Electrochemical Detection of Different Foodborne Bacteria for Point-of-Care Applications
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Culture Preparation
2.2. Apparatus and Instrumentation
2.3. Aptamer Probe Preparations
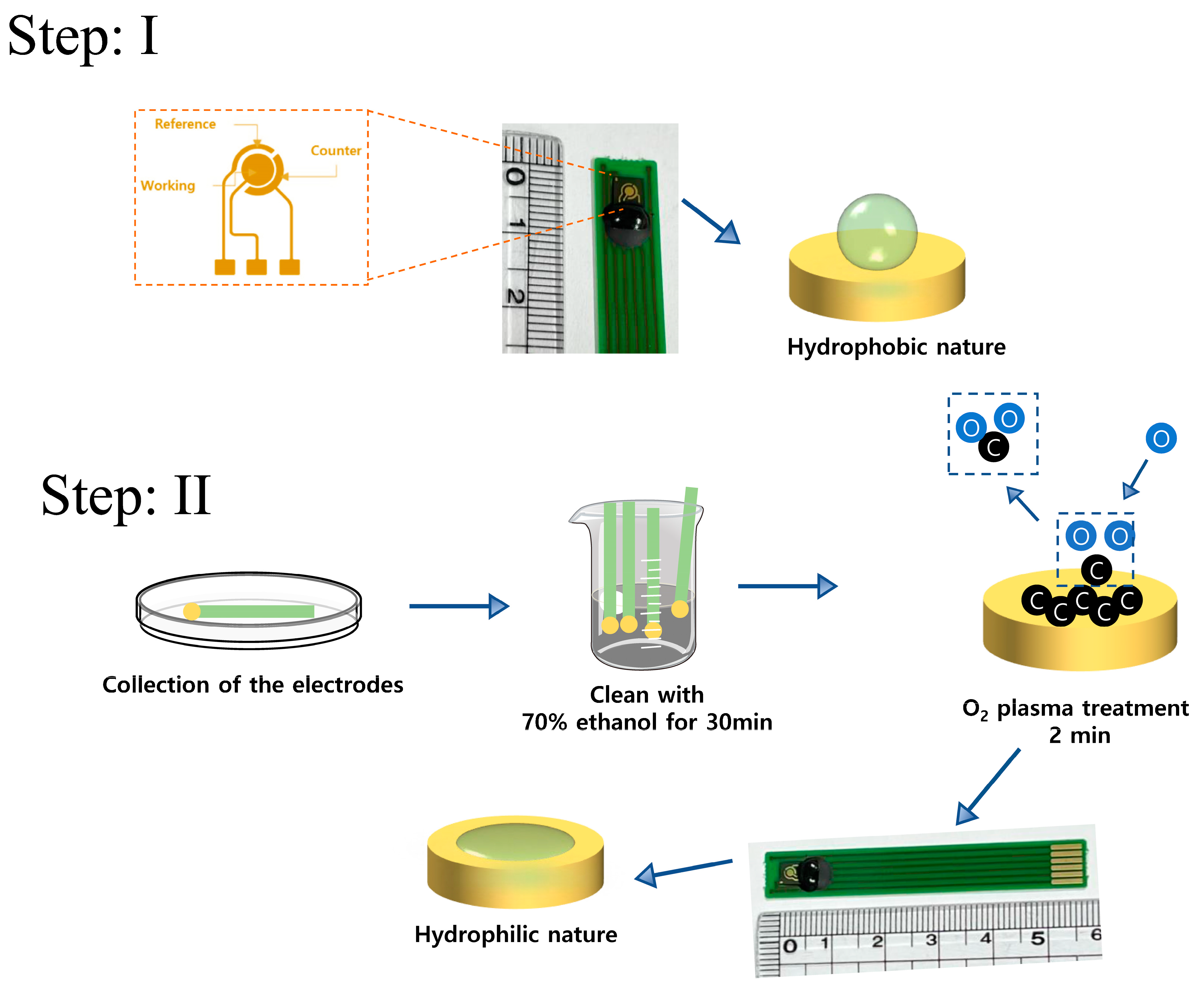
2.4. Gold (Au)-Disk Electrode for Aptamer Immobilization and Bacteria Detection
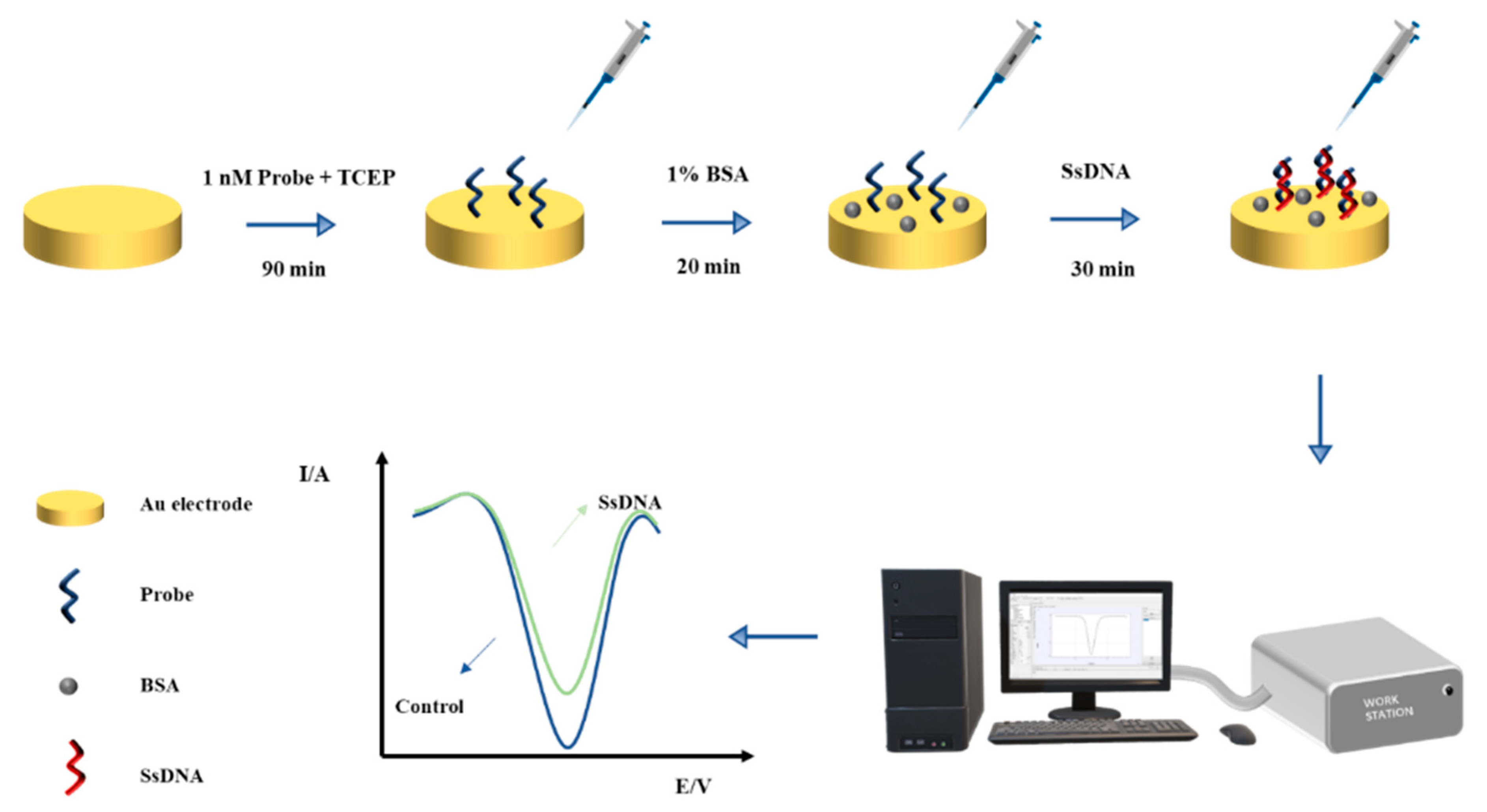
2.5. Construction of Electrochemical Biosensor for Different Types of Bacteria Detection
2.6. Optimization of Conditions for Sensitive Electrochemical Sensor
2.7. Sensitivity and Selectivity Detection
3. Results and Discussion
3.1. Electrochemical Biosensing Assay Principle
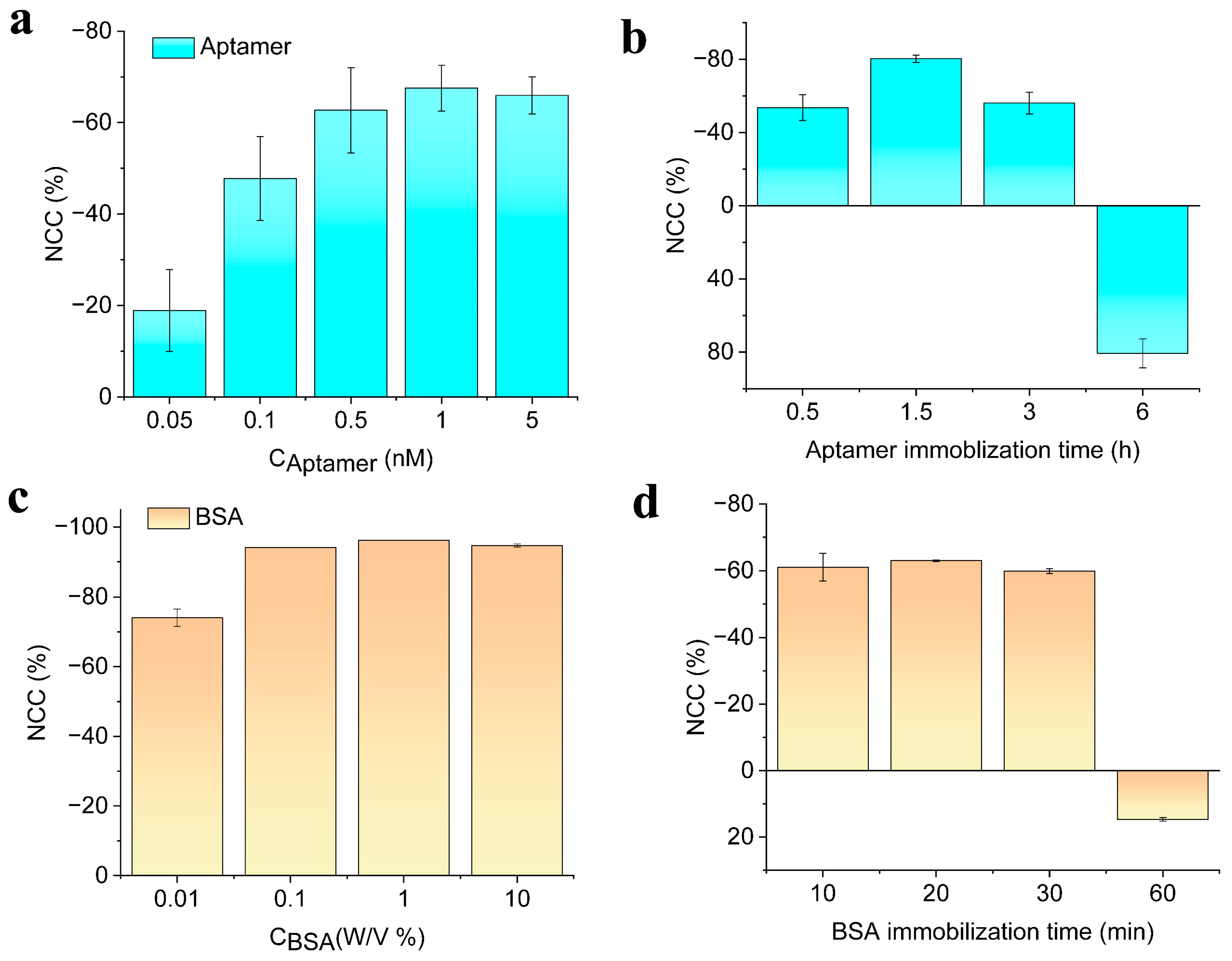
3.2. Optimization of Different Experimental Conditions
Effect of Temperature, Heating Time, and Sonication Time on DNA Hybridization Detection
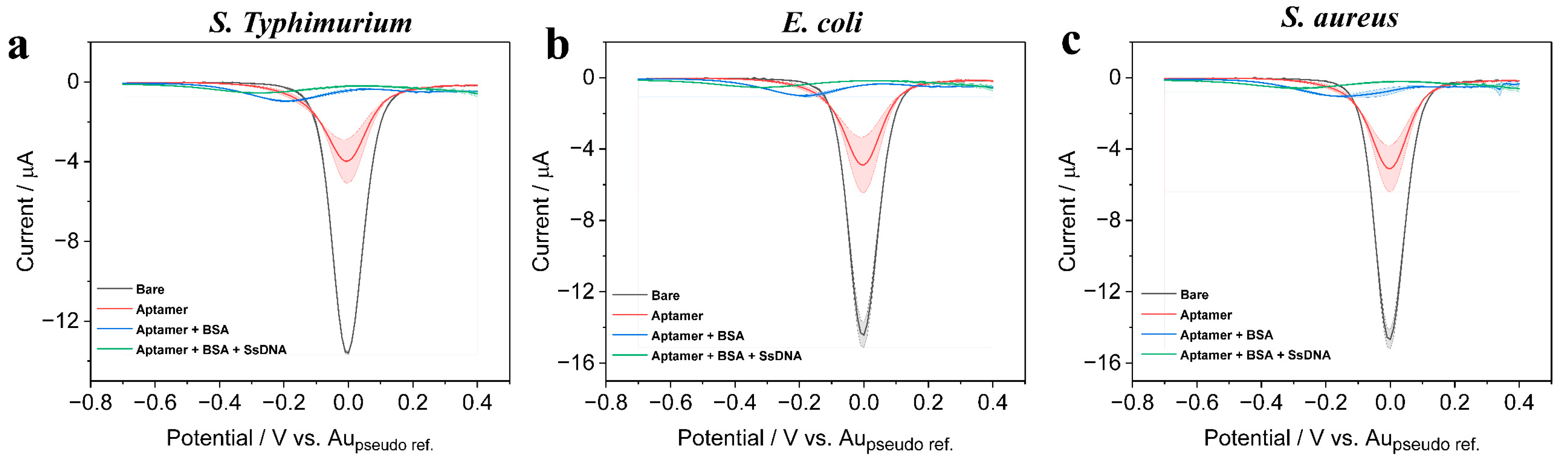
3.3. Characterizing the Biosensing Surface for Bacterial Pathogen Detection
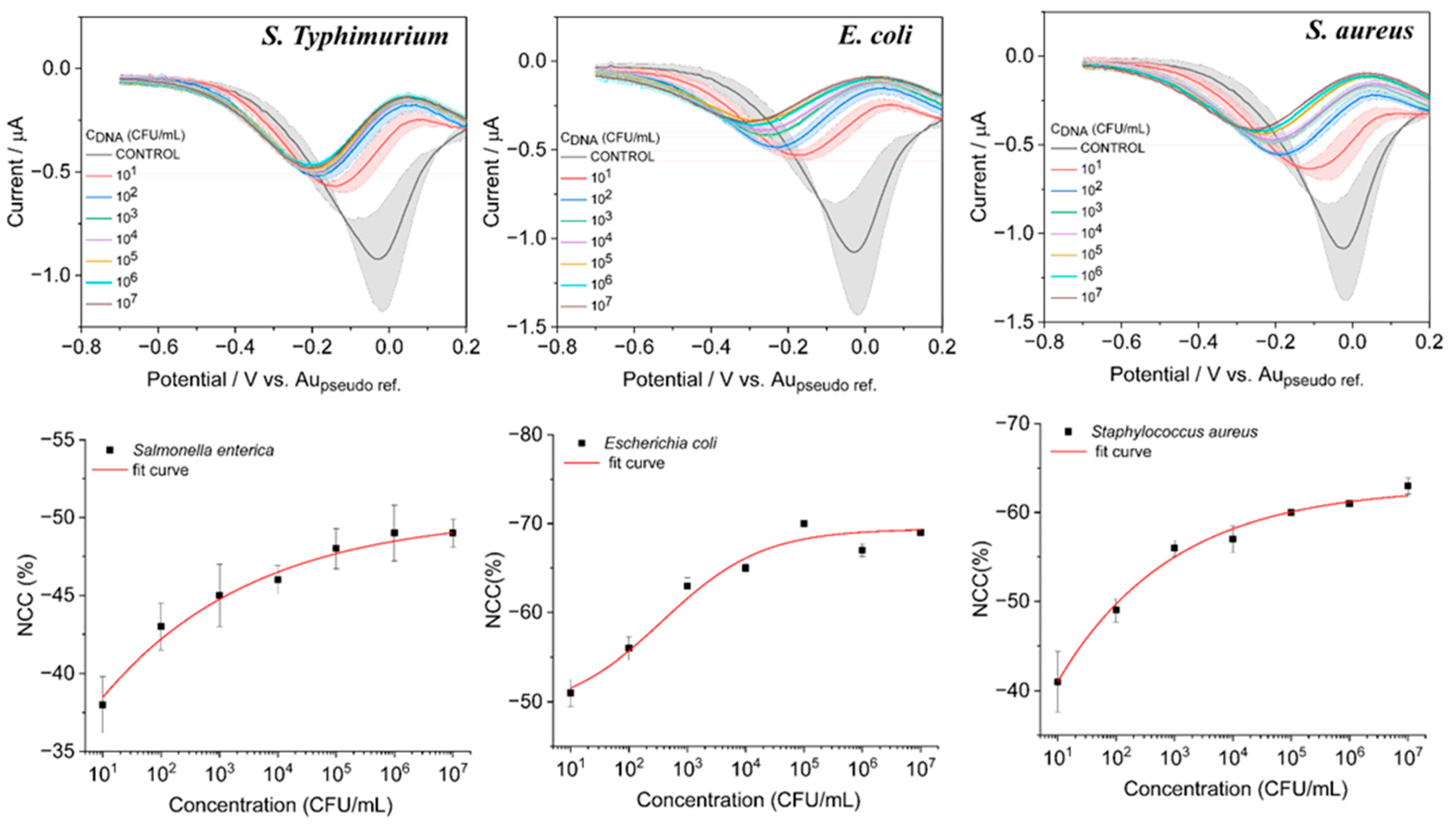
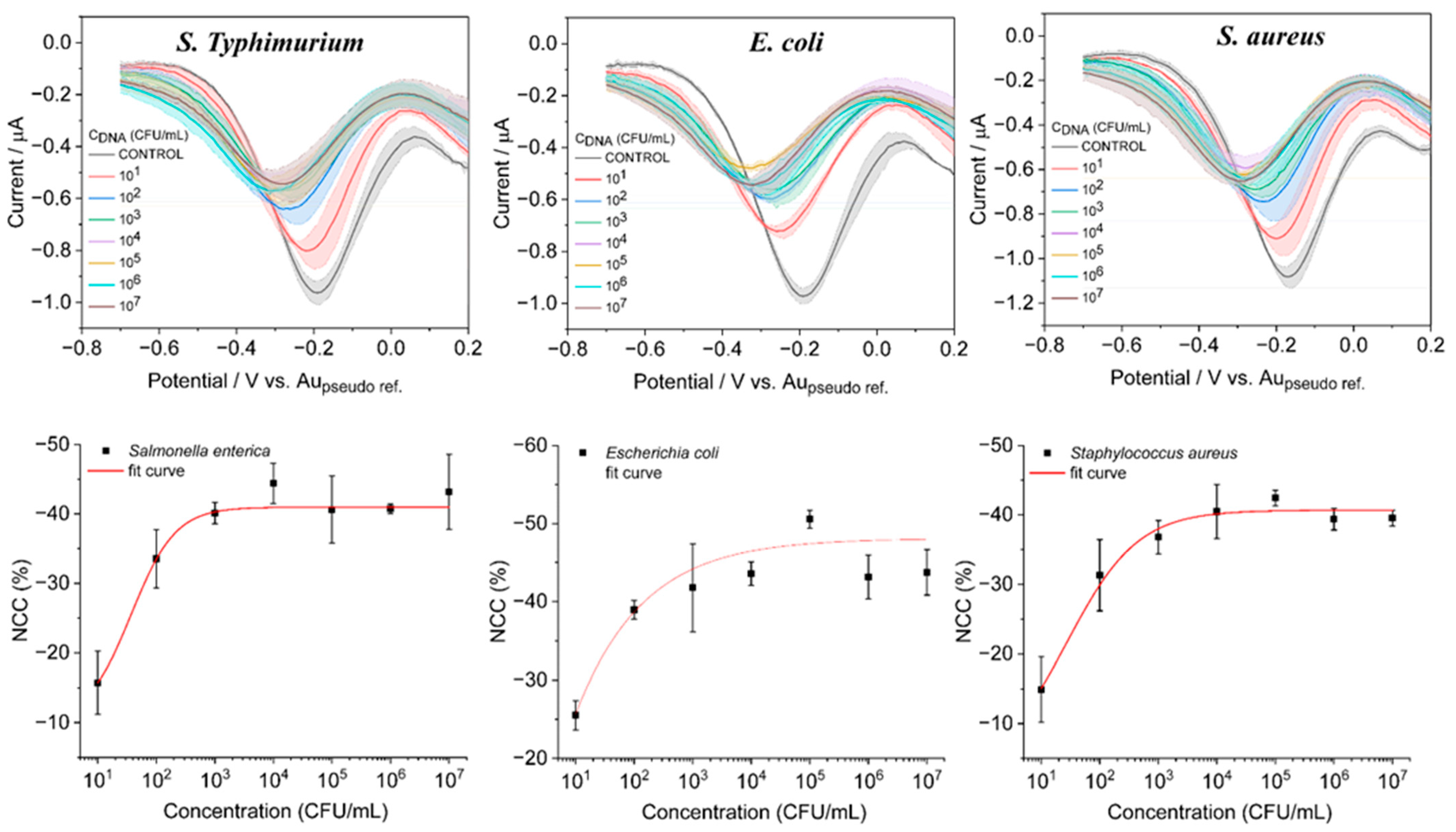
3.4. Analytical Performance of the Sensor for Quantification of Bacterial Types with DPV
3.5. Selectivity, Stability, and Reproducibility of the Assay for Practical Applications
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, J.Y.; Zhao, N.J.; Duan, J.B.; Meng, D.S.; Fang, L.; Yang, R.F.; Xiao, X.; Yin, G.F.; Ma, M.J.; Liu, J.G.; et al. Rapid Quantitative Detection of Bacterial in Water Based on Multi-Wavelength Scattering Spectra. Guang Pu Xue Yu Guang Pu Fen Xi 2017, 37, 333–337. [Google Scholar] [PubMed]
- Ikuta, K.S.; Swetschinski, L.R.; Aguilar, G.R.; Sharara, F.; Mestrovic, T.; Gray, A.P.; Weaver, N.D.; E Wool, E.; Han, C.; Hayoon, A.G.; et al. Global mortality associated with 33 bacterial pathogens in 2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2022, 400, 2221–2248. [Google Scholar] [CrossRef] [PubMed]
- McGowan, J.E.; Carlet, J. Antimicrobial resistance: A worldwide problem for health care institutions. Am. J. Infect. Control. 1998, 26, 541–543. [Google Scholar] [CrossRef] [PubMed]
- Rolain, J.M.; Mallet, M.N.; Fournier, P.E.; Raoult, D. Real-time PCR for universal antibiotic susceptibility testing. J. Antimicrob. Chemother. 2004, 54, 538–541. [Google Scholar] [CrossRef] [PubMed]
- Welker, M.; Van Belkum, A. One System for All: Is Mass Spectrometry a Future Alternative for Conventional Antibiotic Susceptibility Testing? Front. Microbiol. 2019, 10, 2711. [Google Scholar] [CrossRef]
- Dally, S.; Lemuth, K.; Kaase, M.; Rupp, S.; Knabbe, C.; Weile, J. DNA Microarray for Genotyping Antibiotic Resistance Determinants in Acinetobacter baumannii Clinical Isolates. Antimicrob. Agents Chemother. 2013, 57, 4761–4768. [Google Scholar] [CrossRef]
- Zhang, K.; Qin, S.; Wu, S.; Liang, Y.; Li, J. Microfluidic systems for rapid antibiotic susceptibility tests (ASTs) at the single-cell level. Chem. Sci. 2020, 11, 6352–6361. [Google Scholar] [CrossRef]
- Spencer, D.C.; Paton, T.F.; Mulroney, K.T.; Inglis, T.J.J.; Sutton, J.M.; Morgan, H. A fast impedance-based antimicrobial susceptibility test. Nat. Commun. 2020, 11, 5328. [Google Scholar] [CrossRef]
- Davenport, M.; Mach, K.E.; Shortliffe, L.M.D.; Banaei, N.; Wang, T.H.; Liao, J.C. New and developing diagnostic technologies for urinary tract infections. Nat. Rev. Urol. 2017, 14, 296–310. [Google Scholar] [CrossRef]
- Tastanova, A.; Stoffel, C.I.; Dzung, A.; Cheng, P.F.; Bellini, E.; Johansen, P.; Duda, A.; Nobbe, S.; Lienhard, R.; Bosshard, P.P.; et al. A Comparative Study of Real-Time RT-PCR-Based SARS-CoV-2 Detection Methods and Its Application to Human-Derived and Surface Swabbed Material. J. Mol. Diagn. 2021, 23, 796–804. [Google Scholar] [CrossRef]
- Cerda-Kipper, A.S.; Montiel, B.E.; Hosseini, S. Immunoassay|Radioimmunoassays and Enzyme-Linked Immunosorbent Assay. In Encyclopedia of Analytical Science, 3rd ed.; Worsfold, P., Poole, C., Townshend, A., Miró, M., Eds.; Academic Press: Oxford, UK, 2019; pp. 55–75. [Google Scholar]
- Zhao, Y.T.; Song, X. An Electrochemical-Based Point-of-Care Testing Methodology for Uric Acid Measurement. J. Anal. Methods Chem. 2022, 2022, 8555842. [Google Scholar] [CrossRef] [PubMed]
- Biswas, G.C.; Choudhury, S.; Rabbani, M.M.; Das, J. A Review on Potential Electrochemical Point-of-Care Tests Targeting Pandemic Infectious Disease Detection: COVID-19 as a Reference. Chemosensors 2022, 10, 269. [Google Scholar] [CrossRef]
- da Silva, E.T.S.G.; Souto, D.E.P.; Barragan, J.T.C.; Giarola, J.D.; de Moraes, A.C.M.; Kubota, L.T. Electrochemical Biosensors in Point-of-Care Devices: Recent Advances and Future Trends. ChemElectroChem 2017, 4, 778–794. [Google Scholar] [CrossRef]
- Madhurantakam, S.; Muthukumar, S.; Prasad, S. Emerging Electrochemical Biosensing Trends for Rapid Diagnosis of COVID-19 Biomarkers as Point-of-Care Platforms: A Critical Review. ACS Omega 2022, 7, 12467–12473. [Google Scholar] [CrossRef] [PubMed]
- Sinibaldi, A.; Sampaoli, C.; Danz, N.; Munzert, P.; Sonntag, F.; Centola, F.; Occhicone, A.; Tremante, E.; Giacomini, P.; Michelotti, F. Bloch Surface Waves Biosensors for High Sensitivity Detection of Soluble ERBB2 in a Complex Biological Environment. Biosensors 2017, 7, 33. [Google Scholar] [CrossRef]
- Zamora-Olivares, D.; Pridgen, J.R.; Zeng, L.Y.; Kaoud, T.S.; Anslyn, E.V.; Dalby, K.N. Use of differential sensing-based biosensors to quantify ERK kinase activity in complex biological samples. Cancer Res. 2020, 80, 6312. [Google Scholar] [CrossRef]
- Song, K.M.; Lee, S.; Ban, C. Aptamers and Their Biological Applications. Sensors 2012, 12, 612–631. [Google Scholar] [CrossRef]
- Tabatabaei, M.S.; Ahmed, M. Enzyme-Linked Immunosorbent Assay (ELISA). In Cancer Cell Biology: Methods and Protocols; Christian, S.L., Ed.; Springer US: New York, NY, USA, 2022; pp. 115–134. [Google Scholar]
- Kamekawa, N.; Shimomura, Y.; Nakamura, M.; Yamana, K. Pyrene-modified DNA aptamer as a fluorescent biosensor with high affinity and specificity for ATP sensing. Chem. Lett. 2006, 35, 660–661. [Google Scholar] [CrossRef]
- Amirani, M.C.; Tang, T. Electrostatics of DNA nucleotide-carbon nanotube hybrids evaluated from QM:MM simulations. Nanoscale 2015, 7, 19586–19595. [Google Scholar] [CrossRef]
- Xiao, F.; Chen, Z.; Wei, Z.X.; Tian, L.L. Hydrophobic Interaction: A Promising Driving Force for the Biomedical Applications of Nucleic Acids. Adv. Sci. 2020, 7, 2001048. [Google Scholar] [CrossRef]
- Park, J.Y.; Lee, T.S.; Song, I.H.; Cho, Y.L.; Chae, J.R.; Yun, M.; Kang, H.; Lee, J.H.; Lim, J.H.; Cho, W.G.; et al. Hybridization-based aptamer labeling using complementary oligonucleotide platform for PET and optical imaging. Biomaterials 2016, 100, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Mei, H.C.; Bing, T.; Yang, X.J.; Qi, C.; Chang, T.J.; Liu, X.J.; Cao, Z.H.; Shangguan, D.H. Functional-Group Specific Aptamers Indirectly Recognizing Compounds with Alkyl Amino Group. Anal. Chem. 2012, 84, 7323–7329. [Google Scholar] [CrossRef] [PubMed]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Al Bawab, A.; Ismail, S.I. Aptamers Chemistry: Chemical Modifications and Conjugation Strategies. Molecules 2020, 25, 3. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.X.; Zhao, S.Q.; Li, D.S.; Yang, J.L.; Yang, L. Portable chemiluminescence optical fiber aptamer-based biosensors for analysis of multiple mycotoxins. Food Control 2023, 144, 109361. [Google Scholar] [CrossRef]
- Urmann, K.; Walter, J.G.; Scheper, T.; Segal, E. Label-Free Optical Biosensors Based on Aptamer-Functionalized Porous Silicon Scaffolds. Anal. Chem. 2015, 87, 1999–2006. [Google Scholar] [CrossRef]
- Preuss, J.A.; Reich, P.; Bahner, N.; Bahnemann, J. Impedimetric Aptamer-Based Biosensors: Applications. Adv. Biochem. Eng. Biot. 2020, 174, 43–91. [Google Scholar] [CrossRef]
- Grabowska, I.; Sharma, N.; Vasilescu, A.; Iancu, M.; Badea, G.; Boukherroub, R.; Ogale, S.; Szunerits, S. Electrochemical Aptamer-Based Biosensors for the Detection of Cardiac Biomarkers. ACS Omega 2018, 3, 12010–12018. [Google Scholar] [CrossRef]
- Jayanthi, V.S.P.K.S.A.; Das, A.B.; Saxena, U. Recent advances in biosensor development for the detection of cancer biomarkers. Biosens. Bioelectron. 2017, 91, 15–23. [Google Scholar] [CrossRef]
- Goutelle, S.; Maurin, M.; Rougier, F.; Barbaut, X.; Bourguignon, L.; Ducher, M.; Maire, P. The Hill equation: A review of its capabilities in pharmacological modelling. Fund. Clin. Pharmacol. 2008, 22, 633–648. [Google Scholar] [CrossRef]
- Rink, S.; Kaiser, B.; Steiner, M.S.; Duerkop, A.; Baeumner, A.J. Highly sensitive interleukin 6 detection by employing commercially ready liposomes in an LFA format. Anal. Bioanal. Chem. 2022, 414, 3231–3241. [Google Scholar] [CrossRef]
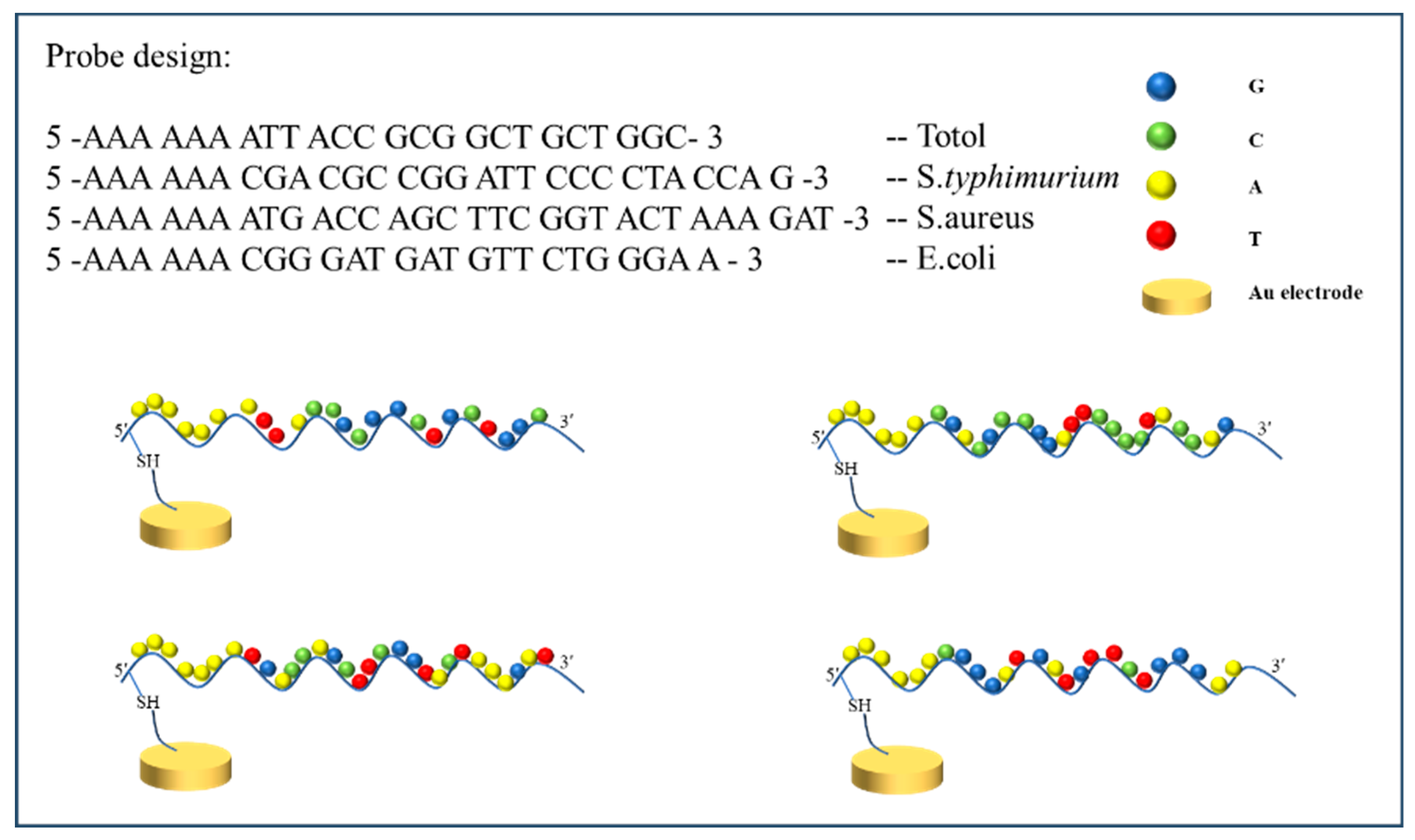


| Aptamer | Thiol-Modified Sequences from 5′ to 3′ |
|---|---|
| Escherichia coli | SH- AAA AAA CGG GAT GAT GTT CTG GGA A |
| Salmonella typhimurium | SH- AAA AAA CGA CGC CGG ATT CCC CTA CCA G |
| Staphylococcus aureus | SH- AAA AAA ATG ACC AGC TTC GGT ACT AAA GAT |
| Total bacteria | SH- AAA AAA ATT ACC GCG GCT GCT GGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, T.; Yagati, A.K.; Min, J. Electrochemical Detection of Different Foodborne Bacteria for Point-of-Care Applications. Biosensors 2023, 13, 641. https://doi.org/10.3390/bios13060641
Wu T, Yagati AK, Min J. Electrochemical Detection of Different Foodborne Bacteria for Point-of-Care Applications. Biosensors. 2023; 13(6):641. https://doi.org/10.3390/bios13060641
Chicago/Turabian StyleWu, Tailin, Ajay Kumar Yagati, and Junhong Min. 2023. "Electrochemical Detection of Different Foodborne Bacteria for Point-of-Care Applications" Biosensors 13, no. 6: 641. https://doi.org/10.3390/bios13060641
APA StyleWu, T., Yagati, A. K., & Min, J. (2023). Electrochemical Detection of Different Foodborne Bacteria for Point-of-Care Applications. Biosensors, 13(6), 641. https://doi.org/10.3390/bios13060641






