highly sensitive detection [32]. The signal amplification is based on in situ formation of particles that can be detected at lower concentrations than initial GNPs.
- Approaches focused on the increase of the label number are aimed at the accumulation of an additional amount of labels (driven by non-covalent binding between labels) or by catalytic conversion of a substrate to the detectable product (usually its oxidation to a colored or fluorescent product) [25]. The signal amplification is based on increasing the concentration of the registered product of the catalytic reaction.
2.1. Modification of Physicochemical Properties of Nanoparticles
2.1.1. Gold Enhancement
2.1.2. Silver Enhancement
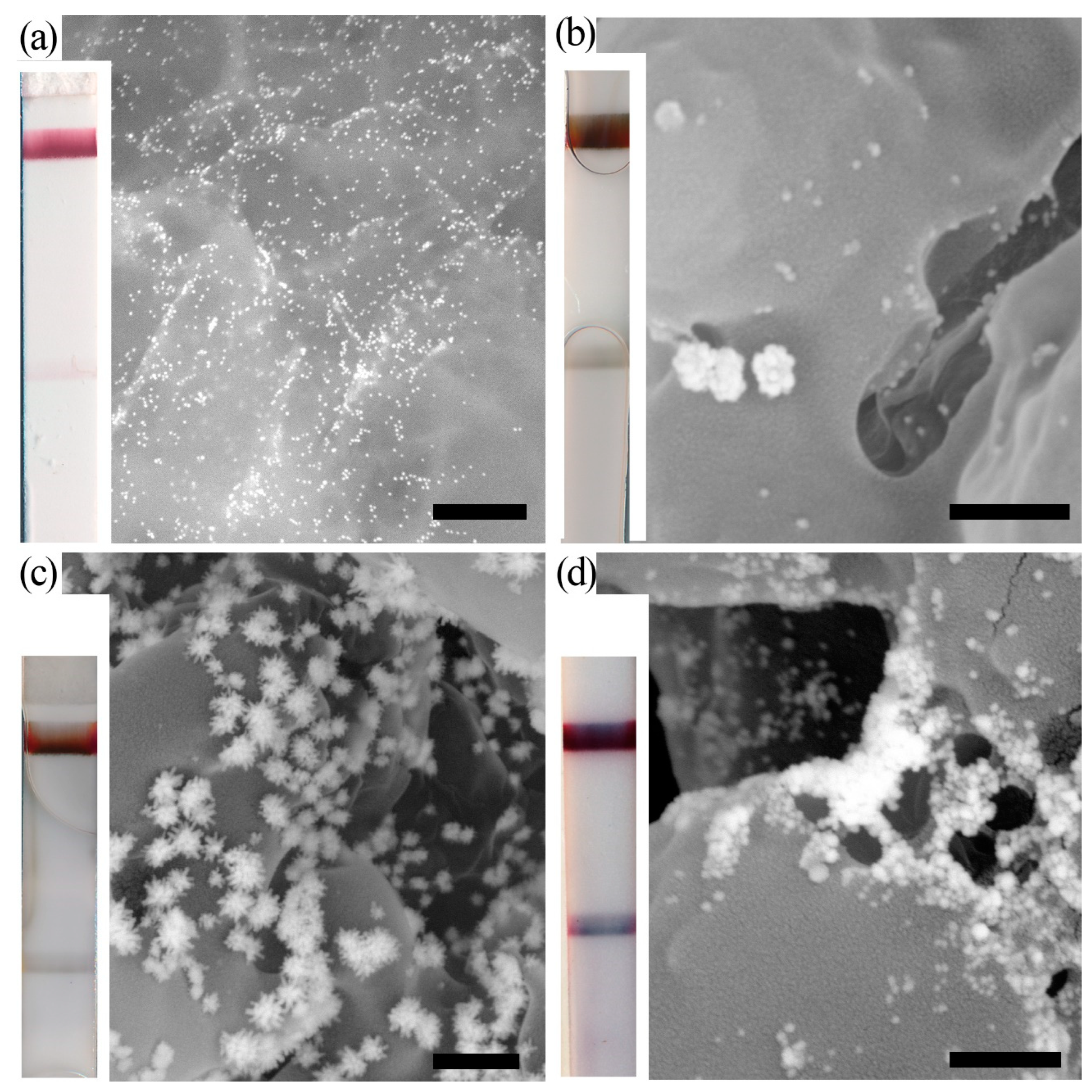
2.1.3. Copper Enhancement
2.1.4. Galvanic Replacement
2.2. Increasing the Number of Labels
2.2.1. Crosslinking of Nanoparticles
2.2.2. Catalytic Accumulation of the Label
Enzymes as the Catalytic Labels
Nanozymes as the Catalytic Label
3. Post-Assay-Integrated Signal Amplification
4. Comparison and Assessment of the Enhancement Approaches
4.1. Quantitative Evaluation of the Improvements Reached by Different Enhancement Approaches
4.2. Discrepancy in LOD-Reduction Values
4.3. Proposal of Additional Studies in LFIA with Post-Assay Enhancement
- (a)
- Comparison of various enhancement approaches performed with the same antigen and immunoreagents.
- (b)
- Comparison of one enhancement approach for various antigens in different matrices performed by the same group.
- (c)
- Quantitative comparison of research (a) and/or (b) performed with an interval of time.
- (d)
- Comparison of the enhancement approach for one target using the same reagents performed by different groups.
5. Development and Validation of Enhanced LFIA
- Determination of the LOD value before enhancement for conventional LFIA in the matrix (Figure 14a). LOD is a quantitative value that is determined as the target concentration that facilitates the coloration higher than the coloration of the blank probe (Ablank) plus three standard deviations of blank (SDblank). From our experience, LFIA with even minimal Ablank is poorly applicable for post-assay signal enhancement, as it results in a high background. Often visual LOD of LFIA is reported. Visual LOD is determined as the lowest analyte concentration facilitating detectable by bare-eye coloration of the test zone. Although visual evaluation of test strips remains common in practice (e.g., pregnancy test screening, SARS-CoV-2 antigen rapid tests), for scientific developments, LOD quantification is recommended. Visual LOD determination will be subjective and depend on the visual acuity, brightness of environment light, matrix coloration, etc. Additional optimization for elimination Ablank before proceeding further with post-assay chemical enhancement. Determining LOD in the matrix, not the buffer, is important, as matrix components may cause higher Ablank. To perform the first stage, the matrix is spiked with the known concentrations of the target. Each concentration is measured at least in three repeats. As a result of this step, the developer must have the LOD value of LFIA in the matrix.
- Optimization of chemical enhancement (Figure 14b). At least two parameters need to be optimized—concentrations of enhancing reagents (Figure 14b(I)) and reaction time (Figure 14b(II)). For the particular enhancement, other variables (e.g., pH of the solution, presence of stabilized agents, method of signal enhancing solution introduction, temperature, etc.) need to be optimized, but here we focus on general parameters. Optimization of the signal enhancing reagents can be performed within the recommended ranges of concentrations and reaction time (Figure 11c,d). For these experiments, at least two samples are used. The first sample should contain the target in a concentration close to the LOD value. The second sample should be an unspiked matrix (blank probe). The goal of the optimization is to find the conditions that facilitate the highest signal enhancement for the first sample (marked with an asterisk) while keeping no signal for the second sample.
- Determination of the LOD value after enhancement (Figure 14c). The LOD value for the optimized procedure (second stage) is determined as described in the first stage. Importantly, the developers should use Ablank and SDblank values for enhanced LFIA, not for conventional, otherwise, the LOD value will be miscalculated. LOD reduction is calculated by dividing LOD values before and after enhancement. It is crucial to understand which LOD values can be compared for reporting enhancement effect. Often authors compare enhanced LFIA with “conventional” GNP-based LFIA, commercial test systems, or even LFIA published in other publications. We claim that LOD values should be compared exclusively between LFIA assembled with the same membranes and immunoreagents. Thus, all comparisons with literature and commercial test systems are eligible to show the superior analytical performance of developed LFIA but not the enhancement strategy. For reporting the benefits of the enhancement approach, it is necessary to compare LOD values within a single study. Also, one needs to understand the principles of signal amplification to compare LOD values before and after accurately. For example, for gold/silver/copper enhancement, one needs to compare the LOD values of LFIA with GNPs before and after the reduction of corresponding salts. Ideally, the same test strips should be used. For nanozyme signal amplification, one needs to compare LOD values before and after the addition of the substrate. The comparison of LOD values for nanozymes after catalysis with GNP will not be accurate because nanozymes as the optical label (before catalysis) may have different LOD values compared to GNPs. Thus, the developers need to clearly understand the principle of signal amplification and only compare LOD values before and after amplification within one strategy. To perform this stage, the developer spikes the matrix with the known concentration of the target and prepares the number of consequent dilutions (titration) as described for stage one. Ideally, calibration plots for LFIA before (stage 1) and after enhancement (stage 3) and LOD calculations should be performed in parallel using the same stock solution. Such performance in parallel will reduce the impact of determinate and indeterminate errors.
- Determination of the accuracy of LFIA before and after enhancement using the spiked matrix (Figure 14d). Using the calibration plots (stages 1 and 3 for LFIAs before and after enhancement, respectively), the developer determines the linear range based on correlation coefficient (R2 ≥ 0.9×) [276]. After that, multiple artificially spiked matrix samples are prepared with the target concentration within the linear range. Then, conventional and enhanced LFIAs are used to quantify the target concentrations. The results can be shown as added–detected (in percent) or in a graphical way (added concentration vs. detected concentration). Values close to 100% in added–detected or R2 ≥ 0.99 for a graphical representation are expected for accurate LFIA.
- Validation of conventional and enhanced LFIAs using real samples (Figure 14e). This stage aims to confirm the practical benefits of the enhanced LFIA over conventional LFIA. As the practical benefits, we understand the ability of enhanced LFIA to detect low-positive samples, while conventional LFIA report false negative results due to insufficiently low LOD value. To perform this stage, real samples containing the target in a wide range of concentrations (true positive) and without the target (true negative) are used. The concentration of the target (or at least the qualitative results) should be confirmed by an independent method (PCR, ELISA). The results of the independent method are considered the reference—i.e., true positive (presence of the target) and true negative (absence of the target). Ideally, the developer should be blinded and perform LFIA without knowing the results of the independent method. The qualitative (number of true positive/negative and number of false positive/negative) and quantitative (concentration of the target) results of conventional and enhanced LFIAs are compared with the independent method. One expects a higher number of true positives (lower number of false negatives) for enhanced LFIA compared with conventional LFIA because of lower LOD. Also, the number of false positives should be the same, meaning that the enhancement procedure does not sacrifice the specificity of the assay.
6. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Di Nardo, F.; Chiarello, M.; Cavalera, S.; Baggiani, C.; Anfossi, L. Ten Years of Lateral Flow Immunoassay Technique Applications: Trends, Challenges and Future Perspectives. Sensors 2021, 21, 5185. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, J.; Xu, X.; Guo, L.; Xu, L.; Sun, M.; Hu, S.; Kuang, H.; Xu, C.; Li, A. An Overview for the Nanoparticles-Based Quantitative Lateral Flow Assay. Small Methods 2022, 6, 1–29. [Google Scholar] [CrossRef]
- Huang, Y.; Xu, T.; Wang, W.; Wen, Y.; Li, K.; Qian, L.; Zhang, X.; Liu, G. Lateral Flow Biosensors Based on the Use of Micro- and Nanomaterials: A Review on Recent Developments. Microchim. Acta 2020, 187, 70. [Google Scholar] [CrossRef] [PubMed]
- Urusov, A.E.; Zherdev, A.V.; Dzantiev, B.B. Towards Lateral Flow Quantitative Assays: Detection Approaches. Biosensors 2019, 9, 89. [Google Scholar] [CrossRef]
- Sena-Torralba, A.; Álvarez-Diduk, R.; Parolo, C.; Piper, A.; Merkoçi, A. Toward Next Generation Lateral Flow Assays: Integration of Nanomaterials. Chem. Rev. 2022, 122, 14881–14910. [Google Scholar] [CrossRef] [PubMed]
- Hristov, D.; Rodriguez-Quijada, C.; Gomez-Marquez, J.; Hamad-Schifferli, K. Designing Paper-Based Immunoassays for Biomedical Applications. Sensors 2019, 19, 554. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhan, L.; Qin, Z.; Sackrison, J.; Bischof, J.C. Ultrasensitive and Highly Specific Lateral Flow Assays for Point-of-Care Diagnosis. ACS Nano 2021, 15, 3593–3611. [Google Scholar] [CrossRef] [PubMed]
- Budd, J.; Miller, B.S.; Weckman, N.E.; Cherkaoui, D.; Huang, D.; Decruz, A.T.; Fongwen, N.; Han, G.-R.; Broto, M.; Estcourt, C.S.; et al. Lateral Flow Test Engineering and Lessons Learned from COVID-19. Nat. Rev. Bioeng. 2023, 1, 13–31. [Google Scholar] [CrossRef]
- Dzantiev, B.B.; Byzova, N.A.; Urusov, A.E.; Zherdev, A.V. Immunochromatographic Methods in Food Analysis. TrAC Trends Anal. Chem. 2014, 55, 81–93. [Google Scholar] [CrossRef]
- Bishop, J.D.; Hsieh, H.V.; Gasperino, D.J.; Weigl, B.H. Sensitivity Enhancement in Lateral Flow Assays: A Systems Perspective. Lab Chip 2019, 19, 2486–2499. [Google Scholar] [CrossRef]
- Zherdev, A.V.; Dzantiev, B.B. Detection Limits of Immunoanalytical Systems: Limiting Factors and Methods of Reduction. J. Anal. Chem. 2022, 77, 391–401. [Google Scholar] [CrossRef]
- Anfossi, L.; Di Nardo, F.; Cavalera, S.; Giovannoli, C.; Baggiani, C. Multiplex Lateral Flow Immunoassay: An Overview of Strategies towards High-Throughput Point-of-Need Testing. Biosensors 2018, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Zherdev, A.V.; Dzantiev, B.B. Ways to Reach Lower Detection Limits of Lateral Flow Immunoassays. In Rapid Test—Advances in Design, Format and Diagnostic Applications; IntechOpen: London, UK, 2018; ISBN 978-953-51-5953-7. [Google Scholar]
- Foote, J.; Eisen, H.N. Kinetic and Affinity Limits on Antibodies Produced during Immune Responses. Proc. Natl. Acad. Sci. USA 1995, 92, 1254–1256. [Google Scholar] [CrossRef] [PubMed]
- Batista, F.D.; Neuberger, M.S. Affinity Dependence of the B Cell Response to Antigen: A Threshold, a Ceiling, and the Importance of Off-Rate. Immunity 1998, 8, 751–759. [Google Scholar] [CrossRef]
- Razo, S.; Panferov, V.; Safenkova, I.; Varitsev, Y.; Zherdev, A.; Pakina, E.; Dzantiev, B. How to Improve Sensitivity of Sandwich Lateral Flow Immunoassay for Corpuscular Antigens on the Example of Potato Virus Y? Sensors 2018, 18, 3975. [Google Scholar] [CrossRef]
- Rivas, L.; Medina-Sánchez, M.; de la Escosura-Muñiz, A.; Merkoçi, A. Improving Sensitivity of Gold Nanoparticle-Based Lateral Flow Assays by Using Wax-Printed Pillars as Delay Barriers of Microfluidics. Lab Chip 2014, 14, 4406–4414. [Google Scholar] [CrossRef]
- Shirshahi, V.; Liu, G. Enhancing the Analytical Performance of Paper Lateral Flow Assays: From Chemistry to Engineering. TrAC Trends Anal. Chem. 2021, 136, 116200. [Google Scholar] [CrossRef]
- Kim, K.; Kashefi-Kheyrabadi, L.; Joung, Y.; Kim, K.; Dang, H.; Chavan, S.G.; Lee, M.-H.; Choo, J. Recent Advances in Sensitive Surface-Enhanced Raman Scattering-Based Lateral Flow Assay Platforms for Point-of-Care Diagnostics of Infectious Diseases. Sens. Actuators B Chem. 2021, 329, 129214. [Google Scholar] [CrossRef]
- Gong, X.; Cai, J.; Zhang, B.; Zhao, Q.; Piao, J.; Peng, W.; Gao, W.; Zhou, D.; Zhao, M.; Chang, J. A Review of Fluorescent Signal-Based Lateral Flow Immunochromatographic Strips. J. Mater. Chem. B 2017, 5, 5079–5091. [Google Scholar] [CrossRef]
- Moyano, A.; Serrano-Pertierra, E.; Salvador, M.; Martínez-García, J.C.; Rivas, M.; Blanco-López, M.C. Magnetic Lateral Flow Immunoassays. Diagnostics 2020, 10, 288. [Google Scholar] [CrossRef]
- Qin, Z.; Chan, W.C.W.; Boulware, D.R.; Akkin, T.; Butler, E.K.; Bischof, J.C. Significantly Improved Analytical Sensitivity of Lateral Flow Immunoassays by Using Thermal Contrast. Angew. Chem. Int. Ed. 2012, 51, 4358–4361. [Google Scholar] [CrossRef] [PubMed]
- Khlebtsov, B.N.; Tumskiy, R.S.; Burov, A.M.; Pylaev, T.E.; Khlebtsov, N.G. Quantifying the Numbers of Gold Nanoparticles in the Test Zone of Lateral Flow Immunoassay Strips. ACS Appl. Nano Mater. 2019, 2, 5020–5028. [Google Scholar] [CrossRef]
- Deng, Y.; Jiang, H.; Li, X.; Lv, X. Recent Advances in Sensitivity Enhancement for Lateral Flow Assay. Microchim. Acta 2021, 188, 379. [Google Scholar] [CrossRef] [PubMed]
- Panferov, V.G.; Safenkova, I.V.; Zherdev, A.V.; Dzantiev, B.B. Methods for Increasing Sensitivity of Immunochromatographic Test Systems with Colorimetric Detection (Review). Appl. Biochem. Microbiol. 2021, 57, 143–151. [Google Scholar] [CrossRef]
- Khlebtsov, N.G. Determination of Size and Concentration of Gold Nanoparticles from Extinction Spectra. Anal. Chem. 2008, 80, 6620–6625. [Google Scholar] [CrossRef] [PubMed]
- Zhan, L.; Guo, S.; Song, F.; Gong, Y.; Xu, F.; Boulware, D.R.; McAlpine, M.C.; Chan, W.C.W.; Bischof, J.C. The Role of Nanoparticle Design in Determining Analytical Performance of Lateral Flow Immunoassays. Nano Lett. 2017, 17, 7207–7212. [Google Scholar] [CrossRef] [PubMed]
- Dolinnyi, A.I. Extinction Coefficients of Gold Nanoparticles and Their Dimers. Dependence of Optical Factor on Particle Size. Colloid J. 2017, 79, 611–620. [Google Scholar] [CrossRef]
- Li, J.; Duan, H.; Xu, P.; Huang, X.; Xiong, Y. Effect of Different-Sized Spherical Gold Nanoparticles Grown Layer by Layer on the Sensitivity of an Immunochromatographic Assay. RSC Adv. 2016, 6, 26178–26185. [Google Scholar] [CrossRef]
- Jain, P.K.; Lee, K.S.; El-Sayed, I.H.; El-Sayed, M.A. Calculated Absorption and Scattering Properties of Gold Nanoparticles of Different Size, Shape, and Composition: Applications in Biological Imaging and Biomedicine. J. Phys. Chem. B 2006, 110, 7238–7248. [Google Scholar] [CrossRef]
- de Puig, H.; Tam, J.O.; Yen, C.-W.; Gehrke, L.; Hamad-Schifferli, K. Extinction Coefficient of Gold Nanostars. J. Phys. Chem. C 2015, 119, 17408–17415. [Google Scholar] [CrossRef]
- Chen, X.; Ding, L.; Huang, X.; Xiong, Y. Tailoring Noble Metal Nanoparticle Designs to Enable Sensitive Lateral Flow Immunoassay. Theranostics 2022, 12, 574–602. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.R.; Lyon, L.A.; Fox, A.P.; Reiss, B.D.; Natan, M.J. Hydroxylamine Seeding of Colloidal Au Nanoparticles. 3. Controlled Formation of Conductive Au Films. Chem. Mater. 2000, 12, 314–323. [Google Scholar] [CrossRef]
- Penner, R.M. Brownian Dynamics Simulations of the Growth of Metal Nanocrystal Ensembles on Electrode Surfaces in Solution: 2. The Effect of Deposition Rate on Particle Size Dispersion. J. Phys. Chem. B 2001, 105, 8672–8678. [Google Scholar] [CrossRef]
- Scharifker, B. Theoretical and Experimental Studies of Multiple Nucleation. Electrochim. Acta 1982, 28, 879–889. [Google Scholar] [CrossRef]
- Brown, K.R.; Natan, M.J. Hydroxylamine Seeding of Colloidal Au Nanoparticles in Solution and on Surfaces. Langmuir 1998, 14, 726–728. [Google Scholar] [CrossRef]
- Panferov, V.G.; Safenkova, I.V.; Zherdev, A.V.; Dzantiev, B.B. Post-Assay Growth of Gold Nanoparticles as a Tool for Highly Sensitive Lateral Flow Immunoassay. Application to the Detection of Potato Virus X. Microchim. Acta 2018, 185, 506. [Google Scholar] [CrossRef]
- Panferov, V.G.; Samokhvalov, A.V.; Safenkova, I.V.; Zherdev, A.V.; Dzantiev, B.B. Study of Growth of Bare and Protein-Modified Gold Nanoparticles in the Presence of Hydroxylamine and Tetrachloroaurate. Nanotechnol. Russ. 2018, 13, 614–622. [Google Scholar] [CrossRef]
- Festag, G.; Steinbrück, A.; Csáki, A.; Möller, R.; Fritzsche, W. Single Particle Studies of the Autocatalytic Metal Deposition onto Surface-Bound Gold Nanoparticles Reveal a Linear Growth. Nanotechnology 2007, 18, 015502. [Google Scholar] [CrossRef]
- Panferov, V.G.; Byzova, N.A.; Biketov, S.F.; Zherdev, A.V.; Dzantiev, B.B. Comparative Study of In Situ Techniques to Enlarge Gold Nanoparticles for Highly Sensitive Lateral Flow Immunoassay of SARS-CoV-2. Biosensors 2021, 11, 229. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, P.; Ye, Y.; Hammock, B.D.; Zhang, C. An Integrated Approach to Improve the Assay Performance of Quantum Dot-Based Lateral Flow Immunoassays by Using Silver Deposition. Microchem. J. 2023, 192, 108932. [Google Scholar] [CrossRef]
- Dias, J.T.; Svedberg, G.; Nystrand, M.; Andersson-Svahn, H.; Gantelius, J. Rapid Signal Enhancement Method for Nanoprobe-Based Biosensing. Sci. Rep. 2017, 7, 6837. [Google Scholar] [CrossRef] [PubMed]
- Shu, R.; Liu, S.; Xu, J.; Wang, S.; Ma, Y.; Chen, Y.; Li, Y.; Sun, J.; Zhang, D.; Wang, J. Galvanic Replacement Inspired Signal Amplification: Background-Free and Antibody-Thrift in-Situ Growth Immunochromatography. Chem. Eng. J. 2022, 437, 135362. [Google Scholar] [CrossRef]
- Xia, X.; Wang, Y.; Ruditskiy, A.; Xia, Y. 25th Anniversary Article: Galvanic Replacement: A Simple and Versatile Route to Hollow Nanostructures with Tunable and Well-Controlled Properties. Adv. Mater. 2013, 25, 6313–6333. [Google Scholar] [CrossRef] [PubMed]
- de la Rica, R.; Stevens, M.M. Plasmonic ELISA for the Ultrasensitive Detection of Disease Biomarkers with the Naked Eye. Nat. Nanotechnol. 2012, 7, 821–824. [Google Scholar] [CrossRef]
- Cao, C.; Li, X.; Lee, J.; Sim, S.J. Homogenous Growth of Gold Nanocrystals for Quantification of PSA Protein Biomarker. Biosens. Bioelectron. 2009, 24, 1292–1297. [Google Scholar] [CrossRef]
- Morris, R.E.; Ciraolo, G.M.; Saelinger, C.B. Gold Enhancement of Gold-Labeled Probes: Gold-Intensified Staining Technique (GIST). J. Histochem. Cytochem. 1991, 39, 1585–1591. [Google Scholar] [CrossRef]
- Zayats, M.; Baron, R.; Popov, I.; Willner, I. Biocatalytic Growth of Au Nanoparticles: From Mechanistic Aspects to Biosensors Design. Nano Lett. 2005, 5, 21–25. [Google Scholar] [CrossRef]
- Wang, X.; Niessner, R.; Knopp, D. Controlled Growth of Immunogold for Amplified Optical Detection of Aflatoxin B1. Analyst 2015, 140, 1453–1458. [Google Scholar] [CrossRef]
- Cid-Barrio, L.; Ruiz Encinar, J.; Costa-Fernández, J.M. Catalytic Gold Deposition for Ultrasensitive Optical Immunosensing of Prostate Specific Antigen. Sensors 2020, 20, 5287. [Google Scholar] [CrossRef]
- Duan, H.; Ma, T.; Huang, X.; Gao, B.; Zheng, L.; Chen, X.; Xiong, Y.; Chen, X. Avoiding the Self-Nucleation Interference: A PH-Regulated Gold in Situ Growth Strategy to Enable Ultrasensitive Immunochromatographic Diagnostics. Theranostics 2022, 12, 2801–2810. [Google Scholar] [CrossRef]
- Fan, A.; Cai, S.; Cao, Z.; Lau, C.; Lu, J. Hydroxylamine-Amplified Gold Nanoparticles for the Homogeneous Detection of Sequence-Specific DNA. Analyst 2010, 135, 1400–1405. [Google Scholar] [CrossRef]
- Ma, Z.; Sui, S.F. Naked-Eye Sensitive Detection of Immunoglubulin G by Enlargement of Au Nanoparticles in Vitro. Angew. Chem. Int. Ed. 2002, 41, 2176–2179. [Google Scholar] [CrossRef]
- Habib, A.; Tabata, M.; Wu, Y.G. Formation of Gold Nanoparticles by Good’s Buffers. Bull. Chem. Soc. Jpn. 2005, 78, 262–269. [Google Scholar] [CrossRef]
- Panda, B.R.; Chattopadhyay, A. Synthesis of Au Nanoparticles at “All” PH by H2O2 Reduction of HAuCl4. J. Nanosci. Nanotechnol. 2007, 7, 1911–1915. [Google Scholar] [CrossRef]
- Presnova, G.V.; Zhdanov, G.A.; Filatova, L.Y.; Ulyashova, M.M.; Presnov, D.E.; Rubtsova, M.Y. Improvement of Seed-Mediated Growth of Gold Nanoparticle Labels for DNA Membrane-Based Assays. Biosensors 2022, 13, 2. [Google Scholar] [CrossRef]
- Ruantip, S.; Pimpitak, U.; Rengpipat, S.; Pasomsub, E.; Seepiban, C.; Gajanandana, O.; Torvorapanit, P.; Hirankarn, N.; Jaru-ampornpan, P.; Siwamogsatham, S.; et al. Self-Enhancement Lateral Flow Immunoassay for COVID-19 Diagnosis. Sens. Actuators B Chem. 2023, 389, 133898. [Google Scholar] [CrossRef]
- Thangavelu, R.M.; Kadirvel, N.; Balasubramaniam, P.; Viswanathan, R. Ultrasensitive Nano-Gold Labelled, Duplex Lateral Flow Immunochromatographic Assay for Early Detection of Sugarcane Mosaic Viruses. Sci. Rep. 2022, 12, 4144. [Google Scholar] [CrossRef]
- Li, J.; Zou, M.; Chen, Y.; Xue, Q.; Zhang, F.; Li, B.; Wang, Y.; Qi, X.; Yang, Y. Gold Immunochromatographic Strips for Enhanced Detection of Avian Influenza and Newcastle Disease Viruses. Anal. Chim. Acta 2013, 782, 54–58. [Google Scholar] [CrossRef]
- Zhao, M.; Yao, X.; Liu, S.; Zhang, H.; Wang, L.; Yin, X.; Su, L.; Xu, B.; Wang, J.; Lan, Q.; et al. Antibiotic and Mammal IgG Based Lateral Flow Assay for Simple and Sensitive Detection of Staphylococcus Aureus. Food Chem. 2021, 339, 127955. [Google Scholar] [CrossRef]
- Bu, T.; Huang, Q.; Yan, L.; Huang, L.; Zhang, M.; Yang, Q.; Yang, B.; Wang, J.; Zhang, D. Ultra Technically-Simple and Sensitive Detection for Salmonella Enteritidis by Immunochromatographic Assay Based on Gold Growth. Food Control 2018, 84, 536–543. [Google Scholar] [CrossRef]
- Wang, J.Y.; Chen, M.H.; Sheng, Z.C.; Liu, D.F.; Wu, S.S.; Lai, W.H. Development of Colloidal Gold Immunochromatographic Signal-Amplifying System for Ultrasensitive Detection of Escherichia coli O157:H7 in Milk. RSC Adv. 2015, 5, 62300–62305. [Google Scholar] [CrossRef]
- Sharma, A.; Tok, A.I.Y.; Alagappan, P.; Liedberg, B. Gold Nanoparticle Conjugated Magnetic Beads for Extraction and Nucleation Based Signal Amplification in Lateral Flow Assaying. Sens. Actuators B Chem. 2020, 312, 127959. [Google Scholar] [CrossRef]
- Oh, H.-K.; Kim, K.; Park, J.; Jang, H.; Kim, M.-G. Advanced Trap Lateral Flow Immunoassay Sensor for the Detection of Cortisol in Human Bodily Fluids. Sci. Rep. 2021, 11, 22580. [Google Scholar] [CrossRef] [PubMed]
- Schulz, F.; Homolka, T.; Bastús, N.G.; Puntes, V.; Weller, H.; Vossmeyer, T. Little Adjustments Significantly Improve the Turkevich Synthesis of Gold Nanoparticles. Langmuir 2014, 30, 10779–10784. [Google Scholar] [CrossRef]
- Danscher, G. Histochemical Demonstration of Heavy Metals—A Revised Version of the Sulphide Silver Method Suitable for Both Light and Electronmicroscopy. Histochemistry 1981, 71, 1–16. [Google Scholar] [CrossRef]
- Newman, G.R.; Jasani, B. Silver Development in Microscopy and Bioanalysis: A New Versatile Formulation for Modern Needs. Histochem. J. 1998, 30, 635–646. [Google Scholar] [CrossRef]
- Wada, A.; Sakoda, Y.; Oyamada, T.; Kida, H. Development of a Highly Sensitive Immunochromatographic Detection Kit for H5 Influenza Virus Hemagglutinin Using Silver Amplification. J. Virol. Methods 2011, 178, 82–86. [Google Scholar] [CrossRef]
- Stierhof, Y.D.; Humbel, B.M.; Schwarz, H. Suitability of Different Silver Enhancement Methods Applied to 1 Nm Colloidal Gold Particles: An Immunoelectron Microscopic Study. J. Electron Microsc. Tech. 1991, 17, 336–343. [Google Scholar] [CrossRef]
- Seopsi, L.; Larsson, L.-I.; Bastholm, L.; Nielsen, M.H. Silver-Enhanced Colloidal Gold Probes as Markers for Scanning Electron Microscopy. Histochemistry 1986, 86, 35–41. [Google Scholar] [CrossRef]
- Hacker, G.W.; Grimelius, L.; Danscher, G.; Bernatzky, G.; Muss, W.; Adam, H. Silver Acetate Autometallography: An Alternative Enhancement Technique for Immunogold-Silver Staining (Igss) and Silver Amplification of Gold, Silver, Mercury and Zinc in Tissues. J. Histotechnol. 1988, 11, 213–221. [Google Scholar] [CrossRef]
- Chevallet, M.; Luche, S.; Rabilloud, T. Silver Staining of Proteins in Polyacrylamide Gels. Nat. Protoc. 2006, 1, 1852–1858. [Google Scholar] [CrossRef] [PubMed]
- Danscher, G.; Hacker, G.W.; Grimelius, L.; Nørgaard, J.O.R. Autometallographic Silver Amplification of Colloidal Gold. J. Histotechnol. 1993, 16, 201–207. [Google Scholar] [CrossRef]
- Newman, G.R.; Jasani, B. Silver Development in Microscopy and Bioanalysis: Past and Present. J. Pathol. 1998, 186, 119–125. [Google Scholar] [CrossRef]
- Skutelsky, E.; Goyal, V.; Alroy, J. The Use of Avidin-Gold Complex for Light Microscopic Localization of Lectin Receptors. Histochemistry 1987, 86, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Liu, X.; Tang, Y.; Wu, L.; Hou, X.; Lv, Y. Highly Sensitive Immunoassay Based on Immunogold-Silver Amplification and Inductively Coupled Plasma Mass Spectrometric Detection. Anal. Chem. 2011, 83, 2330–2336. [Google Scholar] [CrossRef] [PubMed]
- Burry, R.W.; Vandre, D.D.; Hayes, D.M. Silver Enhancement of Gold Antibody Probes in Pre-Embedding Electron Microscopic Immunocytochemistry. J. Histochem. Cytochem. 1992, 40, 1849–1856. [Google Scholar] [CrossRef]
- Panferov, V.G.; Safenkova, I.V.; Byzova, N.A.; Varitsev, Y.A.; Zherdev, A.V.; Dzantiev, B.B. Silver-Enhanced Lateral Flow Immunoassay for Highly-Sensitive Detection of Potato Leafroll Virus. Food Agric. Immunol. 2018, 29, 445–457. [Google Scholar] [CrossRef]
- Mitamura, K.; Shimizu, H.; Yamazaki, M.; Ichikawa, M.; Nagai, K.; Katada, J.; Wada, A.; Kawakami, C.; Sugaya, N. Clinical Evaluation of Highly Sensitive Silver Amplification Immunochromatography Systems for Rapid Diagnosis of Influenza. J. Virol. Methods 2013, 194, 123–128. [Google Scholar] [CrossRef]
- Bazsefidpar, S.; Serrano-Pertierra, E.; Gutiérrez, G.; Calvo, A.S.; Matos, M.; Blanco-López, M.C. Rapid and Sensitive Detection of E. Coli O157:H7 by Lateral Flow Immunoassay and Silver Enhancement. Microchim. Acta 2023, 190, 264. [Google Scholar] [CrossRef]
- Byzova, N.A.; Zherdev, A.V.; Sveshnikov, P.G.; Sadykhov, E.G.; Dzantiev, B.B. Development of an Immunochromatographic Test System for the Detection of Helicobacter Pylori Antigens. Appl. Biochem. Microbiol. 2015, 51, 608–617. [Google Scholar] [CrossRef]
- Yu, Q.; Li, H.; Li, C.; Zhang, S.; Shen, J.; Wang, Z. Gold Nanoparticles-Based Lateral Flow Immunoassay with Silver Staining for Simultaneous Detection of Fumonisin B1 and Deoxynivalenol. Food Control 2015, 54, 347–352. [Google Scholar] [CrossRef]
- Xing, C.; Kuang, H.; Hao, C.; Liu, L.; Wang, L.; Xu, C. A Silver Enhanced and Sensitive Strip Sensor for Cadmium Detection. Food Agric. Immunol. 2014, 25, 287–300. [Google Scholar] [CrossRef]
- Anfossi, L.; Di Nardo, F.; Giovannoli, C.; Passini, C.; Baggiani, C. Increased Sensitivity of Lateral Flow Immunoassay for Ochratoxin A through Silver Enhancement. Anal. Bioanal. Chem. 2013, 405, 9859–9867. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.O.; Covián, L.B.; García, A.C.; Blanco-López, M.C. Silver and Gold Enhancement Methods for Lateral Flow Immunoassays. Talanta 2016, 148, 272–278. [Google Scholar] [CrossRef]
- Apilux, A.; Rengpipat, S.; Suwanjang, W.; Chailapakul, O. Development of Competitive Lateral Flow Immunoassay Coupled with Silver Enhancement for Simple and Sensitive Salivary Cortisol Detection. EXCLI J. 2018, 17, 1198–1209. [Google Scholar] [CrossRef]
- Poosinuntakul, N.; Chanmee, T.; Porntadavity, S.; Chailapakul, O.; Apilux, A. Silver-Enhanced Colloidal Gold Dip Strip Immunoassay Integrated with Smartphone-Based Colorimetry for Sensitive Detection of Cardiac Marker Troponin I. Sci. Rep. 2022, 12, 19866. [Google Scholar] [CrossRef] [PubMed]
- Shyu, R.H.; Shyu, H.F.; Liu, H.W.; Tang, S.S. Colloidal Gold-Based Immunochromatographic Assay for Detection of Ricin. Toxicon 2002, 40, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Broger, T.; Sossen, B.; du Toit, E.; Kerkhoff, A.D.; Schutz, C.; Ivanova Reipold, E.; Ward, A.; Barr, D.A.; Macé, A.; Trollip, A.; et al. Novel Lipoarabinomannan Point-of-Care Tuberculosis Test for People with HIV: A Diagnostic Accuracy Study. Lancet Infect. Dis. 2019, 19, 852–861. [Google Scholar] [CrossRef]
- Miyakawa, K.; Funabashi, R.; Yamaoka, Y.; Jeremiah, S.S.; Katada, J.; Wada, A.; Takei, T.; Shimizu, K.; Ozawa, H.; Kawakami, C.; et al. SARS-CoV-2 Antigen Rapid Diagnostic Test Enhanced with Silver Amplification Technology. medRxiv 2021. [Google Scholar] [CrossRef]
- Couturier, C.; Wada, A.; Louis, K.; Mistretta, M.; Beitz, B.; Povogui, M.; Ripaux, M.; Mignon, C.; Werle, B.; Lugari, A.; et al. Characterization and Analytical Validation of a New Antigenic Rapid Diagnostic Test for Ebola Virus Disease Detection. PLoS Negl. Trop. Dis. 2020, 14, e0007965. [Google Scholar] [CrossRef]
- Phan, L.M.T.; Rafique, R.; Baek, S.H.; Nguyen, T.P.; Park, K.Y.; Kim, E.B.; Kim, J.G.; Park, J.P.; Kailasa, S.K.; Kim, H.-J.; et al. Gold-Copper Nanoshell Dot-Blot Immunoassay for Naked-Eye Sensitive Detection of Tuberculosis Specific CFP-10 Antigen. Biosens. Bioelectron. 2018, 121, 111–117. [Google Scholar] [CrossRef]
- Wei, X.; Liang, A.; Zhang, S.S.; Jiang, Z.L. A Selective Resonance Scattering Assay for Immunoglobulin G Using Cu(II)-Ascorbic Acid-Immunonanogold Reaction. Anal. Biochem. 2008, 380, 223–228. [Google Scholar] [CrossRef]
- Kim, J.; Park, J.; Lin, M.; Kim, S.; Kim, G.; Park, S.; Ko, G.; Nam, J. Sensitive, Quantitative Naked-Eye Biodetection with Polyhedral Cu Nanoshells. Adv. Mater. 2017, 29, 1702945. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, Y.; Liu, Y.; Fang, H.; Huang, X.; Leng, Y.; Liu, Z.; Hou, L.; Zhang, W.; Lai, W.; et al. Controlled Copper in Situ Growth-Amplified Lateral Flow Sensors for Sensitive, Reliable, and Field-Deployable Infectious Disease Diagnostics. Biosens. Bioelectron. 2021, 171, 112753. [Google Scholar] [CrossRef]
- Shao, Y.; Xu, W.; Zheng, Y.; Wang, J.; Xie, J.; Zhu, Z.; Xiang, X.; Ye, Q.; Zhang, Y.; Xue, L.; et al. Controlled PAH-Mediated Method with Enhanced Optical Properties for Simple, Stable Immunochromatographic Assays. Biosens. Bioelectron. 2022, 206, 114150. [Google Scholar] [CrossRef]
- Phan, L.M.T.; Kim, E.B.; Cheon, S.A.; Shim, T.S.; Kim, H.J.; Park, T.J. Reliable Naked-Eye Detection of Mycobacterium Tuberculosis Antigen 85B Using Gold and Copper Nanoshell-Enhanced Immunoblotting Techniques. Sens. Actuators B Chem. 2020, 317, 128220. [Google Scholar] [CrossRef]
- Kim, M.W.; Park, H.-J.; Park, C.Y.; Kim, J.H.; Cho, C.H.; Phan, L.M.T.; Park, J.P.; Kailasa, S.K.; Lee, C.-H.; Park, T.J. Fabrication of a Paper Strip for Facile and Rapid Detection of Bovine Viral Diarrhea Virus via Signal Enhancement by Copper Polyhedral Nanoshells. RSC Adv. 2020, 10, 29759–29764. [Google Scholar] [CrossRef]
- Tian, M.; Lei, L.; Xie, W.; Yang, Q.; Li, C.M.; Liu, Y. Copper Deposition-Induced Efficient Signal Amplification for Ultrasensitive Lateral Flow Immunoassay. Sens. Actuators B Chem. 2019, 282, 96–103. [Google Scholar] [CrossRef]
- Peng, T.; Jiao, X.; Liang, Z.; Zhao, H.; Zhao, Y.; Xie, J.; Jiang, Y.; Yu, X.; Fang, X.; Dai, X. Lateral Flow Immunoassay Coupled with Copper Enhancement for Rapid and Sensitive SARS-CoV-2 Nucleocapsid Protein Detection. Biosensors 2021, 12, 13. [Google Scholar] [CrossRef]
- Panferov, V.G.; Byzova, N.A.; Zherdev, A.V.; Dzantiev, B.B. Peroxidase-Mimicking Nanozyme with Surface-Dispersed Pt Atoms for the Colorimetric Lateral Flow Immunoassay of C-Reactive Protein. Microchim. Acta 2021, 188, 309. [Google Scholar] [CrossRef]
- Bai, T.; Wang, L.; Wang, M.; Zhu, Y.; Li, W.; Guo, Z.; Zhang, Y. Strategic Synthesis of Trimetallic Au@Ag–Pt Nanorattles for Ultrasensitive Colorimetric Detection in Lateral Flow Immunoassay. Biosens. Bioelectron. 2022, 208, 114218. [Google Scholar] [CrossRef] [PubMed]
- Bai, T.; Wang, M.; Cao, M.; Zhang, J.; Zhang, K.; Zhou, P.; Liu, Z.; Liu, Y.; Guo, Z.; Lu, X. Functionalized Au@Ag-Au Nanoparticles as an Optical and SERS Dual Probe for Lateral Flow Sensing. Anal. Bioanal. Chem. 2018, 410, 2291–2303. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Ye, H.; Wang, Q.; Kim, M.J.; Tang, D.; Xi, Z.; Wei, Z.; Shao, S.; Xia, X. Template Regeneration in Galvanic Replacement: A Route to Highly Diverse Hollow Nanostructures. ACS Nano 2020, 14, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wan, J.; Peng, X.; Zhao, H.; Shi, D.; Mai, L.; Yang, H.; Zhao, Y.; Yang, X. Calorimetric Lateral Flow Immunoassay Detection Platform Based on the Photothermal Effect of Gold Nanocages with High Sensitivity, Specificity, and Accuracy. Int. J. Nanomed. 2019, 14, 7695–7705. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, L.; Huang, Y.; Dandapat, A.; Dai, L.; Zhang, G.; Lu, X.; Zhang, J.; Lai, W.; Chen, T. Hollow Au-Ag Nanoparticles Labeled Immunochromatography Strip for Highly Sensitive Detection of Clenbuterol. Sci. Rep. 2017, 7, 41419. [Google Scholar] [CrossRef]
- Merkoçi, F.; Patarroyo, J.; Russo, L.; Piella, J.; Genç, A.; Arbiol, J.; Bastús, N.G.; Puntes, V. Understanding Galvanic Replacement Reactions: The Case of Pt and Ag. Mater. Today Adv. 2020, 5, 100037. [Google Scholar] [CrossRef]
- Chen, J.; Wiley, B.; McLellan, J.; Xiong, Y.; Li, Z.Y.; Xia, Y. Optical Properties of Pd-Ag and Pt-Ag Nanoboxes Synthesized via Galvanic Replacement Reactions. Nano Lett. 2005, 5, 2058–2062. [Google Scholar] [CrossRef]
- Taranova, N.A.; Urusov, A.E.; Sadykhov, E.G.; Zherdev, A.V.; Dzantiev, B.B. Bifunctional Gold Nanoparticles as an Agglomeration-Enhancing Tool for Highly Sensitive Lateral Flow Tests: A Case Study with Procalcitonin. Microchim. Acta 2017, 184, 4189–4195. [Google Scholar] [CrossRef]
- Taranova, N.A.; Slobodenuyk, V.D.; Zherdev, A.V.; Dzantiev, B.B. Network of Gold Conjugates for Enhanced Sensitive Immunochromatographic Assays of Troponins. RSC Adv. 2021, 11, 16445–16452. [Google Scholar] [CrossRef]
- Panferov, V.G.; Ivanov, N.A.; Mazzulli, T.; Brinc, D.; Kulasingam, V.; Krylov, S.N. Electrophoresis-Assisted Multilayer Assembly of Nanoparticles for Sensitive Lateral Flow Immunoassay**. Angew. Chem. Int. Ed. 2023, 62, e202215548. [Google Scholar] [CrossRef]
- Hendrickson, O.D.; Zvereva, E.A.; Zherdev, A.V.; Dzantiev, B.B. Cascade-Enhanced Lateral Flow Immunoassay for Sensitive Detection of Okadaic Acid in Seawater, Fish, and Seafood. Foods 2022, 11, 1691. [Google Scholar] [CrossRef]
- Huang, X.; Zhou, Y.; Ding, L.; Yu, G.; Leng, Y.; Lai, W.; Xiong, Y.; Chen, X. Supramolecular Recognition-Mediated Layer-by-Layer Self-Assembled Gold Nanoparticles for Customized Sensitivity in Paper-Based Strip Nanobiosensors. Small 2019, 15, 1903861. [Google Scholar] [CrossRef]
- Sena-Torralba, A.; Alvarez-Diduk, R.; Parolo, C.; Torné-Morató, H.; Müller, A.; Merkoçi, A. Paper-Based Electrophoretic Bioassay: Biosensing in Whole Blood Operating via Smartphone. Anal. Chem. 2021, 93, 3112–3121. [Google Scholar] [CrossRef]
- Nanthasurasak, P.; Cabot, J.M.; See, H.H.; Guijt, R.M.; Breadmore, M.C. Electrophoretic Separations on Paper: Past, Present, and Future-A Review. Anal. Chim. Acta 2017, 985, 7–23. [Google Scholar] [CrossRef]
- Panferov, V.G.; Ivanov, N.A.; Brinc, D.; Fabros, A.; Krylov, S.N. Electrophoretic Assembly of Antibody–Antigen Complexes Facilitates 1000 Times Improvement in the Limit of Detection of Serological Paper-Based Assay. ACS Sens. 2023, 8, 1792–1798. [Google Scholar] [CrossRef]
- Rao, J.; Lahiri, J.; Isaacs, L.; Weis, R.M.; Whitesides, G.M. A Trivalent System from Vancomycin.D-Ala-D-Ala with Higher Affinity than Avidin.Biotin. Science 1998, 280, 708–711. [Google Scholar] [CrossRef]
- Wu, P.; Song, J.; Zuo, W.; Zhu, J.; Meng, X.; Yang, J.; Liu, X.; Jiang, H.; Zhang, D.; Dai, J.; et al. A Universal Boronate Affinity Capture-Antibody-Independent Lateral Flow Immunoassay for Point-of-Care Glycoprotein Detection. Talanta 2023, 265, 124927. [Google Scholar] [CrossRef]
- Javani, A.; Javadi-Zarnaghi, F.; Rasaee, M.J. A Multiplex Protein-Free Lateral Flow Assay for Detection of MicroRNAs Based on Unmodified Molecular Beacons. Anal. Biochem. 2017, 537, 99–105. [Google Scholar] [CrossRef]
- You, Q.; Zhang, X.; Wu, F.-G.; Chen, Y. Colorimetric and Test Stripe-Based Assay of Bacteria by Using Vancomycin-Modified Gold Nanoparticles. Sens. Actuators B Chem. 2019, 281, 408–414. [Google Scholar] [CrossRef]
- Deev, S.M.; Lebedenko, E.N. Antibody Engineering: Molecular Constructor on the Basis of Barnase-Barstar Module. Russ. J. Bioorganic Chem. 2009, 35, 685–701. [Google Scholar] [CrossRef]
- Favalli, N.; Bassi, G.; Pellegrino, C.; Millul, J.; De Luca, R.; Cazzamalli, S.; Yang, S.; Trenner, A.; Mozaffari, N.L.; Myburgh, R.; et al. Stereo- and Regiodefined DNA-Encoded Chemical Libraries Enable Efficient Tumour-Targeting Applications. Nat. Chem. 2021, 13, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Calabria, D.; Calabretta, M.M.; Zangheri, M.; Marchegiani, E.; Trozzi, I.; Guardigli, M.; Michelini, E.; Di Nardo, F.; Anfossi, L.; Baggiani, C.; et al. Recent Advancements in Enzyme-Based Lateral Flow Immunoassays. Sensors 2021, 21, 3358. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Duan, D.; Gao, L.; Zhou, M.; Fan, K.; Tang, Y.; Xi, J.; Bi, Y.; Tong, Z.; Gao, G.F.; et al. Standardized Assays for Determining the Catalytic Activity and Kinetics of Peroxidase-like Nanozymes. Nat. Protoc. 2018, 13, 1506–1520. [Google Scholar] [CrossRef]
- Han, G.-R.; Ki, H.; Kim, M.-G. Automated, Universal, and Mass-Producible Paper-Based Lateral Flow Biosensing Platform for High-Performance Point-of-Care Testing. ACS Appl. Mater. Interfaces 2020, 12, 1885–1894. [Google Scholar] [CrossRef]
- Zandieh, M.; Liu, J. Nanozyme Catalytic Turnover and Self-Limited Reactions. ACS Nano 2021, 15, 15645–15655. [Google Scholar] [CrossRef]
- Zuk, R.F.; Ginsberg, V.K.; Houts, T.; Rabble, J.; Merrick, H.; Ullman, E.F.; Fischer, M.M.; Chung Slzto, C.; Stiso, S.N.; Utman, D.J. Enzyme Immunochromatography—A Quantitative Immunoassay Requinng No Instrumentation. Clin. Chem. 1985, 3117, 1144–1150. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Y.; Wang, S. Development of Multianalyte Flow-through and Lateral-Flow Assays Using Gold Particles and Horseradish Peroxidase as Tracers for the Rapid Determination of Carbaryl and Endosulfan in Agricultural Products. J. Agric. Food Chem. 2006, 54, 2502–2507. [Google Scholar] [CrossRef]
- Kim, H.-S.; Ko, H.; Kang, M.-J.; Pyun, J.-C. Highly Sensitive Rapid Test with Chemiluminescent Signal Bands. BioChip J. 2010, 4, 155–160. [Google Scholar] [CrossRef]
- Cho, I.-H.; Irudayaraj, J. Lateral-Flow Enzyme Immunoconcentration for Rapid Detection of Listeria Monocytogenes. Anal. Bioanal. Chem. 2013, 405, 3313–3319. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Satake, A.; Kido, Y.; Tsuji, A. Monoclonal-Based Enzyme-Linked Immunosorbent Assay and Immunochromatographic Rapid Assay for Dihydrostreptomycin in Milk. Anal. Chim. Acta 2002, 472, 45–53. [Google Scholar] [CrossRef]
- Panferov, V.G.; Safenkova, I.V.; Varitsev, Y.A.; Zherdev, A.V.; Dzantiev, B.B. Enhancement of Lateral Flow Immunoassay by Alkaline Phosphatase: A Simple and Highly Sensitive Test for Potato Virus X. Microchim. Acta 2018, 185, 25. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Kawamura, M.; Arao, S.; Nariuchi, H. A Highly Sensitive Quantitative Immunochromatography Assay for Antigen-Specific IgE. J. Immunol. Methods 2003, 272, 211–218. [Google Scholar] [CrossRef]
- Kim, H.T.; Jin, E.; Lee, M.H. Portable Chemiluminescence-Based Lateral Flow Assay Platform for the Detection of Cortisol in Human Serum. Biosensors 2021, 11, 191. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhang, S.; Zhang, X.; Baloda, M.; Gurung, A.S.; Xu, H.; Zhang, X.; Liu, G. Ultrasensitive Nucleic Acid Biosensor Based on Enzyme-Gold Nanoparticle Dual Label and Lateral Flow Strip Biosensor. Biosens. Bioelectron. 2011, 26, 2018–2024. [Google Scholar] [CrossRef]
- Cho, J.-H.; Paek, E.-H.; Cho, I.-H.; Paek, S.-H. An Enzyme Immunoanalytical System Based on Sequential Cross-Flow Chromatography. Anal. Chem. 2005, 77, 4091–4097. [Google Scholar] [CrossRef] [PubMed]
- Samsonova, J.V.; Safronova, V.A.; Osipov, A.P. Pretreatment-Free Lateral Flow Enzyme Immunoassay for Progesterone Detection in Whole Cows’ Milk. Talanta 2015, 132, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, O.D.; Zvereva, E.A.; Panferov, V.G.; Solopova, O.N.; Zherdev, A.V.; Sveshnikov, P.G.; Dzantiev, B.B. Application of Au@Pt Nanozyme as Enhancing Label for the Sensitive Lateral Flow Immunoassay of Okadaic Acid. Biosensors 2022, 12, 1137. [Google Scholar] [CrossRef]
- Hendrickson, O.D.; Zvereva, E.A.; Zherdev, A.V.; Dzantiev, B.B. Ultrasensitive Lateral Flow Immunoassay of Phycotoxin Microcystin-LR in Seafood Based on Magnetic Particles and Peroxidase Signal Amplification. Food Control 2022, 133, 108655. [Google Scholar] [CrossRef]
- Shu, Q.; Wang, L.; Ouyang, H.; Wang, W.; Liu, F.; Fu, Z. Multiplexed Immunochromatographic Test Strip for Time-Resolved Chemiluminescent Detection of Pesticide Residues Using a Bifunctional Antibody. Biosens. Bioelectron. 2017, 87, 908–914. [Google Scholar] [CrossRef]
- Wang, J.; Majkova, Z.; Bever, C.R.S.; Yang, J.; Gee, S.J.; Li, J.; Xu, T.; Hammock, B.D. One-Step Immunoassay for Tetrabromobisphenol A Using a Camelid Single Domain Antibody–Alkaline Phosphatase Fusion Protein. Anal. Chem. 2015, 87, 4741–4748. [Google Scholar] [CrossRef]
- Manes, T.; Hoylaerts, M.F.; Müller, R.; Lottspeich, F.; Hölke, W.; Millán, J.L. Genetic Complexity, Structure, and Characterization of Highly Active Bovine Intestinal Alkaline Phosphatases. J. Biol. Chem. 1998, 273, 23353–23360. [Google Scholar] [CrossRef] [PubMed]
- Fosset, M.; Chappelet-Tordo, D.; Lazdunski, M. Intestinal Alkaline Phosphatase. Physical Properties and Quaternary Structure. Biochemistry 1974, 13, 1783–1788. [Google Scholar] [CrossRef] [PubMed]
- Neumann, H.; Lustig, A. The Activation of Alkaline Phosphatase by Effector Molecules. A Combined Kinetic and Hydrodynamic Study. Eur. J. Biochem. 1980, 109, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Rennke, H.G.; Venkatachalam, M.A. Chemical Modification of Horseradish Peroxidase. Preparation and Characterization of Tracer Enzymes with Different Isoelectric Points. J. Histochem. Cytochem. 1979, 27, 1352–1353. [Google Scholar] [CrossRef] [PubMed]
- Sotnikov, D.V.; Berlina, A.N.; Ivanov, V.S.; Zherdev, A.V.; Dzantiev, B.B. Adsorption of Proteins on Gold Nanoparticles: One or More Layers? Colloids Surf. B Biointerfaces 2019, 173, 557–563. [Google Scholar] [CrossRef]
- Kim, L.H.-Y.; Plaza, K.; Thomas, S.R.; Draijer, C.; Radford, K.; Peters-Golden, M.; Mukherjee, M.; Nair, P. Endogenous Peroxidases in Sputum Interfere with Horse-Radish Peroxidase-Based ELISAs. J. Immunol. Methods 2018, 454, 76–79. [Google Scholar] [CrossRef]
- Jiang, S.; Penner, M.H. Overcoming Reductant Interference in Peroxidase-Based Assays for Hydrogen Peroxide Quantification. J. Agric. Food Chem. 2017, 65, 8213–8219. [Google Scholar] [CrossRef]
- Patsoukis, N.; Papapostolou, I.; Georgiou, C.D. Interference of Non-Specific Peroxidases in the Fluorescence Detection of Superoxide Radical by Hydroethidine Oxidation: A New Assay for H2O2. Anal. Bioanal. Chem. 2005, 381, 1065–1072. [Google Scholar] [CrossRef]
- Abraham, A.; Albrechtsen, S.E. Comparison of Penicillinase, Urease and Alkaline Phosphatase as Labels in Enzyme-Linked Immunosorbent Assay (ELISA) for the Detection of Plant Viruses. J. Plant Dis. Prot. 2001, 108, 49–57. [Google Scholar]
- Europe, O. PM 7/125 (1) ELISA Tests for Viruses. EPPO Bull. 2015, 45, 445–449. [Google Scholar] [CrossRef]
- Frey, A.; Meckelein, B.; Externest, D.; Schmidt, M.A. A Stable and Highly Sensitive 3,3′,5,5′-Tetramethylbenzidine-Based Substrate Reagent for Enzyme-Linked Immunosorbent Assays. J. Immunol. Methods 2000, 233, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Fu, E.; Lutz, B.; Yager, P. Long-Term Dry Storage of an Enzyme-Based Reagent System for ELISA in Point-of-Care Devices. Analyst 2014, 139, 1456–1462. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gui, X.; Zheng, Q.; Chen, Y.; Ge, S.; Zhang, J.; Xia, N. An HRP-Labeled Lateral Flow Immunoassay for Rapid Simultaneous Detection and Differentiation of Influenza A and B Viruses. J. Med. Virol. 2019, 91, 503–507. [Google Scholar] [CrossRef] [PubMed]
- Byers, K.M.; Bird, A.R.; Cho, H.D.D.; Linnes, J.C. Fully Dried Two-Dimensional Paper Network for Enzymatically Enhanced Detection of Nucleic Acid Amplicons. ACS Omega 2020, 5, 4673–4681. [Google Scholar] [CrossRef] [PubMed]
- Joung, H.A.; Oh, Y.K.; Kim, M.G. An Automatic Enzyme Immunoassay Based on a Chemiluminescent Lateral Flow Immunosensor. Biosens. Bioelectron. 2014, 53, 330–335. [Google Scholar] [CrossRef]
- Kim, K.; Joung, H.A.; Han, G.R.; Kim, M.G. An Immunochromatographic Biosensor Combined with a Water-Swellable Polymer for Automatic Signal Generation or Amplification. Biosens. Bioelectron. 2016, 85, 422–428. [Google Scholar] [CrossRef]
- Gao, L.; Zhuang, J.; Nie, L.; Zhang, J.; Zhang, Y.; Gu, N.; Wang, T.; Feng, J.; Yang, D.; Perrett, S.; et al. Intrinsic Peroxidase-like Activity of Ferromagnetic Nanoparticles. Nat. Nanotechnol. 2007, 2, 577–583. [Google Scholar] [CrossRef]
- Wu, J.; Wang, X.; Wang, Q.; Lou, Z.; Li, S.; Zhu, Y.; Qin, L.; Wei, H. Nanomaterials with Enzyme-like Characteristics (Nanozymes): Next-Generation Artificial Enzymes (II). Chem. Soc. Rev. 2019, 48, 1004–1076. [Google Scholar] [CrossRef]
- Huang, Y.; Ren, J.; Qu, X. Nanozymes: Classification, Catalytic Mechanisms, Activity Regulation, and Applications. Chem. Rev. 2019, 119, 4357–4412. [Google Scholar] [CrossRef]
- Wei, Z.; Xi, Z.; Vlasov, S.; Ayala, J.; Xia, X. Nanocrystals of Platinum-Group Metals as Peroxidase Mimics for in Vitro Diagnostics. Chem. Commun. 2020, 56, 14962–14975. [Google Scholar] [CrossRef]
- Gao, Z.; Ye, H.; Tang, D.; Tao, J.; Habibi, S.; Minerick, A.; Tang, D.; Xia, X. Platinum-Decorated Gold Nanoparticles with Dual Functionalities for Ultrasensitive Colorimetric in Vitro Diagnostics. Nano Lett. 2017, 17, 5572–5579. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; He, Q.; Pan, J.; Shen, D.; Xiao, H.; Cui, X.; Zhao, S. A Pt-Ir Nanocube Amplified Lateral Flow Immunoassay for Dehydroepiandrosterone. Analyst 2021, 146, 2726–2733. [Google Scholar] [CrossRef]
- He, Q.; Yang, H.; Chen, Y.; Shen, D.; Cui, X.; Zhang, C.; Xiao, H.; Eremin, S.A.; Fang, Y.; Zhao, S. Prussian Blue Nanoparticles with Peroxidase-Mimicking Properties in a Dual Immunoassays for Glycocholic Acid. J. Pharm. Biomed. Anal. 2020, 187, 113317. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Huang, Q.; Dou, L.; Bu, T.; Chen, K.; Yang, Q.; Yan, L.; Wang, J.; Zhang, D. Prussian Blue Nanoparticles Based Lateral Flow Assay for High Sensitive Determination of Clenbuterol. Sens. Actuators B Chem. 2018, 275, 223–229. [Google Scholar] [CrossRef]
- Luo, Y.; Luo, H.; Zou, S.; Jiang, J.; Duan, D.; Chen, L.; Gao, L. An In Situ Study on Nanozyme Performance to Optimize Nanozyme-Strip for Aβ Detection. Sensors 2023, 23, 3414. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Wang, L.; Jia, T.; Lian, T.; Yang, K.; Li, X.; Wang, X.; Xue, C. Au/Fe3O4-Based Nanozymes with Peroxidase-like Activity Integrated in Immunochromatographic Strips for Highly-Sensitive Biomarker Detection. Anal. Methods 2023, 15, 663–674. [Google Scholar] [CrossRef]
- Jiang, T.; Song, Y.; Du, D.; Liu, X.; Lin, Y. Detection of P53 Protein Based on Mesoporous Pt-Pd Nanoparticles with Enhanced Peroxidase-like Catalysis. ACS Sens. 2016, 1, 717–724. [Google Scholar] [CrossRef]
- Zhang, J.; Yu, Q.; Qiu, W.; Li, K.; Qian, L.; Zhang, X.; Liu, G. Gold-Platinum Nanoflowers as a Label and as an Enzyme Mimic for Use in Highly Sensitive Lateral Flow Immunoassays: Application to Detection of Rabbit IgG. Microchim. Acta 2019, 186, 357. [Google Scholar] [CrossRef]
- Duan, D.; Fan, K.; Zhang, D.; Tan, S.; Liang, M.; Liu, Y.; Zhang, J.; Zhang, P.; Liu, W.; Qiu, X.; et al. Nanozyme-Strip for Rapid Local Diagnosis of Ebola. Biosens. Bioelectron. 2015, 74, 134–141. [Google Scholar] [CrossRef]
- Panferov, V.G.; Safenkova, I.V.; Zherdev, A.V.; Dzantiev, B.B. Urchin Peroxidase-Mimicking Au@Pt Nanoparticles as a Label in Lateral Flow Immunoassay: Impact of Nanoparticle Composition on Detection Limit of Clavibacter Michiganensis. Microchim. Acta 2020, 187, 268. [Google Scholar] [CrossRef]
- Park, J.-M.; Jung, H.-W.; Chang, Y.W.; Kim, H.-S.; Kang, M.-J.; Pyun, J.-C. Chemiluminescence Lateral Flow Immunoassay Based on Pt Nanoparticle with Peroxidase Activity. Anal. Chim. Acta 2015, 853, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xi, X.; Zhu, J.; Wu, X.; Zhang, X.; Wang, S.; Wen, W. High Sensitivity and Rapid Detection of Hepatitis B Virus DNA Using Lateral Flow Biosensors Based on Au@Pt Nanorods in the Absence of Hydrogen Peroxide. Analyst 2022, 147, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.Y.; Heo, N.S.; Kang, J.W.; Lee, J.B.; Kim, H.J.; Kim, M. Il Nanoceria-Based Lateral Flow Immunoassay for Hydrogen Peroxide-Free Colorimetric Biosensing for C-Reactive Protein. Anal. Bioanal. Chem. 2022, 414, 3257–3265. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Liang, M.; Ma, F.; Zhang, Z.; Tang, X.; Jiang, J.; Guo, C.; Ramzy Mohamed, S.; Abdel Goda, A.; Dawood, D.H.; et al. Nanozyme-Strip Based on MnO2 Nanosheets as a Catalytic Label for Multi-Scale Detection of Aflatoxin B1 with an Ultrabroad Working Range. Food Chem. 2022, 377, 131965. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Guan, Z.; Song, Y.; Song, E.; Lu, Z.; Liu, D.; An, Y.; Zhu, Z.; Zhou, L.; Yang, C. Lateral Flow Assay with Pressure Meter Readout for Rapid Point-of-Care Detection of Disease-Associated Protein. Lab Chip 2018, 18, 965–970. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Lin, B.; Song, Y.; Guan, Z.; Cheng, J.; Zhu, Z.; Yang, C. Staining Traditional Colloidal Gold Test Strips with Pt Nanoshell Enables Quantitative Point-of-Care Testing with Simple and Portable Pressure Meter Readout. ACS Appl. Mater. Interfaces 2019, 11, 1800–1806. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, G.; Dou, W. An Unplugged and Quantitative Foam Based Immunochromatographic Assay for Escherichia coli O157:H7 Using Nanozymes to Catalyze Hydrogen Peroxide Decomposition Reaction. Microchem. J. 2020, 152, 104313. [Google Scholar] [CrossRef]
- Liu, L.; Liu, J.; Huang, H.; Li, Y.; Zhao, G.; Dou, W. A Quantitative Foam Immunoassay for Detection of Escherichia coli O157:H7 Based on Bimetallic Nanocatalyst-gold Platinum. Microchem. J. 2019, 148, 702–707. [Google Scholar] [CrossRef]
- Wang, K.-Y.; Bu, S.-J.; Ju, C.-J.; Han, Y.; Ma, C.-Y.; Liu, W.-S.; Li, Z.-Y.; Li, C.-T.; Wan, J.-Y. Disposable Syringe-Based Visual Immunotest for Pathogenic Bacteria Based on the Catalase Mimicking Activity of Platinum Nanoparticle-Concanavalin A Hybrid Nanoflowers. Microchim. Acta 2019, 186, 57. [Google Scholar] [CrossRef]
- Yu, Z.; Cai, G.; Liu, X.; Tang, D. Platinum Nanozyme-Triggered Pressure-Based Immunoassay Using a Three-Dimensional Polypyrrole Foam-Based Flexible Pressure Sensor. ACS Appl. Mater. Interfaces 2020, 12, 40133–40140. [Google Scholar] [CrossRef]
- Wei, Z.; Luciano, K.; Xia, X. Catalytic Gold-Iridium Nanoparticles as Labels for Sensitive Colorimetric Lateral Flow Assay. ACS Nano 2022, 16, 21609–21617. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, B.; Yang, R.; Liu, J. Filling in the Gaps between Nanozymes and Enzymes: Challenges and Opportunities. Bioconjug. Chem. 2017, 28, 2903–2909. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Shao, S.; Gao, W.; Tang, D.; Tang, D.; Zou, S.; Kim, M.J.; Xia, X. Morphology-Invariant Metallic Nanoparticles with Tunable Plasmonic Properties. ACS Nano 2021, 15, 2428–2438. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Ahn, J.; Shi, S.; Wang, P.; Gao, R.; Qin, D. Noble-Metal Nanoframes and Their Catalytic Applications. Chem. Rev. 2021, 121, 796–833. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xianyu, Y. Tuning the Plasmonic and Catalytic Signals of Au@Pt Nanoparticles for Dual-Mode Biosensing. Biosens. Bioelectron. 2023, 237, 115553. [Google Scholar] [CrossRef]
- Jiao, L.; Xu, W.; Wu, Y.; Yan, H.; Gu, W.; Du, D.; Lin, Y.; Zhu, C. Single-Atom Catalysts Boost Signal Amplification for Biosensing. Chem. Soc. Rev. 2021, 50, 750–765. [Google Scholar] [CrossRef]
- Kong, L.; Chen, D.; Zhang, X.; Zhou, L.; Deng, Y.; Wei, S. Controllable Fabrication of Hg–Pd–PdO Heterostructures as Efficient Peroxidase Mimics for Carcinoembryonic Antigen Detection. ACS Appl. Nano Mater. 2023, 6, 3618–3626. [Google Scholar] [CrossRef]
- Huang, Z.; Liu, B.; Liu, J. Enhancing the Peroxidase-like Activity and Stability of Gold Nanoparticles by Coating a Partial Iron Phosphate Shell. Nanoscale 2020, 12, 22467–22472. [Google Scholar] [CrossRef]
- Jin, S.; Wu, C.; Ye, Z.; Ying, Y. Designed Inorganic Nanomaterials for Intrinsic Peroxidase Mimics: A Review. Sens. Actuators B Chem. 2019, 283, 18–34. [Google Scholar] [CrossRef]
- Hermanson, G.T. Antibody Modification and Conjugation. In Bioconjugate Techniques, 3rd ed.; Hermanson, G.-B.T., Ed.; Elsevier: Boston, MA, USA, 2013; pp. 867–920. ISBN 978-0-12-382239-0. [Google Scholar]
- Nakane, K. Peroxidase-Labeled a New Antibody Method Conjugation. J. Histochem. Cytochem. 1974, 22, 1084–1091. [Google Scholar] [CrossRef]
- Tao, X.; Wang, X.; Liu, B.; Liu, J. Conjugation of Antibodies and Aptamers on Nanozymes for Developing Biosensors. Biosens. Bioelectron. 2020, 168, 112537. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Xu, M.; Hou, L.; Chen, G.; Tang, D. Irregular-Shaped Platinum Nanoparticles as Peroxidase Mimics for Highly Efficient Colorimetric Immunoassay. Anal. Chim. Acta 2013, 776, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Xu, M.; Lu, M.; Chen, G.; Tang, D. Urchin-like (Gold Core)@(Platinum Shell) Nanohybrids: A Highly Efficient Peroxidase-Mimetic System for in Situ Amplified Colorimetric Immunoassay. Biosens. Bioelectron. 2015, 70, 194–201. [Google Scholar] [CrossRef]
- Lai, X.; Zhang, G.; Zeng, L.; Xiao, X.; Peng, J.; Guo, P.; Zhang, W.; Lai, W. Synthesis of PDA-Mediated Magnetic Bimetallic Nanozyme and Its Application in Immunochromatographic Assay. ACS Appl. Mater. Interfaces 2021, 13, 1413–1423. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ren, J.; Yin, X.; Li, Y.; Shu, R.; Wang, J.; Zhang, D. Vanadium Disulfide Nanosheet Boosts Optical Signal Brightness as a Superior Enzyme Label to Improve the Sensitivity of Lateral Flow Immunoassay. Anal. Chem. 2022, 94, 8693–8703. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Liu, Z.; Fang, Y.; Shen, X.; Xu, Z.; Lei, H.; Huang, X.; Li, X. Two Kinds of Lateral Flow Immunoassays Based on Multifunctional Magnetic Prussian Blue Nanoenzyme and Colloidal Gold for the Detection of 38 β-Agonists in Swine Urine and Pork. Food Chem. 2023, 417, 135897. [Google Scholar] [CrossRef]
- Xia, X.; Zhang, J.; Lu, N.; Kim, M.J.; Ghale, K.; Xu, Y.; McKenzie, E.; Liu, J.; Ye, H. Pd-Ir Core-Shell Nanocubes: A Type of Highly Efficient and Versatile Peroxidase Mimic. ACS Nano 2015, 9, 9994–10004. [Google Scholar] [CrossRef]
- Ye, H.; Yang, K.; Tao, J.; Liu, Y.; Zhang, Q.; Habibi, S.; Nie, Z.; Xia, X. An Enzyme-Free Signal Amplification Technique for Ultrasensitive Colorimetric Assay of Disease Biomarkers. ACS Nano 2017, 11, 2052–2059. [Google Scholar] [CrossRef]
- Tian, M.; Xie, W.; Zhang, T.; Liu, Y.; Lu, Z.; Li, C.M.; Liu, Y. A Sensitive Lateral Flow Immunochromatographic Strip with Prussian Blue Nanoparticles Mediated Signal Generation and Cascade Amplification. Sens. Actuators B Chem. 2020, 309, 127728. [Google Scholar] [CrossRef]
- Hendrickson, O.D.; Zvereva, E.A.; Pridvorova, S.M.; Dzantiev, B.B.; Zherdev, A. V The Use of Au@Pt Nanozyme to Perform Ultrasensitive Immunochromatographic Detection of Banned Pork Additives in Meat Products. Food Control 2023, 154, 110013. [Google Scholar] [CrossRef]
- Chen, Z.J.; Huang, Z.; Huang, S.; Zhao, J.L.; Sun, Y.; Xu, Z.L.; Liu, J. Effect of Proteins on the Oxidase-like Activity of CeO2nanozymes for Immunoassays. Analyst 2021, 146, 864–873. [Google Scholar] [CrossRef] [PubMed]
- McVey, C.; Logan, N.; Thanh, N.T.K.; Elliott, C.; Cao, C. Unusual Switchable Peroxidase-Mimicking Nanozyme for the Determination of Proteolytic Biomarker. Nano Res. 2019, 12, 509–516. [Google Scholar] [CrossRef]
- Chen, R.; Chen, X.; Zhou, Y.; Lin, T.; Leng, Y.; Huang, X.; Xiong, Y. “Three-in-One” Multifunctional Nanohybrids with Colorimetric Magnetic Catalytic Activities to Enhance Immunochromatographic Diagnosis. ACS Nano 2022, 16, 3351–3361. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Jiang, Q.; Wang, K.; Li, T.; Zheng, W.; Cheng, Y.; Ning, Q.; Cui, D. Nanozyme Enhanced Magnetic Immunoassay for Dual-Mode Detection of Gastrin-17. Analyst 2022, 147, 1678–1687. [Google Scholar] [CrossRef]
- Kim, M.S.; Kweon, S.H.; Cho, S.; An, S.S.A.; Kim, M.I.; Doh, J.; Lee, J. Pt-Decorated Magnetic Nanozymes for Facile and Sensitive Point-of-Care Bioassay. ACS Appl. Mater. Interfaces 2017, 9, 35133–35140. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Cai, X.; Gao, Y.; Yan, H.; Fu, J.; Tang, X.; Zhang, Q.; Li, P. A Versatile Nanozyme Integrated Colorimetric and Photothermal Lateral Flow Immunoassay for Highly Sensitive and Reliable Aspergillus Flavus Detection. Biosens. Bioelectron. 2022, 213, 114435. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, X.; Li, Y.; Hai, X.; Bi, S. A Colorimetric and Photothermal Dual-Mode Biosensing Platform Based on Nanozyme-Functionalized Flower-like DNA Structures for Tumor-Derived Exosome Detection. Talanta 2023, 258, 124456. [Google Scholar] [CrossRef]
- Guteneva, N.V.; Znoyko, S.L.; Orlov, A.V.; Nikitin, M.P.; Nikitin, P.I. Rapid Lateral Flow Assays Based on the Quantification of Magnetic Nanoparticle Labels for Multiplexed Immunodetection of Small Molecules: Application to the Determination of Drugs of Abuse. Microchim. Acta 2019, 186, 621. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhao, R.; Feng, L.; Wang, C.; Dong, S.; Zyuzin, M.V.; Timin, A.; Hu, N.; Liu, B.; Yang, P. Dual Nanozyme-Driven PtSn Bimetallic Nanoclusters for Metal-Enhanced Tumor Photothermal and Catalytic Therapy. ACS Nano 2023, 17, 6833–6848. [Google Scholar] [CrossRef]
- Gao, Z.; Lv, S.; Xu, M.; Tang, D. High-Index {: Hk 0} Faceted Platinum Concave Nanocubes with Enhanced Peroxidase-like Activity for an Ultrasensitive Colorimetric Immunoassay of the Human Prostate-Specific Antigen. Analyst 2017, 142, 911–917. [Google Scholar] [CrossRef]
- Chen, T.M.; Tian, X.M.; Huang, L.; Xiao, J.; Yang, G.W. Nanodiamonds as PH-Switchable Oxidation and Reduction Catalysts with Enzyme-like Activities for Immunoassay and Antioxidant Applications. Nanoscale 2017, 9, 15673–15684. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; He, Q.; Chen, Y.; Shen, D.; Xiao, H.; Eremin, S.A.; Cui, X.; Zhao, S. Platinum Nanoflowers with Peroxidase-like Property in a Dual Immunoassay for Dehydroepiandrosterone. Microchim. Acta 2020, 187, 592. [Google Scholar] [CrossRef] [PubMed]
- Panferov, V.G.; Safenkova, I.V.; Zherdev, A.V.; Dzantiev, B.B. The Steadfast Au@Pt Soldier: Peroxide-Tolerant Nanozyme for Signal Enhancement in Lateral Flow Immunoassay of Peroxidase-Containing Samples. Talanta 2021, 225, 121961. [Google Scholar] [CrossRef] [PubMed]
- Khramtsov, P.; Kropaneva, M.; Minin, A.; Bochkova, M.; Timganova, V.; Maximov, A.; Puzik, A.; Zamorina, S.; Rayev, M. Prussian Blue Nanozymes with Enhanced Catalytic Activity: Size Tuning and Application in ELISA-like Immunoassay. Nanomaterials 2022, 12, 1630. [Google Scholar] [CrossRef]
- Asati, A.; Santra, S.; Kaittanis, C.; Nath, S.; Perez, J.M. Oxidase-Like Activity of Polymer-Coated Cerium Oxide Nanoparticles. Angew. Chem. Int. Ed. 2009, 48, 2308–2312. [Google Scholar] [CrossRef]
- Fu, Y.; Zhang, H.; Dai, S.; Zhi, X.; Zhang, J.; Li, W. Glutathione-Stabilized Palladium Nanozyme for Colorimetric Assay of Silver(I) Ions. Analyst 2015, 140, 6676–6683. [Google Scholar] [CrossRef]
- Singh, N.; Savanur, M.A.; Srivastava, S.; D’Silva, P.; Mugesh, G. A Redox Modulatory Mn3O4 Nanozyme with Multi-Enzyme Activity Provides Efficient Cytoprotection to Human Cells in a Parkinson’s Disease Model. Angew. Chemie Int. Ed. 2017, 56, 14267–14271. [Google Scholar] [CrossRef]
- Tian, R.; Sun, J.; Qi, Y.; Zhang, B.; Guo, S.; Zhao, M. Influence of VO2 Nanoparticle Morphology on the Colorimetric Assay of H2O2 and Glucose. Nanomaterials 2017, 7, 347. [Google Scholar] [CrossRef]
- Liu, S.; Lu, F.; Xing, R.; Zhu, J.J. Structural Effects of Fe3O4 Nanocrystals on Peroxidase-like Activity. Chem. Eur. J. 2011, 17, 620–625. [Google Scholar] [CrossRef]
- Robert, A.; Meunier, B. How to Define a Nanozyme. ACS Nano 2022, 16, 6956–6959. [Google Scholar] [CrossRef]
- Zandieh, M.; Liu, J. Nanozymes: Definition, Activity, and Mechanisms. Adv. Mater. 2023, 2211041. [Google Scholar] [CrossRef] [PubMed]
- Loynachan, C.N.; Thomas, M.R.; Gray, E.R.; Richards, D.A.; Kim, J.; Miller, B.S.; Brookes, J.C.; Agarwal, S.; Chudasama, V.; McKendry, R.A.; et al. Platinum Nanocatalyst Amplification: Redefining the Gold Standard for Lateral Flow Immunoassays with Ultrabroad Dynamic Range. ACS Nano 2018, 12, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Wang, Y.; Zhao, M.; Zhang, L. Intrinsic Peroxidase-like Activity and Catalase-like Activity of Co3O4 Nanoparticles. Chem. Commun. 2012, 48, 2540. [Google Scholar] [CrossRef] [PubMed]
- Punekar, N.S. Good Kinetic Practices. In ENZYMES: Catalysis, Kinetics and Mechanisms; Punekar, N.S., Ed.; Springer: Singapore, 2018; pp. 131–142. ISBN 978-981-13-0785-0. [Google Scholar]
- Jin, X.; Chen, L.; Zhang, Y.; Wang, X.; Zhou, N. A Lateral Flow Strip for On-Site Detection of Tobramycin Based on Dual-Functional Platinum-Decorated Gold Nanoparticles. Analyst 2021, 146, 3608–3616. [Google Scholar] [CrossRef]
- Zheng, X.; Liu, Q.; Jing, C.; Li, Y.; Li, D.; Luo, W.; Wen, Y.; He, Y.; Huang, Q.; Long, Y.-T.; et al. Catalytic Gold Nanoparticles for Nanoplasmonic Detection of DNA Hybridization. Angew. Chem. 2011, 123, 12200–12204. [Google Scholar] [CrossRef]
- Li, J.; Liu, F.; Zhu, Z.; Liu, D.; Chen, X.; Song, Y.; Zhou, L.; Yang, C. In Situ Pt Staining Method for Simple, Stable, and Sensitive Pressure-Based Bioassays. ACS Appl. Mater. Interfaces 2018, 10, 13390–13396. [Google Scholar] [CrossRef]
- Fu, J.; Zhou, Y.; Huang, X.; Zhang, W.; Wu, Y.; Fang, H.; Zhang, C.; Xiong, Y. Dramatically Enhanced Immunochromatographic Assay Using Cascade Signal Amplification for Ultrasensitive Detection of Escherichia coli O157:H7 in Milk. J. Agric. Food Chem. 2020, 68, 1118–1125. [Google Scholar] [CrossRef]
- Sun, Y.; Xie, Z.; Pei, F.; Hu, W.; Feng, S.; Hao, Q.; Liu, B.; Mu, X.; Lei, W.; Tong, Z. Trimetallic Au@Pd@Pt Nanozyme-Enhanced Lateral Flow Immunoassay for the Detection of SARS-CoV-2 Nucleocapsid Protein. Anal. Methods 2022, 14, 5091–5099. [Google Scholar] [CrossRef]
- Lyu, Z.; Ding, S.; Tieu, P.; Fang, L.; Li, X.; Li, T.; Pan, X.; Engelhard, M.H.; Ruan, X.; Du, D.; et al. Single-Atomic Site Catalyst Enhanced Lateral Flow Immunoassay for Point-of-Care Detection of Herbicide. Research 2022, 2022, 9823290. [Google Scholar] [CrossRef]
- Cai, X.; Ma, F.; Jiang, J.; Yang, X.; Zhang, Z.; Jian, Z.; Liang, M.; Li, P.; Yu, L. Fe-N-C Single-Atom Nanozyme for Ultrasensitive, on-Site and Multiplex Detection of Mycotoxins Using Lateral Flow Immunoassay. J. Hazard. Mater. 2023, 441, 129853. [Google Scholar] [CrossRef]
- Xia, X.; Figueroa-Cosme, L.; Tao, J.; Peng, H.-C.; Niu, G.; Zhu, Y.; Xia, Y. Facile Synthesis of Iridium Nanocrystals with Well-Controlled Facets Using Seed-Mediated Growth. J. Am. Chem. Soc. 2014, 136, 10878–10881. [Google Scholar] [CrossRef] [PubMed]
- Lori, O.; Elbaz, L. Recent Advances in Synthesis and Utilization of Ultra-low Loading of Precious Metal-based Catalysts for Fuel Cells. ChemCatChem 2020, 12, 3434–3446. [Google Scholar] [CrossRef]
- Cho, J.-H.; Han, S.; Paek, E.-H.; Cho, I.; Paek, S. Plastic ELISA-on-a-Chip Based on Sequential Cross-Flow Chromatography. Anal. Chem. 2006, 78, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.H.; Paek, E.H.; Kim, Y.K.; Kim, J.H.; Paek, S.H. Chemiluminometric Enzyme-Linked Immunosorbent Assays (ELISA)-on-a-Chip Biosensor Based on Cross-Flow Chromatography. Anal. Chim. Acta 2009, 632, 247–255. [Google Scholar] [CrossRef]
- Cho, J.H.; Kim, M.H.; Mok, R.S.; Jeon, J.W.; Lim, G.S.; Chai, C.Y.; Paek, S.H. Two-Dimensional Paper Chromatography-Based Fluorescent Immunosensor for Detecting Acute Myocardial Infarction Markers. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014, 967, 139–146. [Google Scholar] [CrossRef]
- Lim, G.-S.; Seo, S.-M.; Paek, S.-H.; Kim, S.-W.; Jeon, J.-W.; Kim, D.-H.; Cho, I.-H.; Paek, S.-H. Chemiluminometric Immunosensor for High-Sensitivity Cardiac Troponin I Employing a Polymerized Enzyme Conjugate as a Tracer. Sci. Rep. 2015, 5, 14848. [Google Scholar] [CrossRef]
- Cho, I.H.; Seo, S.M.; Paek, E.H.; Paek, S.H. Immunogold-Silver Staining-on-a-Chip Biosensor Based on Cross-Flow Chromatography. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2010, 878, 271–277. [Google Scholar] [CrossRef]
- Shin, J.H.; Hong, J.; Go, H.; Park, J.; Kong, M.; Ryu, S.; Kim, K.-P.; Roh, E.; Park, J.-K. Multiplexed Detection of Foodborne Pathogens from Contaminated Lettuces Using a Handheld Multistep Lateral Flow Assay Device. J. Agric. Food Chem. 2018, 66, 290–297. [Google Scholar] [CrossRef]
- Panraksa, Y.; Apilux, A.; Jampasa, S.; Puthong, S.; Henry, C.S.; Rengpipat, S.; Chailapakul, O. A Facile One-Step Gold Nanoparticles Enhancement Based on Sequential Patterned Lateral Flow Immunoassay Device for C-Reactive Protein Detection. Sens. Actuators B Chem. 2021, 329, 129241. [Google Scholar] [CrossRef]
- Deng, J.; Yang, M.; Wu, J.; Zhang, W.; Jiang, X. A Self-Contained Chemiluminescent Lateral Flow Assay for Point-of-Care Testing. Anal. Chem. 2018, 90, 9132–9137. [Google Scholar] [CrossRef]
- Yang, W.; Li, X.; Liu, G.; Zhang, B.; Zhang, Y.; Kong, T.; Tang, J.; Li, D.; Wang, Z. A Colloidal Gold Probe-Based Silver Enhancement Immunochromatographic Assay for the Rapid Detection of Abrin-A. Biosens. Bioelectron. 2011, 26, 3710–3713. [Google Scholar] [CrossRef] [PubMed]
- Panferov, V.G.; Safenkova, I.V.; Varitsev, Y.A.; Drenova, N.V.; Kornev, K.P.; Zherdev, A.V.; Dzantiev, B.B. Development of the Sensitive Lateral Flow Immunoassay with Silver Enhancement for the Detection of Ralstonia Solanacearum in Potato Tubers. Talanta 2016, 152, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Zangheri, M.; Di Nardo, F.; Mirasoli, M.; Anfossi, L.; Nascetti, A.; Caputo, D.; De Cesare, G.; Guardigli, M.; Baggiani, C.; Roda, A. Chemiluminescence Lateral Flow Immunoassay Cartridge with Integrated Amorphous Silicon Photosensors Array for Human Serum Albumin Detection in Urine Samples. Anal. Bioanal. Chem. 2016, 408, 8869–8879. [Google Scholar] [CrossRef]
- Zangheri, M.; Mirasoli, M.; Guardigli, M.; Di Nardo, F.; Anfossi, L.; Baggiani, C.; Simoni, P.; Benassai, M.; Roda, A. Chemiluminescence-Based Biosensor for Monitoring Astronauts’ Health Status during Space Missions: Results from the International Space Station. Biosens. Bioelectron. 2019, 129, 260–268. [Google Scholar] [CrossRef]
- Zangheri, M.; Di Nardo, F.; Calabria, D.; Marchegiani, E.; Anfossi, L.; Guardigli, M.; Mirasoli, M.; Baggiani, C.; Roda, A. Smartphone Biosensor for Point-of-Need Chemiluminescence Detection of Ochratoxin A in Wine and Coffee. Anal. Chim. Acta 2021, 1163, 338515. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Park, J.K. Functional Packaging of Lateral Flow Strip Allows Simple Delivery of Multiple Reagents for Multistep Assays. Anal. Chem. 2016, 88, 10374–10378. [Google Scholar] [CrossRef]
- Panraksa, Y.; Jang, I.; Carrell, C.S.; Amin, A.G.; Chailapakul, O.; Chatterjee, D.; Henry, C.S. Simple Manipulation of Enzyme-Linked Immunosorbent Assay (ELISA) Using an Automated Microfluidic Interface. Anal. Methods 2022, 114, 1774–1781. [Google Scholar] [CrossRef]
- Han, K.N.; Choi, J.-S.; Kwon, J. Three-Dimensional Paper-Based Slip Device for One-Step Point-of-Care Testing. Sci. Rep. 2016, 6, 25710. [Google Scholar] [CrossRef]
- Park, J.; Shin, J.H.; Park, J.K. Pressed Paper-Based Dipstick for Detection of Foodborne Pathogens with Multistep Reactions. Anal. Chem. 2016, 88, 3781–3788. [Google Scholar] [CrossRef]
- Preechakasedkit, P.; Teekayupak, K.; Citterio, D.; Ruecha, N. Improvement in Sensitivity for Lateral Flow Immunoassay of Ferritin Using Novel Device Design Based on Gold-Enhanced Gold Nanoparticles. Sci. Rep. 2022, 12, 7831. [Google Scholar] [CrossRef]
- Fu, E.; Kauffman, P.; Lutz, B.; Yager, P. Chemical Signal Amplification in Two-Dimensional Paper Networks. Sens. Actuators B Chem. 2010, 149, 325–328. [Google Scholar] [CrossRef]
- Fu, E.; Liang, T.; Houghtaling, J.; Ramachandran, S.; Ramsey, S.A.; Lutz, B.; Yager, P. Enhanced Sensitivity of Lateral Flow Tests Using a Two-Dimensional Paper Network Format. Anal. Chem. 2011, 83, 7941–7946. [Google Scholar] [CrossRef] [PubMed]
- Fu, E.; Liang, T.; Spicar-Mihalic, P.; Houghtaling, J.; Ramachandran, S.; Yager, P. Two-Dimensional Paper Network Format That Enables Simple Multistep Assays for Use in Low-Resource Settings in the Context of Malaria Antigen Detection. Anal. Chem. 2012, 84, 4574–4579. [Google Scholar] [CrossRef] [PubMed]
- Lutz, B.; Liang, T.; Fu, E.; Ramachandran, S.; Kauffman, P.; Yager, P. Dissolvable Fluidic Time Delays for Programming Multi-Step Assays in Instrument-Free Paper Diagnostics. Lab Chip 2013, 13, 2840–2847. [Google Scholar] [CrossRef] [PubMed]
- Mulvaney, S.P.; Kidwell, D.A.; Lanese, J.N.; Lopez, R.P.; Sumera, M.E.; Wei, E. Catalytic Lateral Flow Immunoassays (CLFIATM): Amplified Signal in a Self-Contained Assay Format. Sens. Bio-Sens. Res. 2020, 30, 100390. [Google Scholar] [CrossRef]
- Han, D.K.; Oh, J.; Lee, J.; Cho, Y.G.; Park, J.S.; Choi, J.S.; Kim, D.S.; Kwon, J. Paper-Based Multiplex Analytical Device for Simultaneous Detection of Clostridioides Difficile Toxins and Glutamate Dehydrogenase. Biosens. Bioelectron. 2021, 176, 112894. [Google Scholar] [CrossRef]
- Kim, W.; Lee, S.; Jeon, S. Enhanced Sensitivity of Lateral Flow Immunoassays by Using Water-Soluble Nanofibers and Silver-Enhancement Reactions. Sens. Actuators B Chem. 2018, 273, 1323–1327. [Google Scholar] [CrossRef]
- Yang, X.; Huang, R.; Xiong, L.; Chen, F.; Sun, W.; Yu, L. A Colorimetric Aptasensor for Ochratoxin A Detection Based on Tetramethylrhodamine Charge Effect-Assisted Silver Enhancement. Biosensors 2023, 13, 468. [Google Scholar] [CrossRef]
- Komkova, M.A.; Karyakina, E.E.; Karyakin, A.A. Catalytically Synthesized Prussian Blue Nanoparticles Defeating Natural Enzyme Peroxidase. J. Am. Chem. Soc. 2018, 140, 11302–11307. [Google Scholar] [CrossRef]
- Xi, Z.; Wei, K.; Wang, Q.; Kim, M.J.; Sun, S.; Fung, V.; Xia, X. Nickel–Platinum Nanoparticles as Peroxidase Mimics with a Record High Catalytic Efficiency. J. Am. Chem. Soc. 2021, 143, 2660–2664. [Google Scholar] [CrossRef]
- Wu, J.; Qin, K.; Yuan, D.; Tan, J.; Qin, L.; Zhang, X.; Wei, H. Rational Design of Au@Pt Multibranched Nanostructures as Bifunctional Nanozymes. ACS Appl. Mater. Interfaces 2018, 10, 12954–12959. [Google Scholar] [CrossRef]
- Porstmann, B.; Porstmann, T.; Nugel, E.; Evers, U. Which of the Commonly Used Marker Enzymes Gives the Best Results in Colorimetric and Fluorimetric Enzyme Immunoassays: Horseradish Peroxidase, Alkaline Phosphatase or β-Galactosidase? J. Immunol. Methods 1985, 79, 27–37. [Google Scholar] [CrossRef]
- Lakshmi Kumari, G.; Dhir, R.N. Comparative Studies with Penicillinase, Horseradish Peroxidase, and Alkaline Phosphatase as Enzyme Labels in Developing Enzyme Immunoassay of Cortisol. J. Immunoass. Immunochem. 2003, 24, 173–190. [Google Scholar] [CrossRef]
- He, J. Practical Guide to ELISA Development. In The Immunoassay Handbook, 4th ed.; Wild, D.B.T.-T.I.H., Ed.; Elsevier: Oxford, UK, 2013; pp. 381–393. ISBN 978-0-08-097037-0. [Google Scholar]
- Han, G.R.; Koo, H.J.; Ki, H.; Kim, M.G. Paper/Soluble Polymer Hybrid-Based Lateral Flow Biosensing Platform for High-Performance Point-of-Care Testing. ACS Appl. Mater. Interfaces 2020, 12, 34564–34575. [Google Scholar] [CrossRef]
- Dargatz, D.A.; Byrum, B.A.; Collins, M.T.; Goyal, S.M.; Hietala, S.K.; Jacobson, R.H.; Kopral, C.A.; Martin, B.M.; McCluskey, B.J.; Tewari, D. A Multilaboratory Evaluation of a Commercial Enzyme-Linked Immunosorbent Assay Test for the Detection of Antibodies against Mycobacterium Avium Subsp. Paratuberculosis in Cattle. J. Vet. Diagn. Investig. 2004, 16, 509–514. [Google Scholar] [CrossRef]
- Pecson, B.M.; Darby, E.; Haas, C.N.; Amha, Y.M.; Bartolo, M.; Danielson, R.; Dearborn, Y.; Di Giovanni, G.; Ferguson, C.; Fevig, S.; et al. Reproducibility and Sensitivity of 36 Methods to Quantify the SARS-CoV-2 Genetic Signal in Raw Wastewater: Findings from an Interlaboratory Methods Evaluation in the U.S. Environ. Sci. Water Res. Technol. 2021, 7, 504–520. [Google Scholar] [CrossRef]
- Ley, B.; Satyagraha, A.W.; Kibria, M.G.; Armstrong, J.; Bancone, G.; Bei, A.K.; Bizilj, G.; Brito, M.; Ding, X.C.; Domingo, G.J.; et al. Repeatability and Reproducibility of a Handheld Quantitative G6PD Diagnostic. PLoS Negl. Trop. Dis. 2022, 16, 1–17. [Google Scholar] [CrossRef]
- Farka, Z.; Mickert, M.J.; Pastucha, M.; Mikušová, Z.; Skládal, P.; Gorris, H.H. Advances in Optical Single-Molecule Detection: En Route to Supersensitive Bioaffinity Assays. Angew. Chem. Int. Ed. 2020, 59, 10746–10773. [Google Scholar] [CrossRef]
- Parolo, C.; Sena-Torralba, A.; Bergua, J.F.; Calucho, E.; Fuentes-Chust, C.; Hu, L.; Rivas, L.; Álvarez-Diduk, R.; Nguyen, E.P.; Cinti, S.; et al. Tutorial: Design and Fabrication of Nanoparticle-Based Lateral-Flow Immunoassays. Nat. Protoc. 2020, 15, 3788–3816. [Google Scholar] [CrossRef]
- Hsieh, H.; Dantzler, J.; Weigl, B. Analytical Tools to Improve Optimization Procedures for Lateral Flow Assays. Diagnostics 2017, 7, 29. [Google Scholar] [CrossRef]
- Millipore, M. Rapid Lateral Flow Test Strips Considerations for Product Development. Available online: https://www.merckmillipore.com/INTERSHOP/web/WFS/Merck-RU-Site/ru_RU/-/USD/ShowDocument-Pronet?id=201306.15671 (accessed on 24 August 2023).
- Moosavi, S.M.; Ghassabian, S. Linearity of Calibration Curves for Analytical Methods: A Review of Criteria for Assessment of Method Reliability. In Calibration and Validation of Analytical Methods—A Sampling of Current Approaches; Stauffer, M.T., Ed.; InTechOpen: Rijeka, Croatia, 2018; p. Ch. 6. ISBN 978-1-78923-085-7. [Google Scholar]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. The ACT-Accelerator: Two Years of Impact. Available online: https://www.who.int/publications/m/item/the-act-accelerator--two-years-of-impact (accessed on 24 August 2023).
- Davies, B.; Araghi, M.; Moshe, M.; Gao, H.; Bennet, K.; Jenkins, J.; Atchison, C.; Darzi, A.; Ashby, D.; Riley, S.; et al. Acceptability, Usability, and Performance of Lateral Flow Immunoassay Tests for Severe Acute Respiratory Syndrome Coronavirus 2 Antibodies: REACT-2 Study of Self-Testing in Nonhealthcare Key Workers. Open Forum Infect. Dis. 2021, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Møller, I.J.B.; Utke, A.R.; Rysgaard, U.K.; Østergaard, L.J.; Jespersen, S. Diagnostic Performance, User Acceptability, and Safety of Unsupervised SARS-CoV-2 Rapid Antigen-Detecting Tests Performed at Home. Int. J. Infect. Dis. 2022, 116, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Wang, K.; Zheng, W.; Cheng, Y.; Li, T.; Cao, B.; Jin, Q.; Cui, D. Rapid Developments in Lateral Flow Immunoassay for Nucleic Acid Detection. Analyst 2021, 146, 1514–1528. [Google Scholar] [CrossRef]
- Kim, K.; Lee, B.; Park, J.H.; Park, J.-H.; Lee, K.J.; Kwak, T.J.; Son, T.; Shin, Y.-B.; Im, H.; Kim, M.-G. Rapid PCR Kit: Lateral Flow Paper Strip with Joule Heater for SARS-CoV-2 Detection. Mater. Horizons 2023, 10, 1697–1704. [Google Scholar] [CrossRef]
- Bender, A.T.; Sullivan, B.P.; Zhang, J.Y.; Juergens, D.C.; Lillis, L.; Boyle, D.S.; Posner, J.D. HIV Detection from Human Serum with Paper-Based Isotachophoretic RNA Extraction and Reverse Transcription Recombinase Polymerase Amplification. Analyst 2021, 146, 2851–2861. [Google Scholar] [CrossRef]
- Sullivan, B.P.; Chou, Y.S.; Bender, A.T.; Martin, C.D.; Kaputa, Z.G.; March, H.; Song, M.; Posner, J.D. Quantitative Isothermal Amplification on Paper Membranes Using Amplification Nucleation Site Analysis. Lab Chip 2022, 22, 2352–2363. [Google Scholar] [CrossRef]
- Bender, A.T.; Borysiak, M.D.; Levenson, A.M.; Lillis, L.; Boyle, D.S.; Posner, J.D. Semiquantitative Nucleic Acid Test with Simultaneous Isotachophoretic Extraction and Amplification. Anal. Chem. 2018, 90, 7221–7229. [Google Scholar] [CrossRef]
- Chang, L.; Li, J.; Wang, L. Immuno-PCR: An Ultrasensitive Immunoassay for Biomolecular Detection. Anal. Chim. Acta 2016, 910, 12–24. [Google Scholar] [CrossRef]
- Kim, S.; Sikes, H.D. Radical Polymerization Reactions for Amplified Biodetection Signals. Polym. Chem. 2020, 11, 1424–1444. [Google Scholar] [CrossRef]



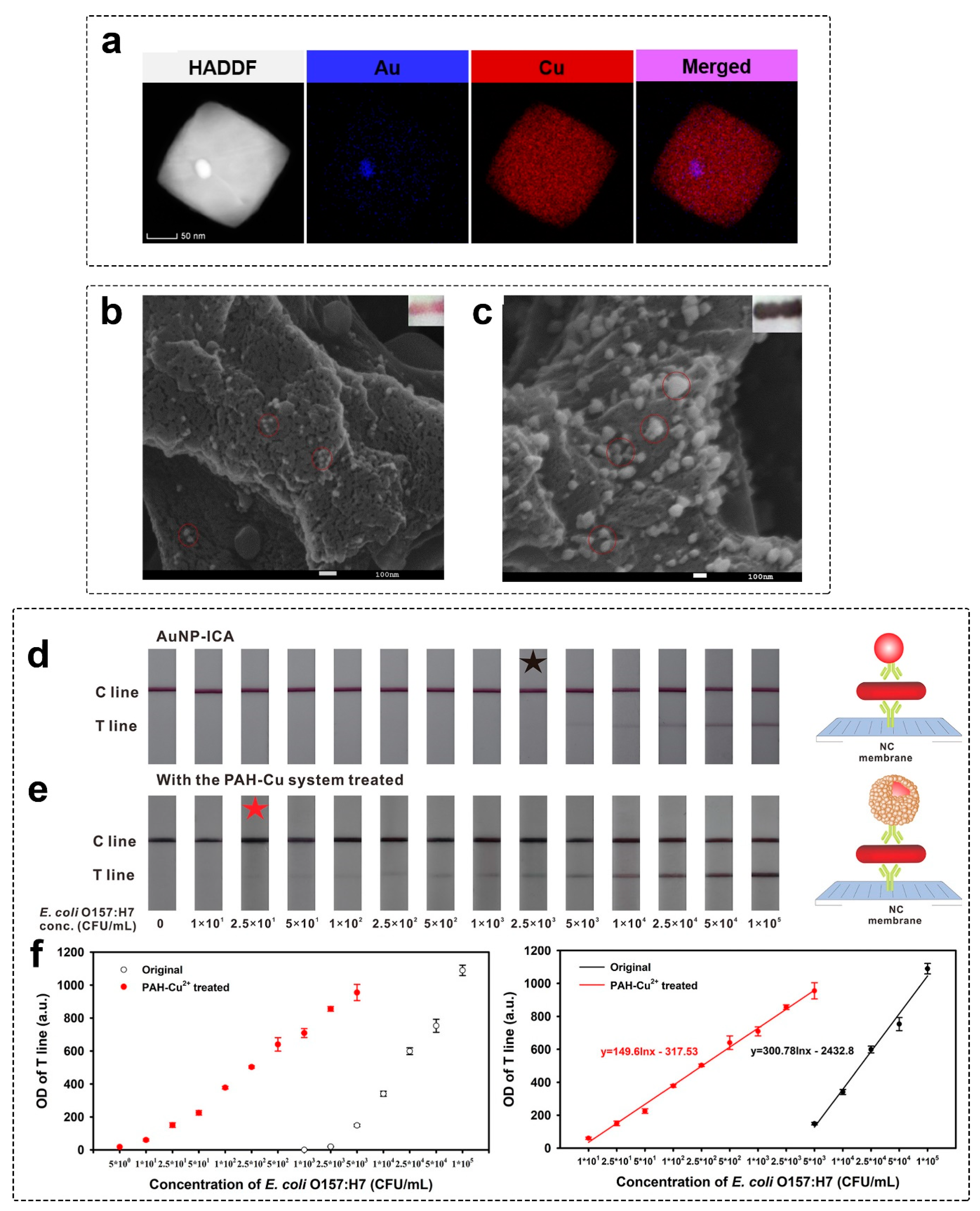
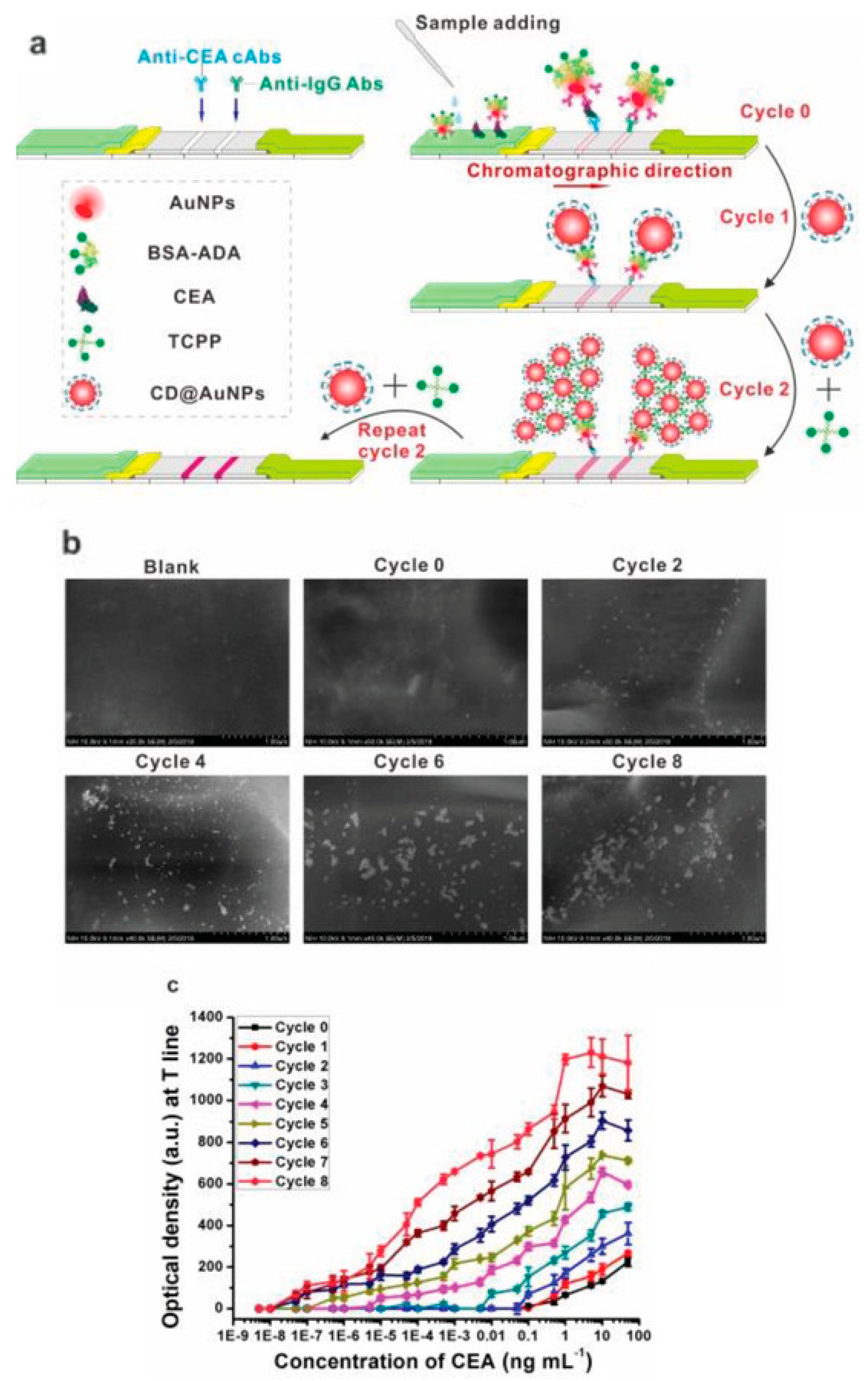
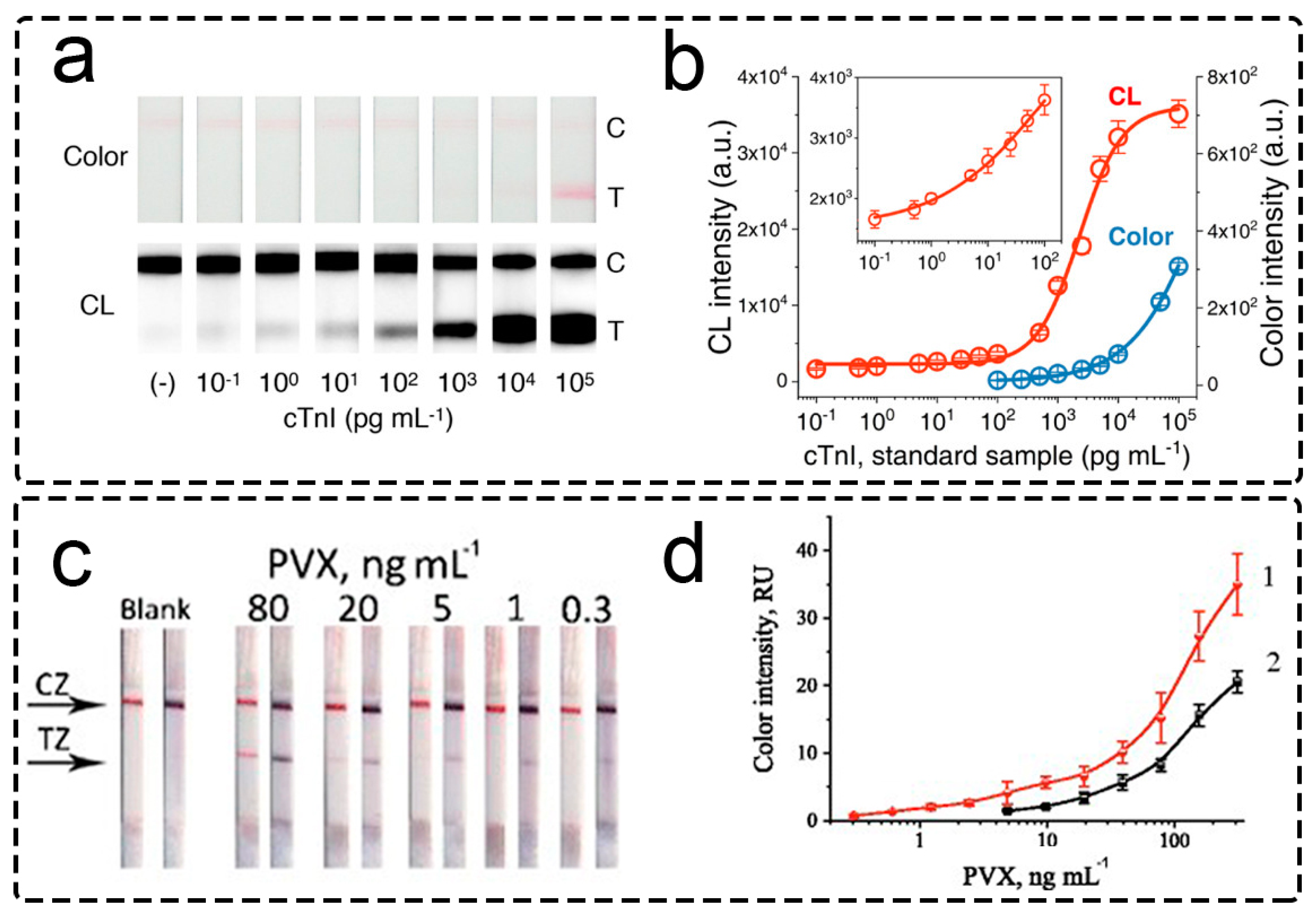
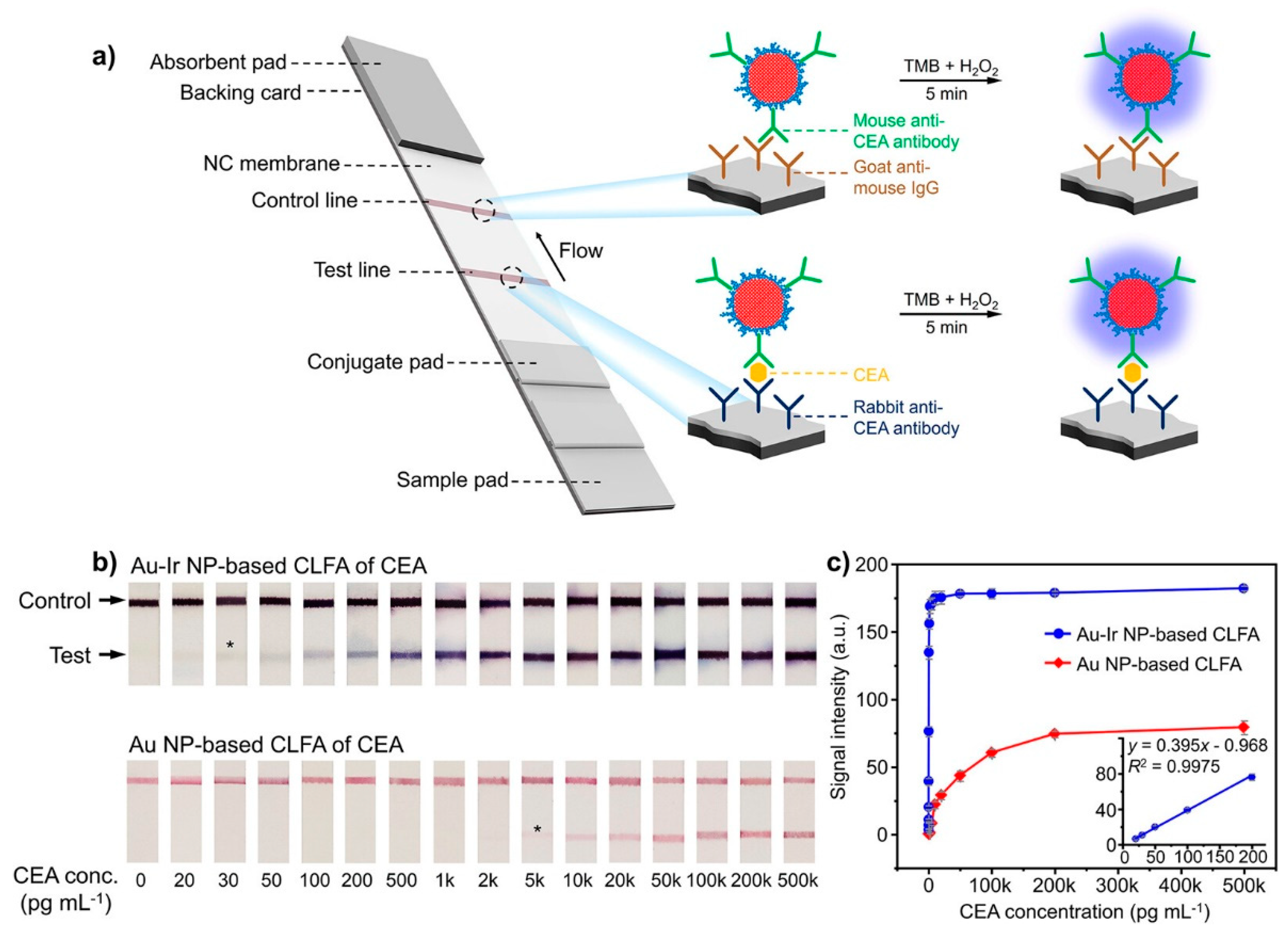
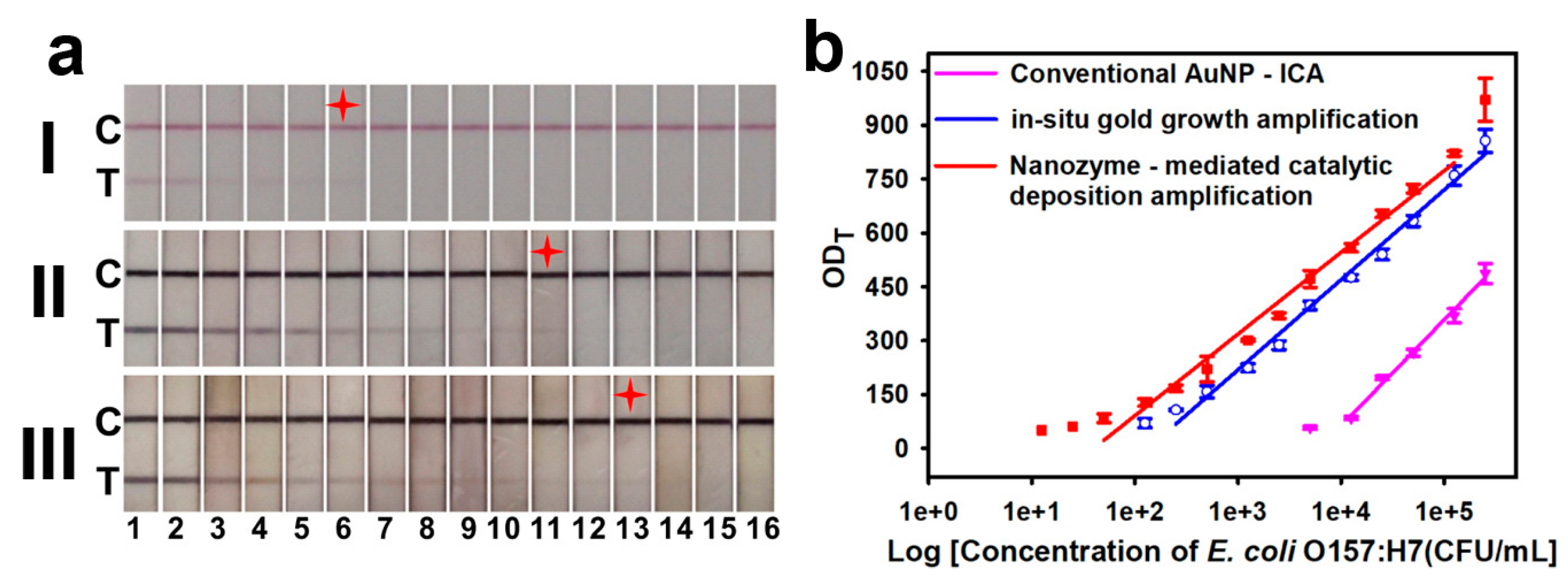
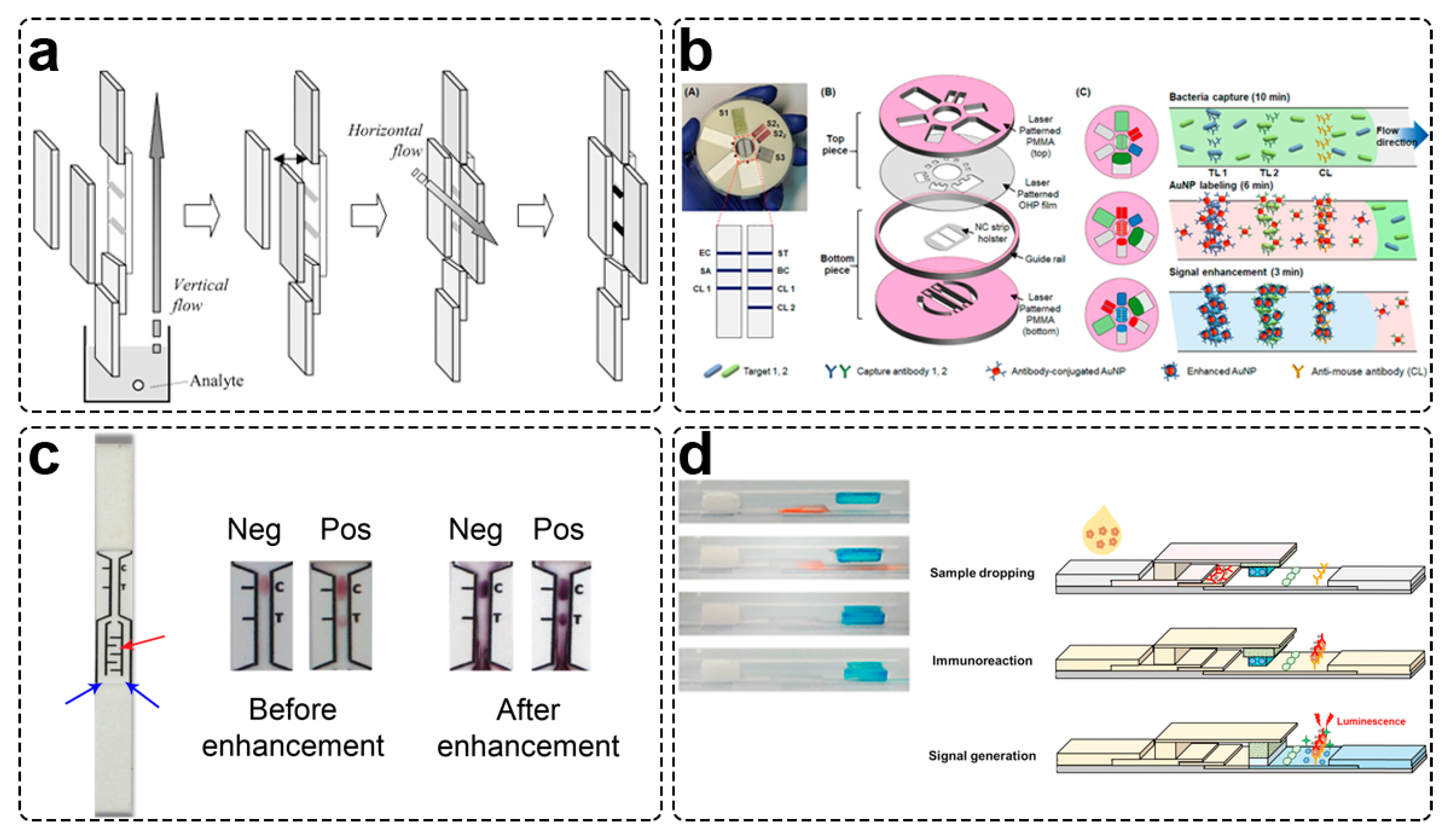
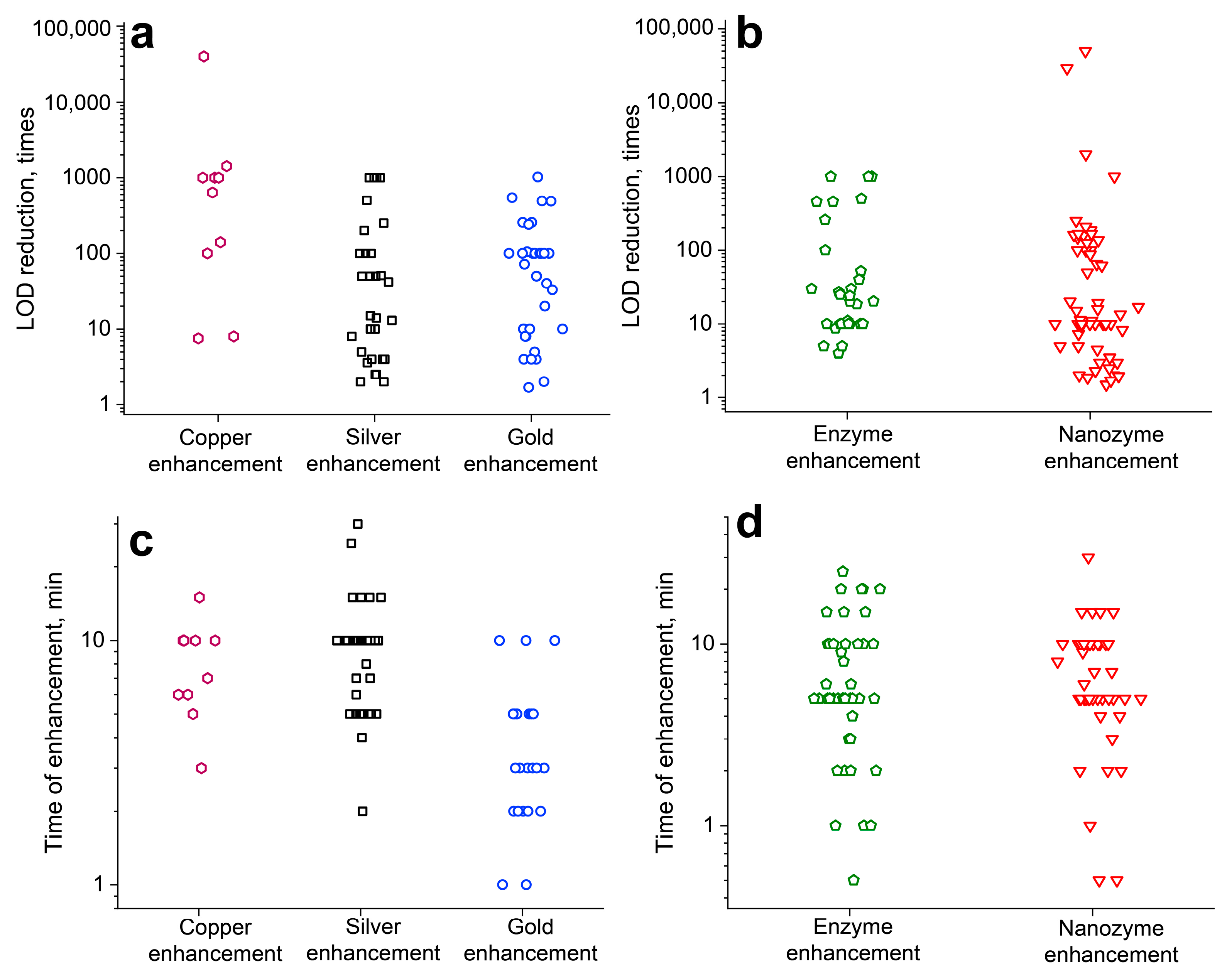
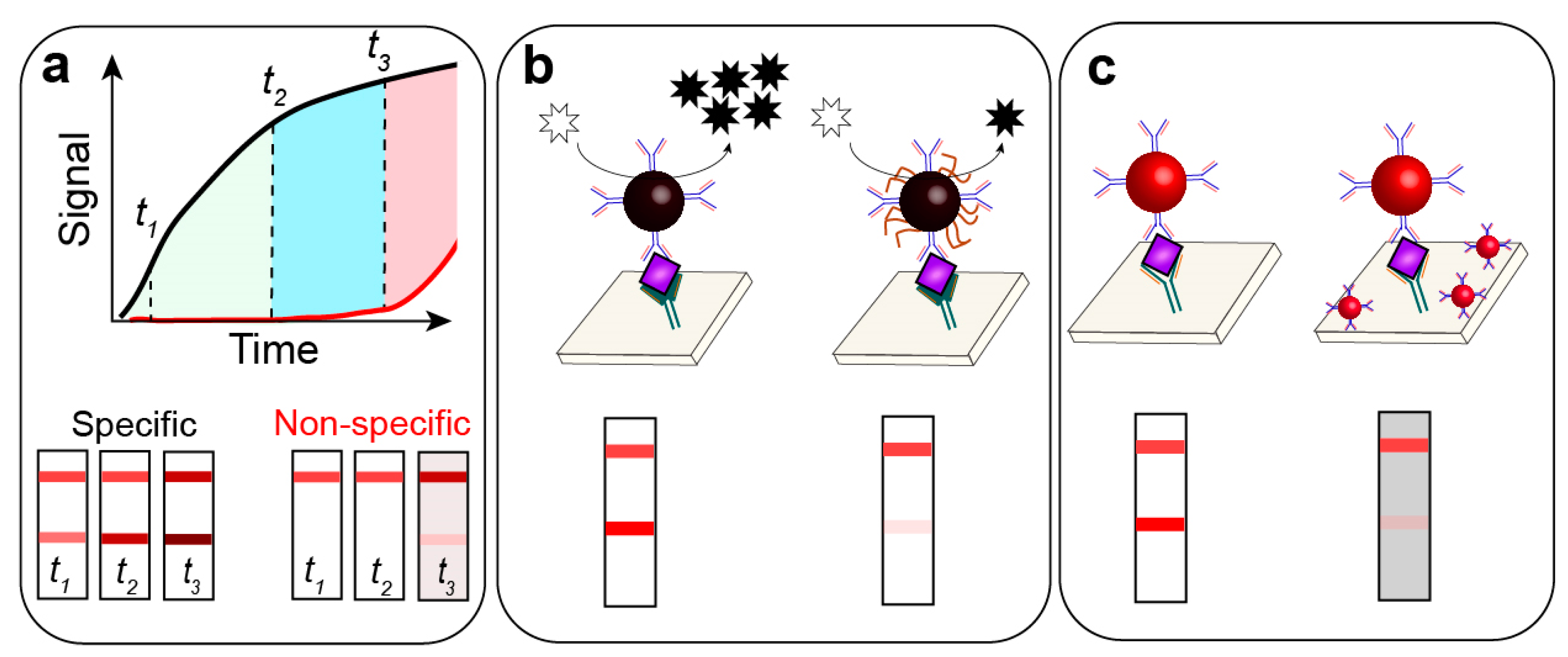
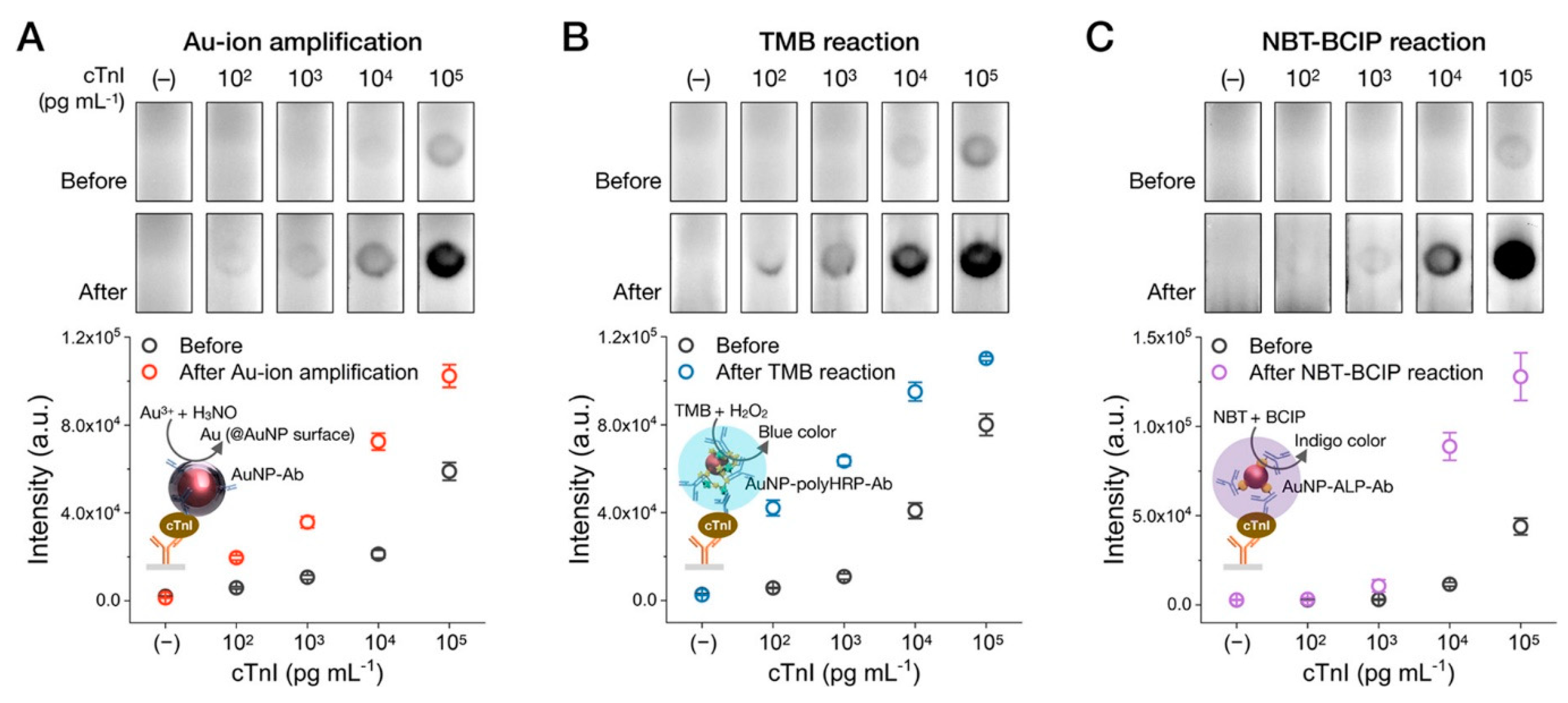
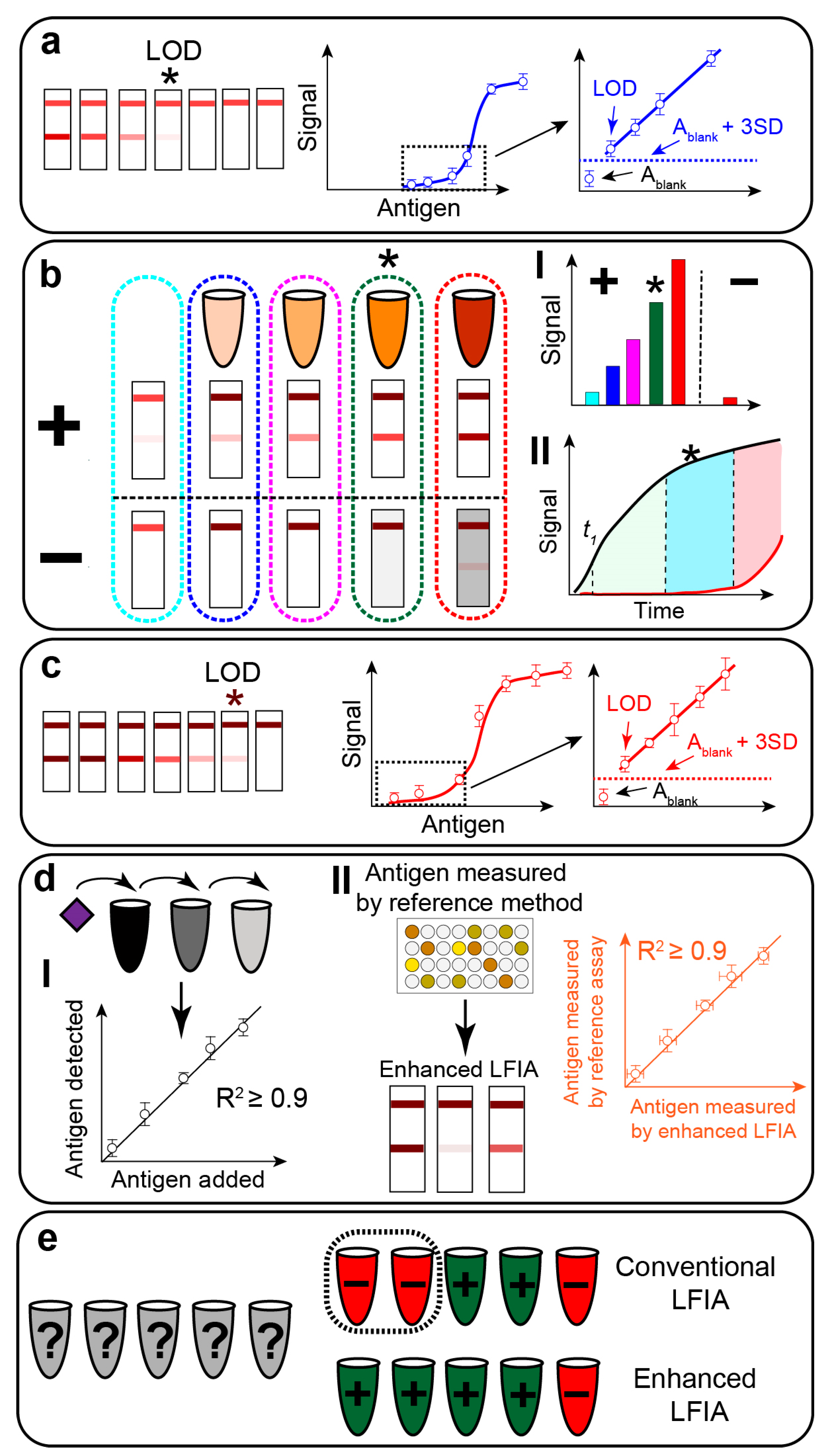
| Enhancement Strategy | Range of LOD Reduction, Times | Duration of the Enhancement Stage, Min | Comments |
|---|---|---|---|
| Copper enhancement | 7.5–40,000 | 3–15 |
|
| Silver enhancement | 2–1000 | 2–30 |
|
| Gold enhancement | 1.68–1024 | 1–10 |
|
| Enzyme enhancement | 4–1000 | 0.5–25 |
|
| Nanozyme enhancement | 1.7–50,000 | 0.5–30 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panferov, V.G.; Zherdev, A.V.; Dzantiev, B.B. Post-Assay Chemical Enhancement for Highly Sensitive Lateral Flow Immunoassays: A Critical Review. Biosensors 2023, 13, 866. https://doi.org/10.3390/bios13090866
Panferov VG, Zherdev AV, Dzantiev BB. Post-Assay Chemical Enhancement for Highly Sensitive Lateral Flow Immunoassays: A Critical Review. Biosensors. 2023; 13(9):866. https://doi.org/10.3390/bios13090866
Chicago/Turabian StylePanferov, Vasily G., Anatoly V. Zherdev, and Boris B. Dzantiev. 2023. "Post-Assay Chemical Enhancement for Highly Sensitive Lateral Flow Immunoassays: A Critical Review" Biosensors 13, no. 9: 866. https://doi.org/10.3390/bios13090866
APA StylePanferov, V. G., Zherdev, A. V., & Dzantiev, B. B. (2023). Post-Assay Chemical Enhancement for Highly Sensitive Lateral Flow Immunoassays: A Critical Review. Biosensors, 13(9), 866. https://doi.org/10.3390/bios13090866







