Magnetically Assisted Immobilization-Free Detection of microRNAs Based on the Signal Amplification of Duplex-Specific Nuclease
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Procedures for miRNA Detection
2.3. Assays of miRNAs in Cell Lysates
3. Results and Discussion
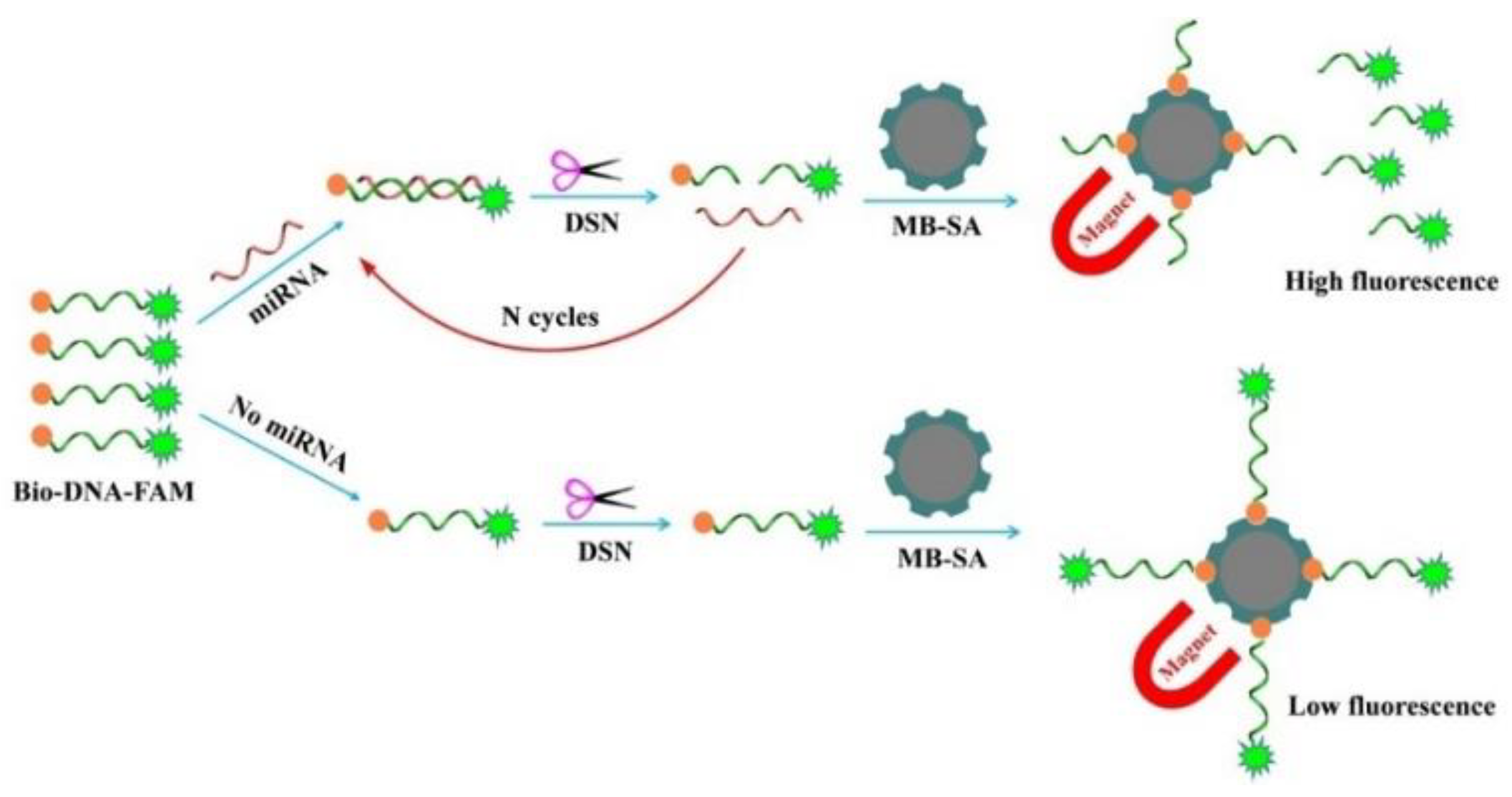
3.1. Detection Principle
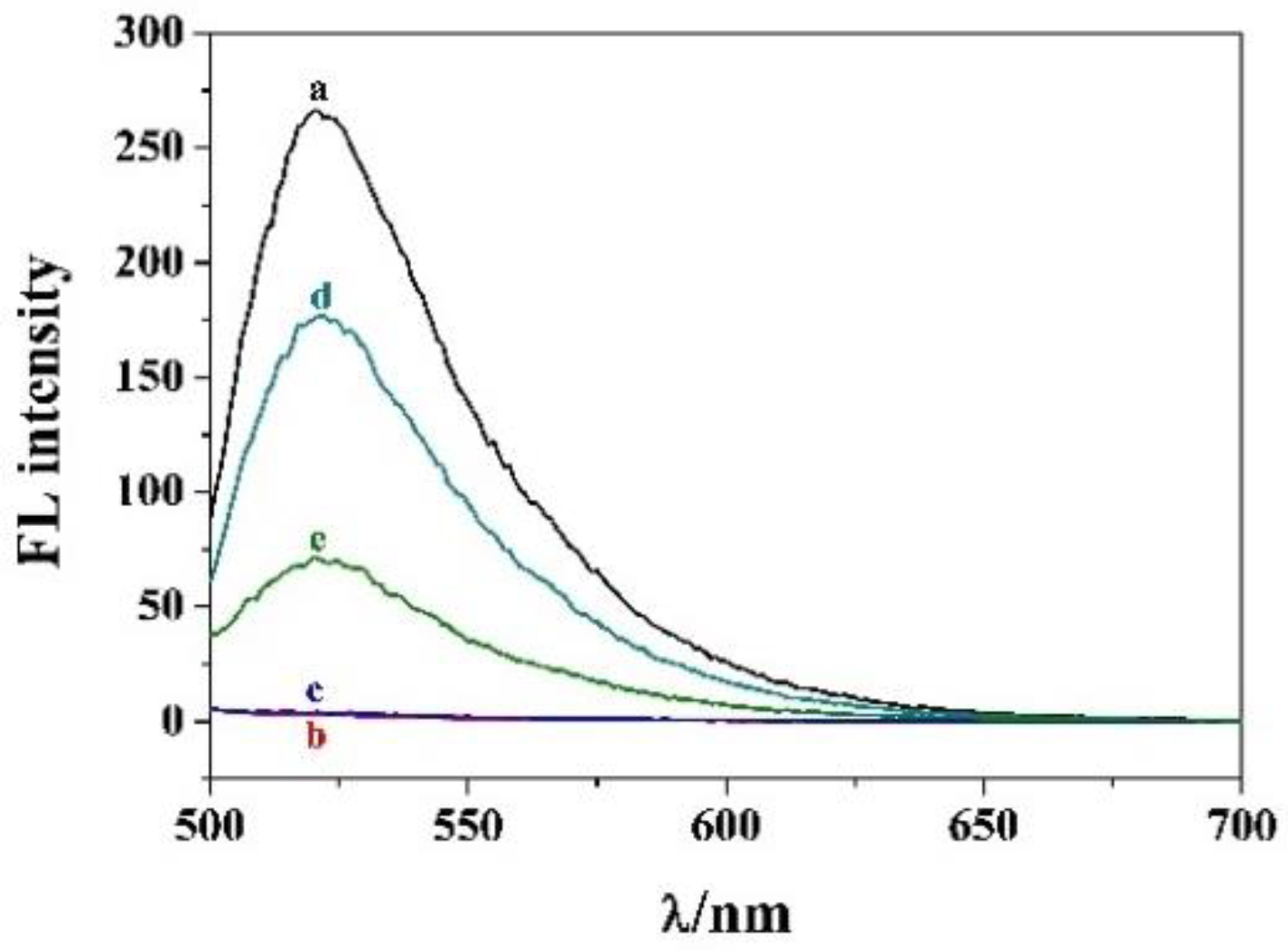
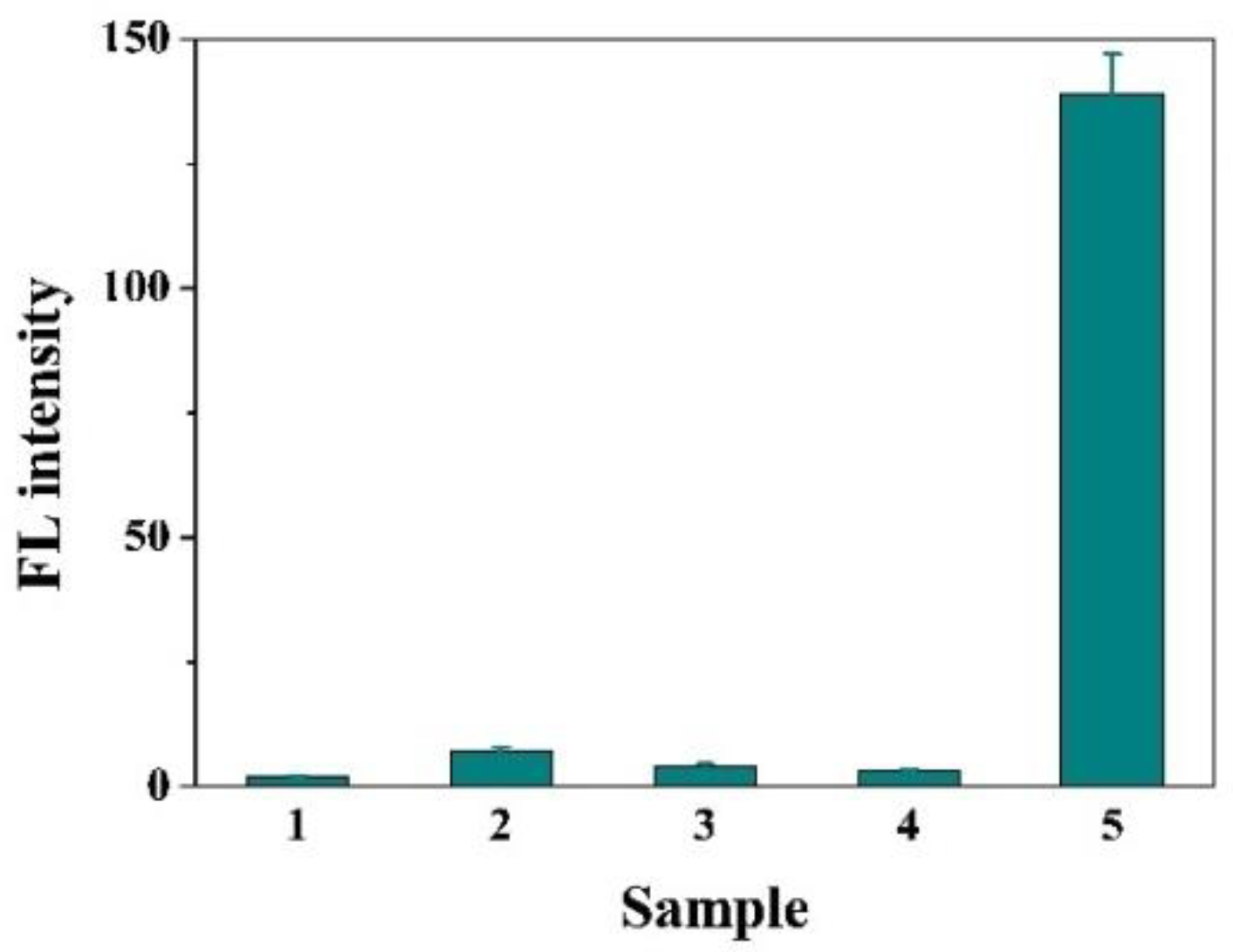
3.2. Feasibility of this Method
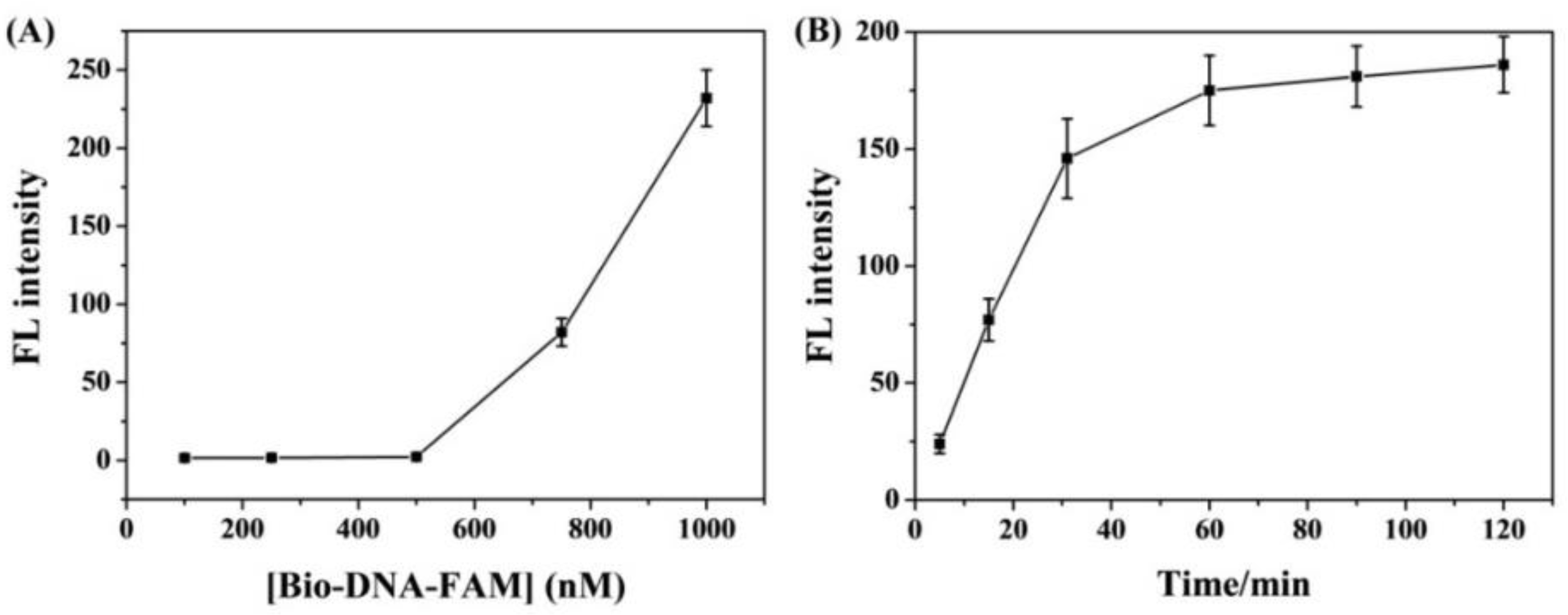
3.3. Optimization of Experimental Conditions
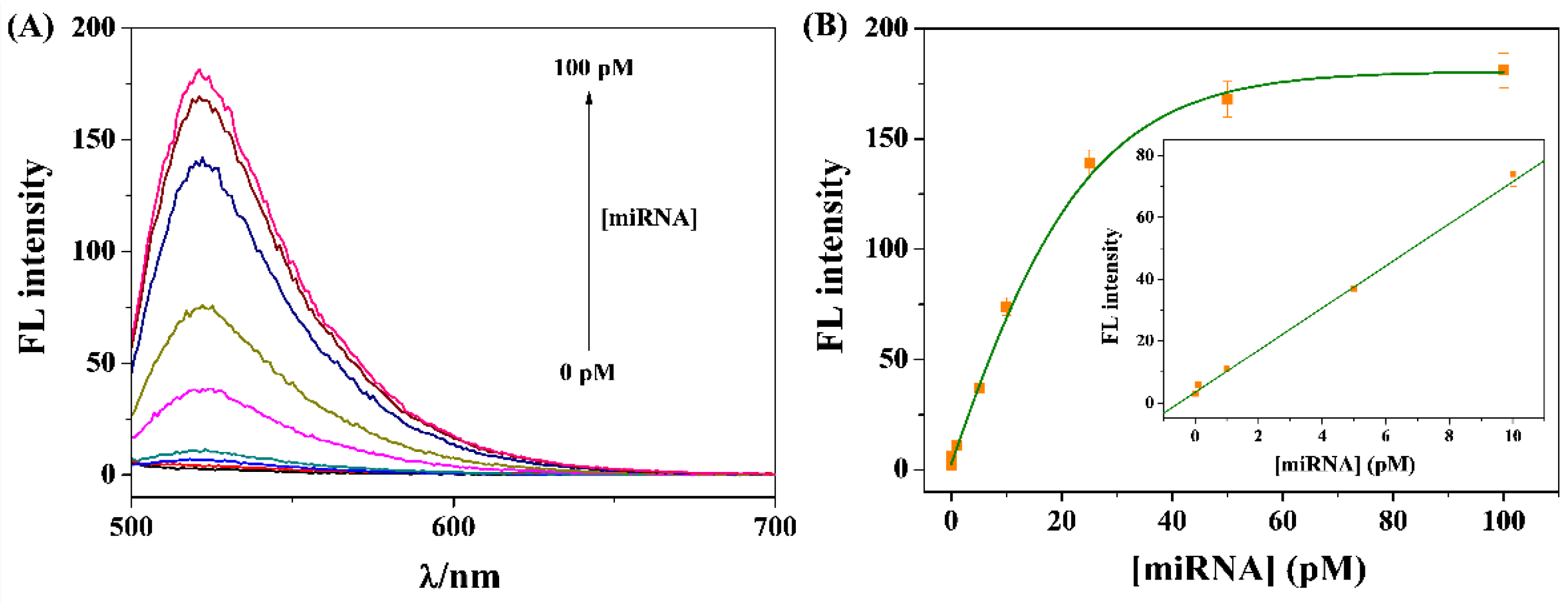
3.4. Sensitivity for miRNA-21 Detection
3.5. Selectivity
3.6. Assays of miRNA-21 in Cellular Lysates
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dong, H.; Lei, J.; Ding, L.; Wen, Y.; Ju, H.; Zhang, X. MicroRNA: Function, detection, and bioanalysis. Chem. Rev. 2013, 113, 6207–6233. [Google Scholar] [CrossRef] [PubMed]
- Imas, J.J.; Zamarreño, C.R.; Zubiate, P.; Sanchez-Martín, L.; Campión, J.; Matías, I.R. Optical biosensors for the detection of Rheumatoid Arthritis (RA) biomarkers: A comprehensive review. Sensors 2020, 20, 6289. [Google Scholar] [CrossRef] [PubMed]
- Chai, H.; Tang, Y.; Miao, P. Tetrahedral DNA supported walking nanomachine for ultrasensitive miRNA detection in cancer cells and serums. Anal. Chem. 2022, 94, 9975–9980. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Cui, S.; Li, Q.; Zhang, R.; Song, Z.; Gao, Y.; Chen, W.; Xing, D. Recent advances in duplex-specific nuclease-based signal amplification strategies for microRNA detection. Biosens. Bioelectron. 2020, 165, 112449. [Google Scholar] [CrossRef]
- Ashraf, G.; Zhong, Z.T.; Asif, M.; Aziz, A.; Iftikhar, T.; Chen, W.; Zhao, Y.D. State-of-the-art fluorescent probes: Duplex-specific nuclease-based strategies for early disease diagnostics. Biosensors 2022, 12, 1172. [Google Scholar] [CrossRef]
- Wu, D.; Yi, X.; Xia, N. Electrochemical biosensors for microRNA detection using duplex-specific nuclease based signal amplification strategies. Int. J. Electrochem. Sci. 2020, 15, 12136–12148. [Google Scholar] [CrossRef]
- Qiu, X.; Liu, X.; Zhang, W.; Zhang, H.; Jiang, T.; Fan, D.; Luo, Y. Dynamic monitoring of microRNA-DNA hybridization using DNAase-triggered signal amplification. Anal. Chem. 2015, 87, 6303–6310. [Google Scholar] [CrossRef]
- Ashraf, G.; Zhong, Z.-T.; Asif, M.; Aziz, A.; Song, L.; Zhang, S.; Liu, B.; Chen, W.; Zhao, Y.-D. Extension of duplex specific nuclease sensing application with RNA aptamer. Talanta 2022, 242, 123314. [Google Scholar] [CrossRef]
- Zhu, J.; Guo, Z.; Cui, J.; Miao, P. Partial collapse of DNA tetrahedron for miRNA assay with duplex-specific nuclease-assisted amplification. Analyst 2023, 148, 512–515. [Google Scholar] [CrossRef]
- Castañeda, A.D.; Brenes, N.J.; Kondajji, A.; Crooks, R.M. Detection of microRNA by electrocatalytic amplification: A general approach for aingle-particle biosensing. J. Am. Chem. Soc. 2017, 139, 7657–7664. [Google Scholar] [CrossRef]
- Yin, B.-C.; Liu, Y.-Q.; Ye, B.-C. One-step, multiplexed fluorescence detection of microRNAs based on duplex-specific nuclease signal amplification. J. Am. Chem. Soc. 2012, 134, 5064–5067. [Google Scholar] [CrossRef]
- Zhou, H.; Yang, C.; Chen, H.; Li, X.; Li, Y.; Fan, X. A simple G-quadruplex molecular beacon-based biosensor for highly selective detection of microRNA. Biosens. Bioelectron. 2017, 87, 552–557. [Google Scholar] [CrossRef]
- Hu, Z.; Chen, J.; Li, W.; Wang, Y.; Li, Y.; Sang, L.; Li, J.; Zhang, Q.; Ibupoto, Z.H.; Yu, C. Label-free fluorescence turn-on detection of microRNA based on duplex-specific nuclease and a perylene probe. Anal. Chim. Acta 2015, 895, 89–94. [Google Scholar] [CrossRef]
- Lu, W.; Chen, Y.; Liu, Z.; Tang, W.; Feng, Q.; Sun, J.; Jiang, X. Quantitative detection of microRNA in one step via next generation magnetic relaxation switch sensing. ACS Nano 2016, 10, 6685–6692. [Google Scholar] [CrossRef]
- Pang, Y.; Wang, C.; Wang, J.; Sun, Z.; Xiao, R.; Wang, S. Fe3O4@Ag magnetic nanoparticles for microRNA capture and duplex-specific nuclease signal amplification based SERS detection in cancer cells. Biosens. Bioelectron. 2016, 79, 574–580. [Google Scholar] [CrossRef]
- Li, M.; Xiong, C.; Zheng, Y.; Liang, W.; Yuan, R.; Chai, Y. Ultrasensitive photoelectrochemical biosensor based on DNA tetrahedron as nanocarrier for efficient immobilization of CdTe QDs-methylene blue as signal probe with near-zero background noise. Anal. Chem. 2018, 90, 8211–8216. [Google Scholar] [CrossRef]
- Bai, Y.-Y.; Wu, Z.; Xu, C.-M.; Zhang, L.; Feng, J.; Pang, D.-W.; Zhang, Z.-L. One-to-many single entity electrochemistry biosensing for ultrasensitive detection of microRNA. Anal. Chem. 2020, 92, 853–858. [Google Scholar] [CrossRef]
- Tian, B.; Ma, J.; Qiu, Z.; Zardán Gómez de la Torre, T.; Donolato, M.; Hansen, M.F.; Svedlindh, P.; Strömberg, M. Optomagnetic detection of microRNA based on duplex-specific nuclease-assisted target recycling and multilayer core-satellite magnetic superstructures. ACS Nano 2017, 11, 1798–1806. [Google Scholar] [CrossRef]
- Kim, E.; Howes, P.D.; Crowder, S.W.; Stevens, M.M. Multi-amplified sensing of microRNA by a small DNA fragment-driven enzymatic cascade reaction. ACS Sens. 2017, 2, 111–118. [Google Scholar] [CrossRef]
- Deng, H.; Ren, Y.; Shen, W.; Gao, Z. An ultrasensitive homogeneous chemiluminescent assay for microRNAs. Chem. Commun. 2013, 49, 9401–9403. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, D.; Chen, Q.; Chen, M.; Xia, Y.; Cai, S.; Zhang, X.; Wu, F.; Chen, J. Label-free microRNA detection based on terbium and duplex-specific nuclease assisted target recycling. Analyst 2015, 140, 5082–5089. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Xu, Z.; Liu, J.-H.; Yu, B.-Y.; Tian, J. Dual signal amplification for microRNA-21 detection based on duplex-specific nuclease and invertase. RSC Adv. 2020, 10, 11257–11262. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tong, Y.; Sun, Z.; Chen, Y.; Wang, Y.; Zhou, L.; Jiang, Y.; Li, P.; Wang, C.; Du, L. A duplex-specific nuclease assisted photoelectrochemical biosensor based on MoS2@ReS2/Ti3C2 hybrid for ultrasensitive detection of colorectal cancer-related piRNA-31,143. Acta Biomater. 2022, 149, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Yeo, K.H.; Gao, Z. A simple and highly sensitive fluorescence assay for microRNAs. Analyst 2015, 140, 1932–1938. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Shen, W.; Ren, Y.; Gao, Z. A highly sensitive and selective homogenous assay for profiling microRNA expression. Biosens. Bioelectron. 2014, 54, 650–655. [Google Scholar] [CrossRef]
- Fu, C.; Liu, C.; Wang, S.; Luo, F.; Lin, Z.; Chen, G. A signal-on homogeneous electrochemical biosensor for sequence-specific microRNA based on duplex-specific nuclease-assisted target recycling amplification. Anal. Methods 2016, 8, 7034–7039. [Google Scholar] [CrossRef]
- Djebbi, K.; Shi, B.; Weng, T.; Bahri, M.; Elaguech, M.A.; Liu, J.; Tlili, C.; Wang, D. Highly sensitive fluorescence assay for miRNA detection: Investigation of the DNA spacer effect on the DSN enzyme activity toward magnetic-bead-tethered probes. ACS Omega 2022, 7, 2224–2233. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, D.Z.; Cai, S.X.; Chen, M.; Xia, Y.K.; Wu, F.; Chen, J.H. An immobilization-free electrochemical impedance biosensor based on duplex-specific nuclease assisted target recycling for amplified detection of microRNA. Biosens. Bioelectron. 2016, 75, 452–457. [Google Scholar] [CrossRef]
- Chang, J.F.; Wang, X.; Wang, J.; Li, H.Y.; Li, F. Nucleic acid functionalized MOFs-based homogeneous electrochemical biosensor for simultaneous detection of multiple tumor biomarkers. Anal. Chem. 2019, 91, 3604–3610. [Google Scholar] [CrossRef]
- Seferos, D.S.; Prigodich, A.; Giljohann, D.A.; Patel, P.C.; Mirkin, C.A. Polyvalent DNA nanoparticle conjugates stabilize nucleic acids. Nano Lett. 2009, 9, 308–311. [Google Scholar] [CrossRef]
- Prigodich, A.E.; Alhasan, A.H.; Mirkin, C.A. Selective enhancement of nucleases by polyvalent DNA-functionalized gold nanoparticles. J. Am. Chem. Soc. 2011, 133, 2120–2123. [Google Scholar] [CrossRef]
- Zhao, G.; Yan, X.; Zhang, Y.; Deng, J.; Liang, X. Sensitive detection of miRNA and circRNA through DSN enzyme cooperating NEase assisted dual signal amplification. Anal. Biochem. 2022, 654, 114744. [Google Scholar] [CrossRef]
- Ma, F.; Liu, W.J.; Zhang, Q.; Zhang, C.Y. Sensitive detection of microRNAs by duplex specific nuclease-assisted target recycling and pyrene excimer switching. Chem. Commun. 2017, 53, 10596–10599. [Google Scholar] [CrossRef]
- Qi, T.; Song, C.; He, J.; Shen, W.; Kong, D.; Shi, H.; Tan, L.; Pan, R.; Tang, S.; Lee, H.K. Highly sensitive detection of multiple microRNAs by high-performance liquid chromatography coupled with long and short probe-based recycling amplification. Anal. Chem. 2020, 92, 5033–5040. [Google Scholar] [CrossRef]
- Yang, L.M.; Yin, X.H.; An, B.; Li, F. Precise capture and direct quantification of tumor exosomes via highly efficient dual-aptamer recognition-assisted ratiometric immobilization-free electrochemical strategy. Anal. Chem. 2021, 93, 1709–1716. [Google Scholar] [CrossRef]
- Sukla, S.; Mondal, P.; Biswas, S.; Ghosh, S. A rapid and easy-to-perform method of nucleic-acid based dengue virus diagnosis using fluorescence-based molecular beacons. Biosensors 2021, 11, 479. [Google Scholar] [CrossRef]
- Wang, L.; Lin, J. Recent advances on magnetic nanobead based biosensors: From separation to detection. TrAC-Trend. Anal. Chem. 2020, 128, 115915. [Google Scholar] [CrossRef]
- Xia, N.; Sun, T.; Liu, L.; Tian, L.; Sun, Z. Heterogeneous sensing of post-translational modification enzymes by integrating the advantage of homogeneous analysis. Talanta 2022, 237, 122949. [Google Scholar] [CrossRef]
- Liu, L.; Deng, D.; Wu, D.; Hou, W.; Wang, L.; Li, N.; Sun, Z. Duplex-specific nuclease-based electrochemical biosensor for the detection of microRNAs by conversion of homogeneous assay into surface-tethered electrochemical analysis. Anal. Chim. Acta 2021, 1149, 338199. [Google Scholar] [CrossRef]
- Huang, Y.; Sun, T.; Liu, L.; Xia, N.; Zhao, Y.; Yi, X. Surface plasmon resonance biosensor for the detection of miRNAs by combining the advantages of homogeneous reaction and heterogeneous detection. Talanta 2021, 234, 122622. [Google Scholar] [CrossRef]
- Degliangeli, F.; Kshirsagar, P.; Brunetti, V.; Pompa, P.P.; Fiammengo, R. Absolute and direct microRNA quantification using DNA—Gold nanoparticle probes. J. Am. Chem. Soc. 2014, 136, 2264–2267. [Google Scholar] [CrossRef] [PubMed]
- Daniels, J.S.; Pourmand, N. Label-free impedance biosensors: Opportunities and challenges. Electroanalysis 2007, 19, 1239–1257. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.; Cheng, Z.; Ma, H.; Li, Z.; Xue, N.; Wang, P. Label-free platform for microRNA detection based on the fluorescence quenching of positively charged gold nanoparticles to silver nanoclusters. Anal. Chem. 2018, 90, 1098–1103. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Li, J.; Yang, Y.; Tong, Y.; Li, H.; Wang, C.; Du, L.; Jiang, Y. Ratiometric fluorescent biosensor based on self-assembled fluorescent gold nanoparticles and duplex-specific nuclease-assisted signal amplification for sensitive detection of exosomal miRNA. Bioconjug. Chem. 2022, 33, 1698–1706. [Google Scholar] [CrossRef]
- Zhao, Q.; Piao, J.; Peng, W.; Wang, Y.; Zhang, B.; Gong, X.; Chang, J. Simple and sensitive quantification of microRNAs via PS@Au microspheres-based DNA probes and DSN-assisted signal amplification platform. ACS Appl. Mater. Interfaces 2018, 10, 3324–3332. [Google Scholar] [CrossRef]
- Ma, H.; Xue, N.; Li, Z.; Xing, K.; Miao, X. Ultrasensitive detection of miRNA-155 using multi-walled carbon nanotube-gold nanocomposites as a novel fluorescence quenching platform. Sens. Actuat. B Chem. 2018, 266, 221–227. [Google Scholar] [CrossRef]
- Guo, S.; Yang, F.; Zhang, Y.; Ning, Y.; Yao, Q.; Zhang, G.-J. Amplified fluorescence sensing of miRNA by combination of graphene oxide with duplex-specific nuclease. Anal. Methods 2014, 6, 3598–3603. [Google Scholar] [CrossRef]
- Yang, Z.; Qin, L.; Yang, D.; Chen, W.; Qian, Y.; Jin, J. Signal amplification method for miR-205 assay through combining graphene oxide with duplex-specific nuclease. RSC Adv. 2019, 9, 27341–27346. [Google Scholar] [CrossRef]
- Xiao, M.; Man, T.; Zhu, C.; Pei, H.; Shi, J.; Li, L.; Qu, X.; Shen, X.; Li, J. MoS2 nanoprobe for microRNA quantification based on duplex-specific nuclease signal amplification. ACS Appl. Mater. Interfaces 2018, 10, 7852–7858. [Google Scholar] [CrossRef]
- Gao, Z.; Yuan, H.; Mao, Y.; Ding, L.; Effah, C.Y.; He, S.; He, L.; Liu, L.-E.; Yu, S.; Wang, Y.; et al. In situ detection of plasma exosomal microRNA for lung cancer diagnosis using duplex-specific nuclease and MoS2 nanosheets. Analyst 2021, 146, 1924–1931. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, J.; Zhao, L.; Li, Y.; Chen, H.; Li, S.; Cheng, Y. Visual detection of multiplex microRNAs using cationic conjugated polymer materials. ACS Appl. Mater. Interfaces 2016, 8, 1520–1526. [Google Scholar] [CrossRef]
- Deng, X.; Wu, S.; Zang, S.; Liu, X.; Ma, Y. PDA—PEI-copolymerized nanodots with tailorable fluorescence emission and quenching properties for the sensitive ratiometric fluorescence sensing of miRNA in serum. Anal. Chem. 2022, 94, 14546–14553. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, C.; Tang, L.; Zhang, Y.; Zhang, G.-J. Magnetic-enhanced fluorescence sensing of tumor miRNA by combination of MNPs@PDA with duplex specific nuclease. RSC Adv. 2021, 11, 2968–2975. [Google Scholar] [CrossRef]
- Wang, J.-J.; Zheng, C.; Jiang, Y.-Z.; Zheng, Z.; Lin, M.; Lin, Y.; Zhang, Z.-L.; Wang, H.; Pang, D.-W. One-step monitoring of multiple enterovirus 71 infection-related microRNAs using core-satellite structure of magnetic nanobeads and multicolor quantum dots. Anal. Chem. 2020, 92, 830–837. [Google Scholar] [CrossRef]
- Yan, T.; Zhang, S.; Yang, Y.; Li, Y.; Xu, L. Biomineralizationinspired magnetic nanoflowers for sensitive miRNA detection based on exonuclease assisted target recycling amplification. Microchim. Acta 2022, 189, 260. [Google Scholar] [CrossRef]
- Kaplan, M.; Kilic, T.; Guler, G.; Mandli, J.; Amine, A.; Ozsoz, M. A novel method for sensitive microRNA detection: Electropolymerization based doping. Biosens. Bioelectron. 2017, 92, 770–778. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, Z.; Chang, Y.; Qing, M.; Yuan, R.; Chai, Y. A novel 2D DNA nanoprobe mediated enzyme-free target recycling amplification for ultrasensitive electrochemical detection of microRNA. Anal. Chem. 2018, 90, 9538–9544. [Google Scholar] [CrossRef]

| Materials/Reporters | Linear Range | Detection Limit | Ref. |
|---|---|---|---|
| AuNPs/dye | 5–200 pM | 5 pM | [41] |
| AuNPs/dye | 0.1–1000 pM | 33.4 fM | [43] |
| AuNPs/dye | 0.1–10 pM | 45 fM | [44] |
| PS@Au/dye | 1–1000 pM | 50 fM | [45] |
| MWCNT-AuNCs/dye | 0.1–1000 pM | 33.4 fM | [46] |
| GO/dye | 0.5–1000 pM | 160 fM | [47] |
| GO/dye | 5–40 nM | 132 pM | [48] |
| MoS2/dye | 1–10,000 pM | 10 fM | [49] |
| MoS2/dye | 0.5–50 nM | 426 pM | [50] |
| CCP/dye | 10–200 pM | 4.6 pM | [51] |
| PDA-PEI/dye | 0.8–50 nM | 0.52 nM | [52] |
| MNPs@PDA/dye | 5–5000 pM | 0.42 pM | [53] |
| MB/QDs | 0.5–10 pM | 0.5 pM | [54] |
| Cu3(PO4)2 NFs/dye | 1–100,000 pM | 0.23 pM | [55] |
| MB-SA/dye | 0.01–10 pM | 0.01 pM | this work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, G.; La, M.; Wang, J.; Liu, J.; Han, Y.; Liu, L. Magnetically Assisted Immobilization-Free Detection of microRNAs Based on the Signal Amplification of Duplex-Specific Nuclease. Biosensors 2023, 13, 699. https://doi.org/10.3390/bios13070699
Liu G, La M, Wang J, Liu J, Han Y, Liu L. Magnetically Assisted Immobilization-Free Detection of microRNAs Based on the Signal Amplification of Duplex-Specific Nuclease. Biosensors. 2023; 13(7):699. https://doi.org/10.3390/bios13070699
Chicago/Turabian StyleLiu, Gang, Ming La, Jiwei Wang, Jiawen Liu, Yongjun Han, and Lin Liu. 2023. "Magnetically Assisted Immobilization-Free Detection of microRNAs Based on the Signal Amplification of Duplex-Specific Nuclease" Biosensors 13, no. 7: 699. https://doi.org/10.3390/bios13070699
APA StyleLiu, G., La, M., Wang, J., Liu, J., Han, Y., & Liu, L. (2023). Magnetically Assisted Immobilization-Free Detection of microRNAs Based on the Signal Amplification of Duplex-Specific Nuclease. Biosensors, 13(7), 699. https://doi.org/10.3390/bios13070699






