A Short Review on Miniaturized Biosensors for the Detection of Nucleic Acid Biomarkers
Abstract
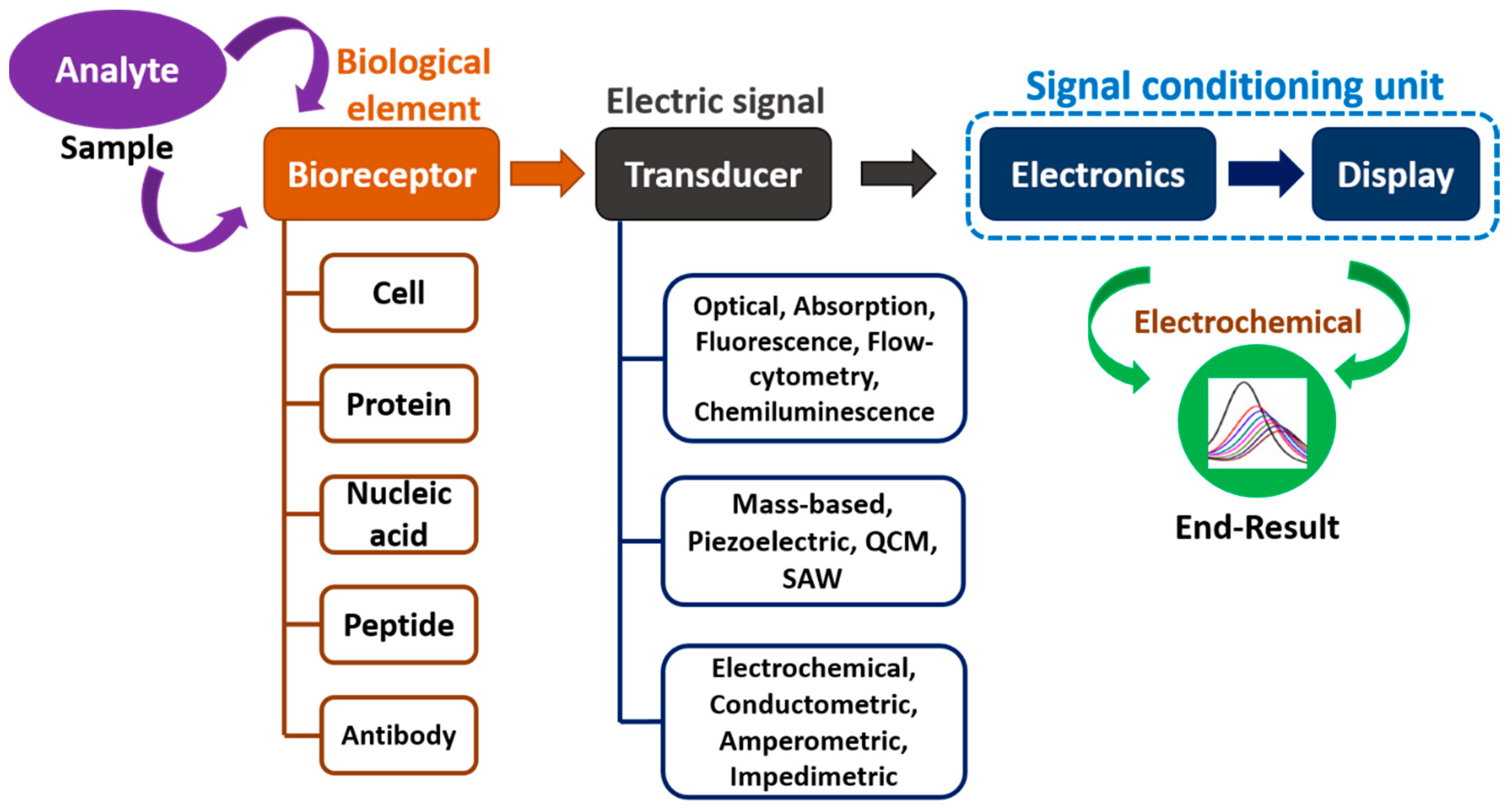
1. Introduction
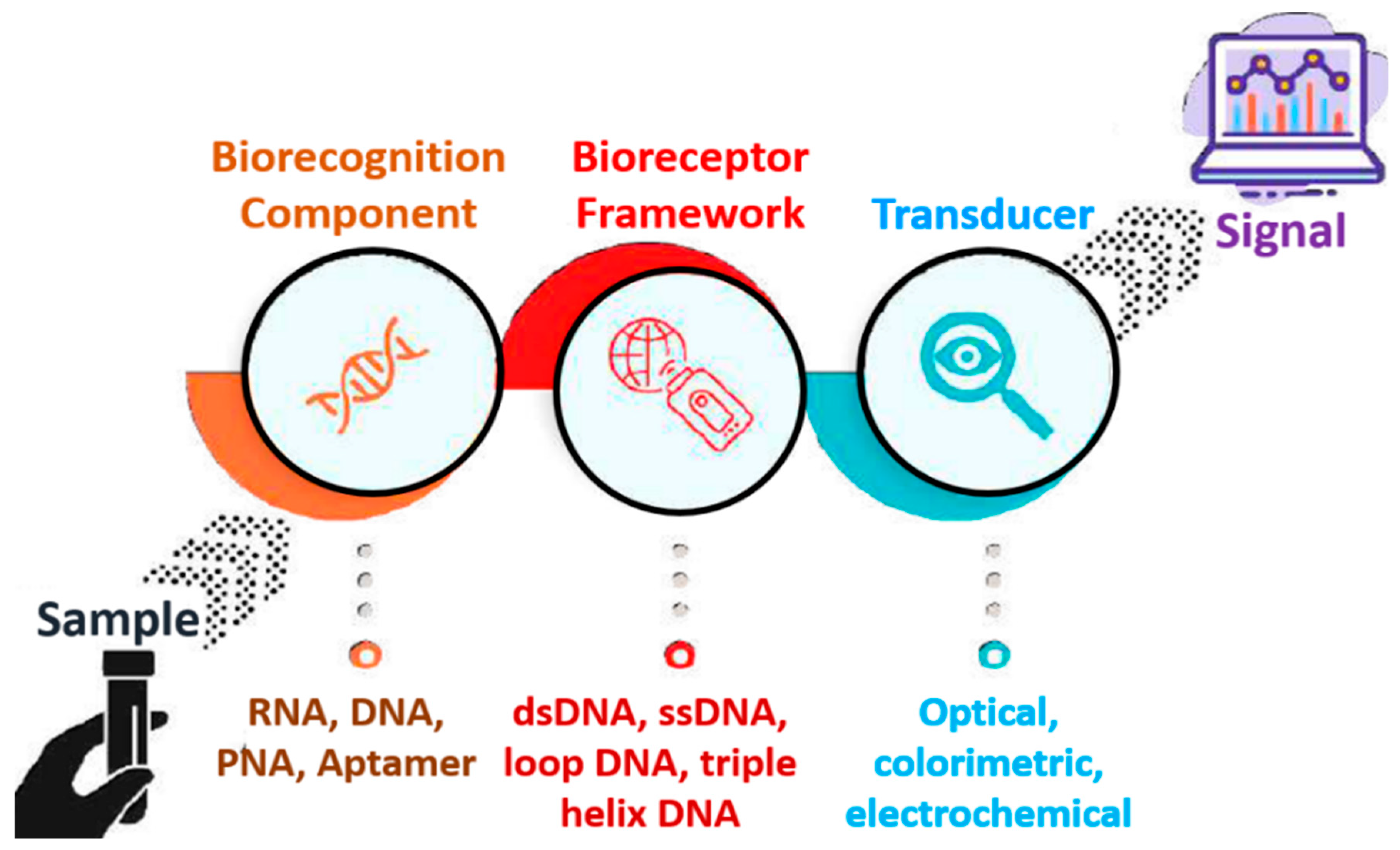
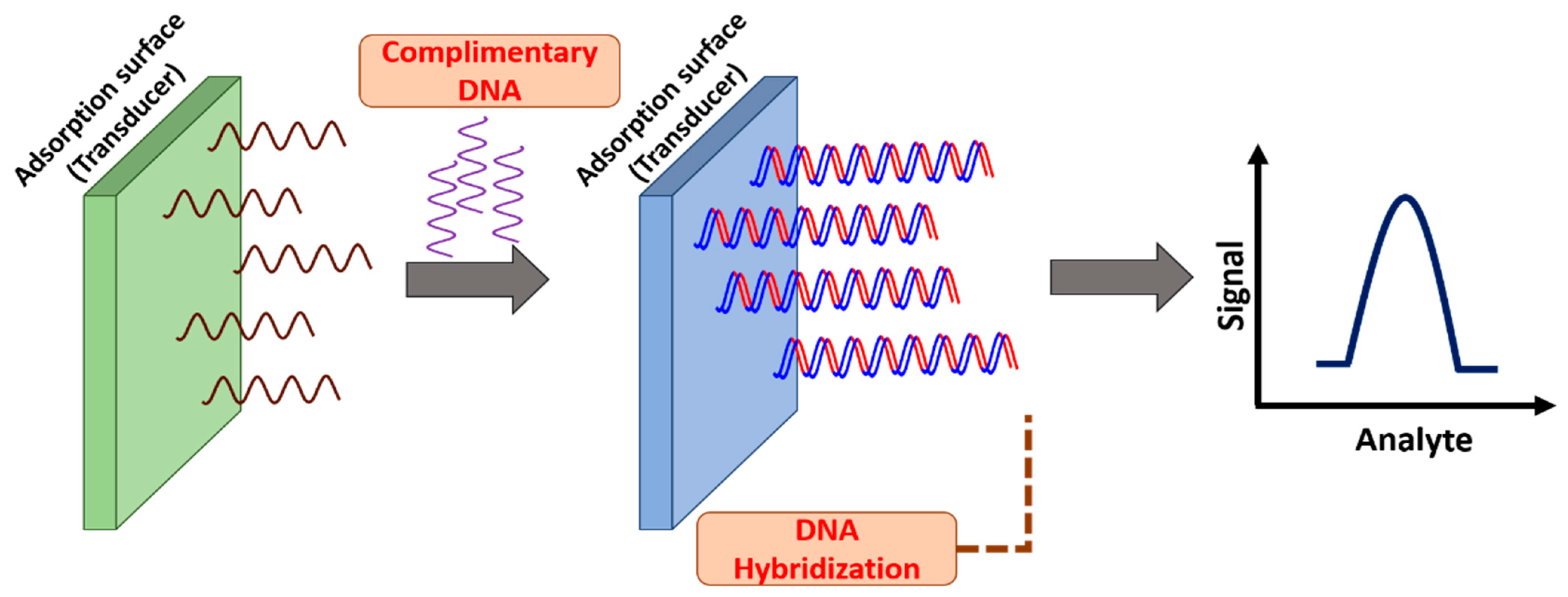
2. Nucleic Acid-Based Biosensors
2.1. Riboswitches
2.2. Aptamers
2.3. Peptide Nucleic Acids (PNA)
3. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dutta, G.; Rainbow, J.; Zupancic, U.; Papamatthaiou, S.; Estrela, P. Microfluidic Devices for Label-Free DNA Detection. Chemosensors 2018, 6, 43. [Google Scholar] [CrossRef]
- Holzinger, M.; LeGoff, A.; Cosnier, S. Nanomaterials for biosensing applications: A review. Front. Chem. 2014, 2, 63. [Google Scholar] [CrossRef]
- Röhlen, D.L.; Pilas, J.; Dahmen, M.; Keusgen, M.; Selmer, T.; Schöning, M.J. Toward a hybrid biosensor system for analysis of organic and volatile fatty acids in fermentation processes. Front. Chem. 2018, 6, 284. [Google Scholar] [CrossRef]
- Kulkarni, M.B.; Ayachit, N.H.; Aminabhavi, T.M. Biosensors and Microfluidic Biosensors: From Fabrication to Application. Biosensors 2022, 12, 543. [Google Scholar] [CrossRef]
- Tricoli, A.; Neri, G. Miniaturized bio-and chemical-sensors for point-of-care monitoring of chronic kidney diseases. Sensors 2018, 18, 942. [Google Scholar] [CrossRef]
- Kulkarni, M.B.; Ayachit, N.H.; Aminabhavi, T.M. Recent Advancements in Nanobiosensors: Current Trends, Challenges, Applications, and Future Scope. Biosensors 2022, 12, 892. [Google Scholar] [CrossRef]
- Kulkarni, M.B.; Upadhyaya, K.; Ayachit, N.H.; Iyer, N. Quantum Dot--Polymer Composites in Light-Emitting Diode Applications. In Quantum Dots and Polymer Nanocomposites; CRC Press: Boca Raton, FL, USA, 2022; pp. 259–279. [Google Scholar]
- Umapathi, R.; Rani, G.M.; Kim, E.; Park, S.-Y.; Cho, Y.; Huh, Y.S. Sowing kernels for food safety: Importance of rapid on-site detction of pesticide residues in agricultural foods. Food Front. 2022, 3, 666–676. [Google Scholar] [CrossRef]
- Zhao, X.; Li, M.; Liu, Y. Microfluidic-Based Approaches for Foodborne Pathogen Detection. Microorganisms 2019, 7, 381. [Google Scholar] [CrossRef]
- Kulkarni, M.B.; Goel, S. Microfluidic devices for synthesizing nanomaterials—A review. Nano Express 2020, 1, 032004. [Google Scholar] [CrossRef]
- Yang, Q.; Li, N.; Li, Q.; Chen, S.; Wang, H.L.; Yang, H. Amperometric sarcosine biosensor based on hollow magnetic Pt–Fe3O4@C nanospheres. Anal. Chim. Acta 2019, 1078, 161–167. [Google Scholar] [CrossRef]
- Fazli, G.; Bahabadi, S.E.; Adlnasab, L.; Ahmar, H. A glassy carbon electrode modified with a nanocomposite prepared from Pd/Al layered double hydroxide and carboxymethyl cellulose for voltammetric sensing of hydrogen peroxide. Microchim. Acta 2019, 186, 821. [Google Scholar] [CrossRef] [PubMed]
- Chin Chwan Chuong, J.J.; Rahman, M.; Ibrahim, N.; Heng, L.Y.; Tan, L.L.; Ahmad, A. Harmful Microalgae Detection: Biosensors versus Some Conventional Methods. Sensors 2022, 22, 3144. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.-J.; Hsu, Y.-C.; Gu, B.-C.; Wu, C.-C. Voltammetric measurement of Escherichia coli concentration through p-APG hydrolysis by endogenous β-galactosidase. Microchem. J. 2020, 154, 104641. [Google Scholar] [CrossRef]
- Kulkarni, M.B.; Salve, M.; Goel, S. Miniaturized Thermal Monitoring Module with CO Laser Ablated Microfluidic Device for Electrochemically Validated DNA Amplification. IEEE Trans. Instrum. Meas. 2021, 70, 1–8. [Google Scholar] [CrossRef]
- Bhaiyya, M.; Kulkarni, M.B.; Pattnaik, P.K.; Goel, S. Internet of things-enabled photomultiplier tube- and smartphone-based electrochemiluminescence platform to detect choline and dopamine using 3D-printed closed bipolar electrodes. Luminescence 2022, 37, 357–365. [Google Scholar] [CrossRef]
- Tamayo, J.; Kosaka, P.M.; Ruz, J.J.; Paulo, Á.S.; Calleja, M. Biosensors based on nanomechanical systems. Chem. Soc. Rev. 2013, 42, 1287–1311. [Google Scholar] [CrossRef]
- Sung, W.J.; Bae, Y.H. Glucose oxidase, lactate oxidase, and galactose oxidase enzyme electrode based on polypyrrole with polyanion/PEG/enzyme conjugate dopant. Sens. Actuators B Chem. 2006, 114, 164–169. [Google Scholar] [CrossRef]
- Kampeera, J.; Pasakon, P.; Karuwan, C.; Arunrut, N.; Sappat, A.; Sirithammajak, S.; Dechokiattawan, N.; Sumranwanich, T.; Chaivisuthangkura, P.; Ounjai, P.; et al. Point-of-care rapid detection of Vibrio parahaemolyticus in seafood using loop-mediated isothermal amplification and graphene-based screen-printed electrochemical sensor. Biosens. Bioelectron. 2019, 132, 271–278. [Google Scholar] [CrossRef]
- Kulkarni, M.B.; Goel, S. A Review on Recent Advancements in Chamber-Based Microfluidic PCR Devices. In Microelectronics and Signal Processing; CRC Press: Boca Raton, FL, USA, 2021; pp. 49–70. [Google Scholar]
- Velmurugan, K.; Kulkarni, M.B.; Gupta, I.; Das, R.; Goel, S.; Nirmal, J. Role of Microfluidics in Drug Delivery. In Microfluidics and Multi Organs on Chip; Mohanan, P.V., Ed.; Springer Nature Singapore: Singapore, 2022; pp. 107–133. ISBN 978-981-19-1379-2. [Google Scholar]
- Rani, G.M.; Wu, C.-M.; Motora, K.G.; Umapathi, R.; Jose, C.R.M. Acoustic-electric conversion and triboelectric properties of nature-driven CF-CNT based triboelectric nanogenerator for mechanical and sound energy harvesting. Nano Energy 2023, 108, 108211. [Google Scholar] [CrossRef]
- Umapathi, R.; Ghoreishian, S.M. Review—Emerging Trends in the Development of Electrochemical Devices for the On-Site Detection of Food Contaminants. ECS Sens. Plus 2022, 1, 044601. [Google Scholar] [CrossRef]
- Davidson, E.M.; Croal, B.L. Introduction of an Albumin-to-Creatinine Ratio Point-of-Care Device: Analytic, Clinical, and Cost-effectiveness Aspects. Point Care 2003, 2, 89–95. [Google Scholar] [CrossRef]
- Kaprou, G.D.; Papadakis, G.; Papageorgiou, D.P.; Kokkoris, G.; Papadopoulos, V.; Kefala, I.; Gizeli, E.; Tserepi, A. Miniaturized devices for isothermal DNA amplification addressing DNA diagnostics. Microsyst. Technol. 2016, 22, 1529–1534. [Google Scholar] [CrossRef]
- Nagai, H.; Fuchiwaki, Y. Portable microfluidic system for rapid genetic testing. Electron. Commun. Jpn. 2015, 98, 1–6. [Google Scholar] [CrossRef]
- Kumar, K.; Kim, E.; Alhammadi, M.; Reddicherla, U.; Aliya, S.; Tiwari, J.N.; Park, H.S.; Choi, J.H.; Son, C.Y.; Vilian, A.T.E.; et al. Recent advances in microfluidic approaches for the isolation and detection of exosomes. TrAC Trends Anal. Chem. 2023, 159, 116912. [Google Scholar] [CrossRef]
- Kulkarni, M.B.; Goel, S. Recent advancements in integrated microthermofluidic systems for biochemical and biomedical applications—A review. Sens. Actuators A Phys. 2022, 341, 113590. [Google Scholar] [CrossRef]
- Kulkarni, M.B.; Goel, S. Advances in continuous-flow based microfluidic PCR devices—A review. Eng. Res. Express 2020, 2, 042001. [Google Scholar] [CrossRef]
- Puneeth, S.B.; Kulkarni, M.B.; Goel, S. Microfluidic viscometers for biochemical and biomedical applications: A review. Eng. Res. Express 2021, 3, 022003. [Google Scholar] [CrossRef]
- Li, X.; Wu, W.; Manz, A. Thermal gradient for fluorometric optimization of droplet PCR in virtual reaction chambers. Microchim. Acta 2017, 184, 3433–3439. [Google Scholar] [CrossRef]
- Aliya, S.; Lee, H.; Alhammadi, M.; Umapathi, R.; Huh, Y.S. An Overview on Single-Cell Technology for Hepatocellular Carcinoma Diagnosis. Int. J. Mol. Sci. 2022, 23, 1402. [Google Scholar] [CrossRef] [PubMed]
- Naji, O.P.; Bessoth, F.G.; Manz, A. Novel Injection Methods for Miniaturised Gas Chromatography. In Micro Total Analysis Systems 2001, Proceedings of the µTAS 2001 Symposium, Monterey, CA, USA, 21–25 October 2001; Springer Netherlands: Dordrecht, The Netherlands, 2001; pp. 655–657. [Google Scholar] [CrossRef]
- Kulkarni, M.B.; Goyal, S.; Dhar, A.; Sriram, D.; Goel, S. Miniaturized and IoT Enabled Continuous-Flow-Based Microfluidic PCR Device for DNA Amplification. IEEE Trans. Nanobiosci. 2022, 21, 97–104. [Google Scholar] [CrossRef]
- Jiang, Y.; Manz, A.; Wu, W. Fully automatic integrated continuous-flow digital PCR device for absolute DNA quantification. Anal. Chim. Acta 2020, 1125, 50–56. [Google Scholar] [CrossRef]
- Pathak, A.K.; Rahman, B.M.A.; Singh, V.K.; Kumari, S. Sensitivity enhancement of a concave shaped optical fiber refractive index sensor covered with multiple au nanowires. Sensors 2019, 19, 4210. [Google Scholar] [CrossRef]
- Haeberle, S.; Zengerle, R. Microfluidic platforms for lab-on-a-chip applications. Lab Chip 2007, 7, 1094–1110. [Google Scholar] [CrossRef]
- Kukkar, D.; Zhang, D.; Jeon, B.H.; Kim, K.-H. Recent advances in wearable biosensors for non-invasive monitoring of specific metabolites and electrolytes associated with chronic kidney disease: Performance evaluation and future challenges. TrAC Trends Anal. Chem. 2022, 150, 116570. [Google Scholar] [CrossRef]
- Mohan, J.M.; Amreen, K.; Kulkarni, M.B.; Javed, A.; Dubey, S.K.; Goel, S. Optimized Ink Jetted Paper Device for Electroanalytical Detection of Picric Acid. Colloids Surfaces B Biointerfaces 2021, 208, 112056. [Google Scholar] [CrossRef]
- Yao, L.; Wang, L.; Huang, F.; Cai, G.; Xi, X.; Lin, J. A microfluidic impedance biosensor based on immunomagnetic separation and urease catalysis for continuous-flow detection of E. coli O157:H7. Sens. Actuators B Chem. 2018, 259, 1013–1021. [Google Scholar] [CrossRef]
- Xu, M.; Wang, R.; Li, Y. An electrochemical biosensor for rapid detection of: E. coli O157:H7 with highly efficient bi-functional glucose oxidase-polydopamine nanocomposites and Prussian blue modified screen-printed interdigitated electrodes. Analyst 2016, 141, 5441–5449. [Google Scholar] [CrossRef]
- Zhu, T.; Wang, X.; Chang, W.; Zhang, Y.; Maruyama, T.; Luo, L.; Zhao, X. Green fabrication of Cu/rGO decorated SWCNT buckypaper as a flexible electrode for glucose detection. Mater. Sci. Eng. C 2021, 120, 111757. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Zine, N.; Baraket, A.; Zabala, M.; Campabadal, F.; Caruso, R.; Trivella, M.G.; Jaffrezic-Renault, N.; Errachid, A. A novel biosensor based on hafnium oxide: Application for early stage detection of human interleukin-10. Sens. Actuators B Chem. 2012, 175, 201–207. [Google Scholar] [CrossRef]
- Kulkarni, M.B.; Velmurugan, K.; Prasanth, E.; Amreen, K.; Nirmal, J.; Goel, S. Smartphone enabled miniaturized temperature controller platform to synthesize nio/cuo nanoparticles for electrochemical sensing and nanomicelles for ocular drug delivery applications. Biomed. Microdevices 2021, 23, 31. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, M.B.; Goel, S. Miniaturized DNA amplification platform with soft-lithographically fabricated continuous-flow PCR microfluidic device on a portable temperature controller. Microfluid. Nanofluid. 2021, 25, 69. [Google Scholar] [CrossRef]
- Kulkarni, M.B.; Jayapiriya, U.S.; Amreen, K.; Goel, S. Portable Thermal Management Platform for Synthesis of ZnO Nanoparticle in a Microfluidic Device: Validated for Electrochemical Sensing and Glucose Fuel Cell Applications. IEEE Trans. Electron. Devices 2021, 68, 4070–4076. [Google Scholar] [CrossRef]
- Kulkarni, M.B.; Channappa Bhyri, D.; Vanjerkhede, K. Brain Tumor Detection using Random Walk Solver Based Segmentation from MRI. Microsc. Res. Tech. 2018, 20, 1–8. [Google Scholar]
- Kulkarni, M.B. Automated and Integrated Microthermofluidic Platforms for DNA Amplification and Nanomaterial Synthesis. Ph.D. Thesis, BITS-Pilani, Hyderabad Campus, Secunderabad, India, 2022. [Google Scholar]
- Justino, C.I.L.; Duarte, A.C.; Rocha-Santos, T.A.P. Critical overview on the application of sensors and biosensors for clinical analysis. TrAC-Trends Anal. Chem. 2016, 85, 36–60. [Google Scholar] [CrossRef]
- Yerlikaya, S.; Broger, T.; MacLean, E.; Pai, M.; Denkinger, C.M. A tuberculosis biomarker database: The key to novel TB diagnostics. Int. J. Infect. Dis. 2017, 56, 253–257. [Google Scholar] [CrossRef]
- Lorenz, H.M. Technology evaluation: Adalimumab, Abbott laboratories. Curr. Opin. Mol. Ther. 2002, 4, 185–190. [Google Scholar] [PubMed]
- McCall, M.N.; Murakami, P.N.; Lukk, M.; Huber, W.; Irizarry, R.A. Assessing affymetrix GeneChip microarray quality. BMC Bioinform. 2011, 12, 137. [Google Scholar] [CrossRef]
- Ondov, B.D.; Varadarajan, A.; Passalacqua, K.D.; Bergman, N.H. Efficient mapping of Applied Biosystems SOLiD sequence data to a reference genome for functional genomic applications. Bioinformatics 2008, 24, 2776–2777. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Nakashima, C.; Komiya, K.; Kitera, K.; Hirai, M.; Kimura, S.; Aragane, N. Mechanisms of acquired resistance to afatinib clarified with liquid biopsy. PLoS ONE 2018, 13, e0209384. [Google Scholar] [CrossRef] [PubMed]
- Rumley, A. Urine dipstick testing: Comparison of results obtained by visual reading and with the Bayer CLINITEK 50. Ann. Clin. Biochem. 2000, 37, 220–221. [Google Scholar] [CrossRef]
- Holmes, K.L.; Otten, G.; Yokoyama, W.M. Flow Cytometry Analysis Using the Becton Dickinson FACS Calibur. Curr. Protoc. Immunol. 2002, 49, 5.4.1–5.4.22. [Google Scholar] [CrossRef]
- Jason-Moller, L.; Murphy, M.; Bruno, J. Overview of Biacore Systems and Their Applications. Curr. Protoc. Protein Sci. 2006, 45, 19.13.1–19.13.14. [Google Scholar] [CrossRef]
- Bose, A.; Lategan, C.A.; Smith, P.J.; Gupta, J.K. Studies on in vitro antiplasmodial activity of cleome rutidosperma. Acta Polanica Pharm. Drug. Res. 2010, 67, 315–318. [Google Scholar]
- Fogh-Andersen, N.; D’Orazio, P. Proposal for standardizing direct-reading biosensors for blood glucose. Clin. Chem. 1998, 44, 655–659. [Google Scholar] [CrossRef]
- Tomlinson, E.; Fu, L.; John, L.; Hultgren, B.; Huang, X.; Renz, M.; Stephan, J.P.; Tsai, S.P.; Powell-Braxton, L.; French, D.; et al. Transgenic Mice Expressing Human Fibroblast Growth Factor-19 Display Increased Metabolic Rate and Decreased Adiposity. Endocrinology 2002, 143, 1741–1747. [Google Scholar] [CrossRef]
- Jeong, H.; Kim, D.; Wang, G.; Park, S.; Lee, H.; Cho, K.; Hwang, W.-T.; Yoon, M.-H.; Jang, Y.H.; Song, H.; et al. Redox-Induced Asymmetric Electrical Characteristics of Ferrocene-Alkanethiolate Molecular Devices on Rigid and Flexible Substrates. Adv. Funct. Mater. 2014, 24, 2472–2480. [Google Scholar] [CrossRef]
- Sassolas, A.; Leca-Bouvier, B.D.; Blum, L.J. DNA Biosensors and Microarrays. Chem. Rev. 2008, 108, 109–139. [Google Scholar] [CrossRef]
- Lee-Lewandrowski, E.; Januzzi, J.L.; Green, S.M.; Tannous, B.; Wu, A.H.B.; Smith, A.; Wong, A.; Murakami, M.M.; Kaczmarek, J.; Apple, F.S.; et al. Multi-center validation of the Response Biomedical Corporation RAMP® NT-proBNP assay with comparison to the Roche Diagnostics GmbH Elecsys® proBNP assay. Clin. Chim. Acta 2007, 386, 20–24. [Google Scholar] [CrossRef]
- Conjeevaram, A.; Anupama, Y.J.; Vincent, L.; Sastry, N.B.; Siddini, V.; Revanasiddappa, M.; Srinivasa, S.; Thimmegeowda, A.B.; Kulkarni, M.J.; Patil, V.S. Clinico-Epidemiological Profile of Dialysis Services in Karnataka, India–A Multicentric Exploratory Study. Indian J. Nephrol. 2022, 32, 223–232. [Google Scholar] [CrossRef]
- Churakova, E.; Peri, K.; Vis, J.S.; Smith, D.W.; Beam, J.M.; Vijverberg, M.P.; Stor, M.C.; Winter, R.T. Accurate analysis of residual lactose in low-lactose milk: Comparing a variety of analytical techniques. Int. Dairy J. 2019, 96, 126–131. [Google Scholar] [CrossRef]
- Xu, L.; Liang, W.; Wen, Y.; Wang, L.; Yang, X.; Ren, S.; Jia, N.; Zuo, X.; Liu, G. An ultrasensitive electrochemical biosensor for the detection of mecA gene in methicillin-resistant Staphylococcus aureus. Biosens. Bioelectron. 2018, 99, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, C.; Xing, D. Fast identification of foodborne pathogenic viruses using continuous-flow reverse transcription-PCR with fluorescence detection. Microfluid. Nanofluid. 2011, 10, 367–380. [Google Scholar] [CrossRef]
- Obeid, P.J.; Christopoulos, T.K.; Crabtree, H.J.; Backhouse, C.J. Microfabricated device for DNA and RNA amplification by continuous-flow polymerase chain reaction and reverse transcription-polymerase chain reaction with cycle number selection. Anal. Chem. 2003, 75, 288–295. [Google Scholar] [CrossRef]
- Venkateswara Raju, C.; Hwan Cho, C.; Mohana Rani, G.; Manju, V.; Umapathi, R.; Suk Huh, Y.; Pil Park, J. Emerging insights into the use of carbon-based nanomaterials for the electrochemical detection of heavy metal ions. Coord. Chem. Rev. 2023, 476, 214920. [Google Scholar] [CrossRef]
- Kulkarni, M.B.; Ayachit, N.H.; Aminabhavi, T.M. Recent Advances in Microfluidics-Based Electrochemical Sensors for Foodborne Pathogen Detection. Biosensors 2023, 13, 246. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, M.B.; Yashas; Enaganti, P.K.; Amreen, K.; Goel, S. Internet of Things enabled portable thermal management system with microfluidic platform to synthesize MnO2 nanoparticles for electrochemical sensing. Nanotechnology 2020, 31, 425504. [Google Scholar] [CrossRef]
- Goto, M.; Honda, E.; Ogura, A.; Nomoto, A.; Hanaki, K.I. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques 2009, 46, 167–172. [Google Scholar] [CrossRef]
- Fernández-Carballo, B.L.; McBeth, C.; McGuiness, I.; Kalashnikov, M.; Baum, C.; Borrós, S.; Sharon, A.; Sauer-Budge, A.F. Continuous-flow, microfluidic, qRT-PCR system for RNA virus detection. Anal. Bioanal. Chem. 2018, 410, 33–43. [Google Scholar] [CrossRef]
- Cui, F.; Xu, Y.; Wang, R.; Liu, H.; Chen, L.; Zhang, Q.; Mu, X. Label-free impedimetric glycan biosensor for quantitative evaluation interactions between pathogenic bacteria and mannose. Biosens. Bioelectron. 2018, 103, 94–98. [Google Scholar] [CrossRef]
- Kaur, H.; Shorie, M.; Sharma, M.; Ganguli, A.K.; Sabherwal, P. Bridged Rebar Graphene functionalized aptasensor for pathogenic E. coli O78:K80:H11 detection. Biosens. Bioelectron. 2017, 98, 486–493. [Google Scholar] [CrossRef]
- Sharma, E.; Rathi, R.; Misharwal, J.; Sinhmar, B.; Kumari, S.; Dalal, J.; Kumar, A. Evolution in Lithography Techniques: Microlithography to Nanolithography. Nanomaterials 2022, 12, 2754. [Google Scholar] [CrossRef] [PubMed]
- Bilan, R.; Fleury, F.; Nabiev, I.; Sukhanova, A. Quantum dot surface chemistry and functionalization for cell targeting and imaging. Bioconjug. Chem. 2015, 26, 609–624. [Google Scholar] [CrossRef]
- Tian, H.; Sun, Y.; Liu, C.; Duan, X.; Tang, W.; Li, Z. Precise quantitation of MicroRNA in a single cell with droplet digital PCR based on ligation reaction. Anal. Chem. 2016, 88, 11384–11389. [Google Scholar] [CrossRef] [PubMed]
- Dutta, P.; Lu, Y.-J.; Hsieh, H.-Y.; Lee, T.-Y.; Lee, Y.-T.; Cheng, C.-M.; Fan, Y.-J. Detection of Candida albicans Using a Manufactured Electrochemical Sensor. Micromachines 2021, 12, 166. [Google Scholar] [CrossRef]
- Chadha, U.; Bhardwaj, P.; Agarwal, R.; Rawat, P.; Agarwal, R.; Gupta, I.; Panjwani, M.; Singh, S.; Ahuja, C.; Selvaraj, S.K.; et al. Recent progress and growth in biosensors technology: A critical review. J. Ind. Eng. Chem. 2022, 109, 21–51. [Google Scholar] [CrossRef]
- Martin, C.R.; Kohli, P. The emerging field of nanotube biotechnology. Nat. Rev. Drug Discov. 2003, 2, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Datta, D.; Panchal, D.S.; Venuganti, V.V.K. Transdermal delivery of vancomycin hydrochloride: Influence of chemical and physical permeation enhancers. Int. J. Pharm. 2021, 602, 120663. [Google Scholar] [CrossRef] [PubMed]
- Bell, J. The polymerase chain reaction. Immunol. Today 1989, 10, 351–355. [Google Scholar] [CrossRef]
- Hertneky, B.; Eger, J.; Bailly, M.; Christen, J.B. Mobile and Efficient Temperature and Humidity Control Chamber for Point-of-Care Diagnostics. In Proceedings of the 2019 IEEE Healthcare Innovations and Point of Care Technologies, (HI-POCT), Bethesda, MD, USA, 20–22 November 2019; pp. 159–162. [Google Scholar] [CrossRef]
- Mehrotra, P. Biosensors and their applications—A review. J. Oral Biol. Craniofacial Res. 2016, 6, 153–159. [Google Scholar] [CrossRef]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [CrossRef]
- Diculescu, V.C.; Chiorcea-Paquim, A.M.; Oliveira-Brett, A.M. Applications of a DNA-electrochemical biosensor. TrAC-Trends Anal. Chem. 2016, 79, 23–36. [Google Scholar] [CrossRef]
- Chen, G.; Chen, R.; Ding, S.; Li, M.; Wang, J.; Zou, J.; Du, F.; Dong, J.; Cui, X.; Huang, X.; et al. Recombinase assisted loop-mediated isothermal DNA amplification. Analyst 2020, 145, 440–444. [Google Scholar] [CrossRef]
- Teng, J.; Ye, Y.; Yao, L.; Yan, C.; Cheng, K.; Xue, F.; Pan, D.; Li, B.; Chen, W. Rolling circle amplification based amperometric aptamer/immuno hybrid biosensor for ultrasensitive detection of Vibrio parahaemolyticus. Microchim. Acta 2017, 184, 3477–3485. [Google Scholar] [CrossRef]
- Jiang, X.; Shao, N.; Jing, W.; Tao, S.; Liu, S.; Sui, G. Microfluidic chip integrating high throughput continuous-flow PCR and DNA hybridization for bacteria analysis. Talanta 2014, 122, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Garst, A.D.; Edwards, A.L.; Batey, R.T. Riboswitches: Structures and mechanisms. Cold Spring Harb. Perspect. Biol. 2011, 3, a003533. [Google Scholar] [CrossRef]
- Roth, A.; Breaker, R.R. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 2009, 78, 305–334. [Google Scholar] [CrossRef]
- Liss, M.; Petersen, B.; Wolf, H.; Prohaska, E. An aptamer-based quartz crystal protein biosensor. Anal. Chem. 2002, 74, 4488–4495. [Google Scholar] [CrossRef]
- Mishra, A.; Pilloton, R.; Jain, S.; Roy, S.; Khanuja, M.; Mathur, A.; Narang, J. Paper-Based Electrodes Conjugated with Tungsten Disulfide Nanostructure and Aptamer for Impedimetric Detection of Listeria monocytogenes. Biosensors 2022, 12, 88. [Google Scholar] [CrossRef]
- Cai, R.; Zhang, Z.; Chen, H.; Tian, Y.; Zhou, N. A versatile signal-on electrochemical biosensor for Staphylococcus aureus based on triple-helix molecular switch. Sens. Actuators B Chem. 2021, 326, 128842. [Google Scholar] [CrossRef]
- Dave, V.; Tak, K.; Sohgaura, A.; Gupta, A.; Sadhu, V.; Reddy, K.R. Lipid-polymer hybrid nanoparticles: Synthesis strategies and biomedical applications. J. Microbiol. Methods 2019, 160, 130–142. [Google Scholar] [CrossRef]
- Malvano, F.; Pilloton, R.; Albanese, D. A novel impedimetric biosensor based on the antimicrobial activity of the peptide nisin for the detection of Salmonella spp. Food Chem. 2020, 325, 126868. [Google Scholar] [CrossRef] [PubMed]
- Alava, T.; Berthet-Duroure, N.; Ayela, C.; Trévisiol, E.; Pugnière, M.; Morel, Y.; Rameil, P.; Nicu, L. Parallel acoustic detection of biological warfare agents surrogates by means of piezoelectric immunochips. Sens. Actuators B Chem. 2009, 138, 532–538. [Google Scholar] [CrossRef]
- Muthamma, K.; Sunil, D.; Shetty, P. Carbon dots as emerging luminophores in security inks for anti-counterfeit applications—An up-to-date review. Appl. Mater. Today 2021, 23, 101050. [Google Scholar] [CrossRef]
- Roushani, M.; Rahmati, Z.; Golchin, M.; Lotfi, Z.; Nemati, M. Electrochemical immunosensor for determination of Staphylococcus aureus bacteria by IgY immobilized on glassy carbon electrode with electrodeposited gold nanoparticles. Microchim. Acta 2020, 187, 567. [Google Scholar] [CrossRef] [PubMed]
- Rewatkar, P.; Bandapati, M.; Goel, S. Miniaturized additively manufactured co-laminar microfluidic glucose biofuel cell with optimized grade pencil bioelectrodes. Int. J. Hydrogen Energy 2019, 44, 31434–31444. [Google Scholar] [CrossRef]
- Simonian, A.L.; Grimsley, J.K.; Flounders, A.W.; Schoeniger, J.S.; Cheng, T.C.; DeFrank, J.J.; Wild, J.R. Enzyme-based biosensor for the direct detection of fluorine-containing organophosphates. Anal. Chim. Acta 2001, 442, 15–23. [Google Scholar] [CrossRef]
- Roy, A.; Ray, A.; Saha, S.; Ghosh, M.; Das, T.; Satpati, B.; Nandi, M.; Das, S. NiO-CNT composite for high performance supercapacitor electrode and oxygen evolution reaction. Electrochim. Acta 2018, 283, 327–337. [Google Scholar] [CrossRef]
- Knob, R.; Hanson, R.L.; Tateoka, O.B.; Wood, R.L.; Guerrero-Arguero, I.; Robison, R.A.; Pitt, W.G.; Woolley, A.T. Sequence-specific sepsis-related DNA capture and fluorescent labeling in monoliths prepared by single-step photopolymerization in microfluidic devices. J. Chromatogr. A 2018, 1562, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Rhouati, A.; Hayat, A.; Hernandez, D.B.; Meraihi, Z.; Munoz, R.; Marty, J.L. Development of an automated flow-based electrochemical aptasensor for on-line detection of Ochratoxin A. Sens. Actuators B Chem. 2013, 176, 1160–1166. [Google Scholar] [CrossRef]
- Ali, S.; Hassan, A.; Hassan, G.; Eun, C.H.; Bae, J.; Lee, C.H.; Kim, I.J. Disposable all-printed electronic biosensor for instantaneous detection and classification of pathogens. Sci. Rep. 2018, 8, 5920. [Google Scholar] [CrossRef]
- Gou, T.; Hu, J.; Wu, W.; Ding, X.; Zhou, S.; Fang, W.; Mu, Y. Smartphone-based mobile digital PCR device for DNA quantitative analysis with high accuracy. Biosens. Bioelectron. 2018, 120, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.C.; Duong, L.H. Novel solvent bonding method for thermoplastic microfluidic chips. Sens. Actuators B Chem. 2016, 237, 556–562. [Google Scholar] [CrossRef]
- Wei, Y.P.; Zhang, Y.W.; Mao, C.J. A silver nanoparticle-assisted signal amplification electrochemiluminescence biosensor for highly sensitive detection of mucin 1. J. Mater. Chem. B 2020, 8, 2471–2475. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jang, J.; Choi, B.; Yoon, J.; Kim, J.Y.; Choi, Y.K.; Myong Kim, D.; Hwan Kim, D.; Choi, S.J. A Highly Responsive Silicon Nanowire/Amplifier MOSFET Hybrid Biosensor. Sci. Rep. 2015, 5, 12286. [Google Scholar] [CrossRef]
- Qiu, X.; Mauk, M.G. An integrated, cellulose membrane-based PCR chamber. Microsyst. Technol. 2015, 21, 841–850. [Google Scholar] [CrossRef]
- Rutten, I.; Daems, D.; Leirs, K. Highly Sensitive Multiplex Detection of Molecular Biomarkers Using Hybridization Chain Reaction in an Encoded Particle Microfluidic Platform. Biosensors 2023, 13, 100. [Google Scholar] [CrossRef]
- Kaur, A.; Mahmoud, R.; Megalathan, A.; Pettit, S.; Dhakal, S. Multiplexed smFRET Nucleic Acid Sensing Using DNA Nanotweezers. Biosensors 2023, 13, 119. [Google Scholar] [CrossRef]
- Wang, Y.; Han, Y.; Zhou, F.; Fan, T.; Liu, F. Simple Detection of DNA Methyltransferase with an Integrated Padlock Probe. Biosensors 2022, 12, 569. [Google Scholar] [CrossRef]
- Coelho, B.J.; Veigas, B.; Bettencourt, L.; Águas, H.; Fortunato, E.; Martins, R.; Baptista, P.V.; Igreja, R. Digital Microfluidics-Powered Real-Time Monitoring of Isothermal DNA Amplification of Cancer Biomarker. Biosensors 2022, 12, 201. [Google Scholar] [CrossRef]
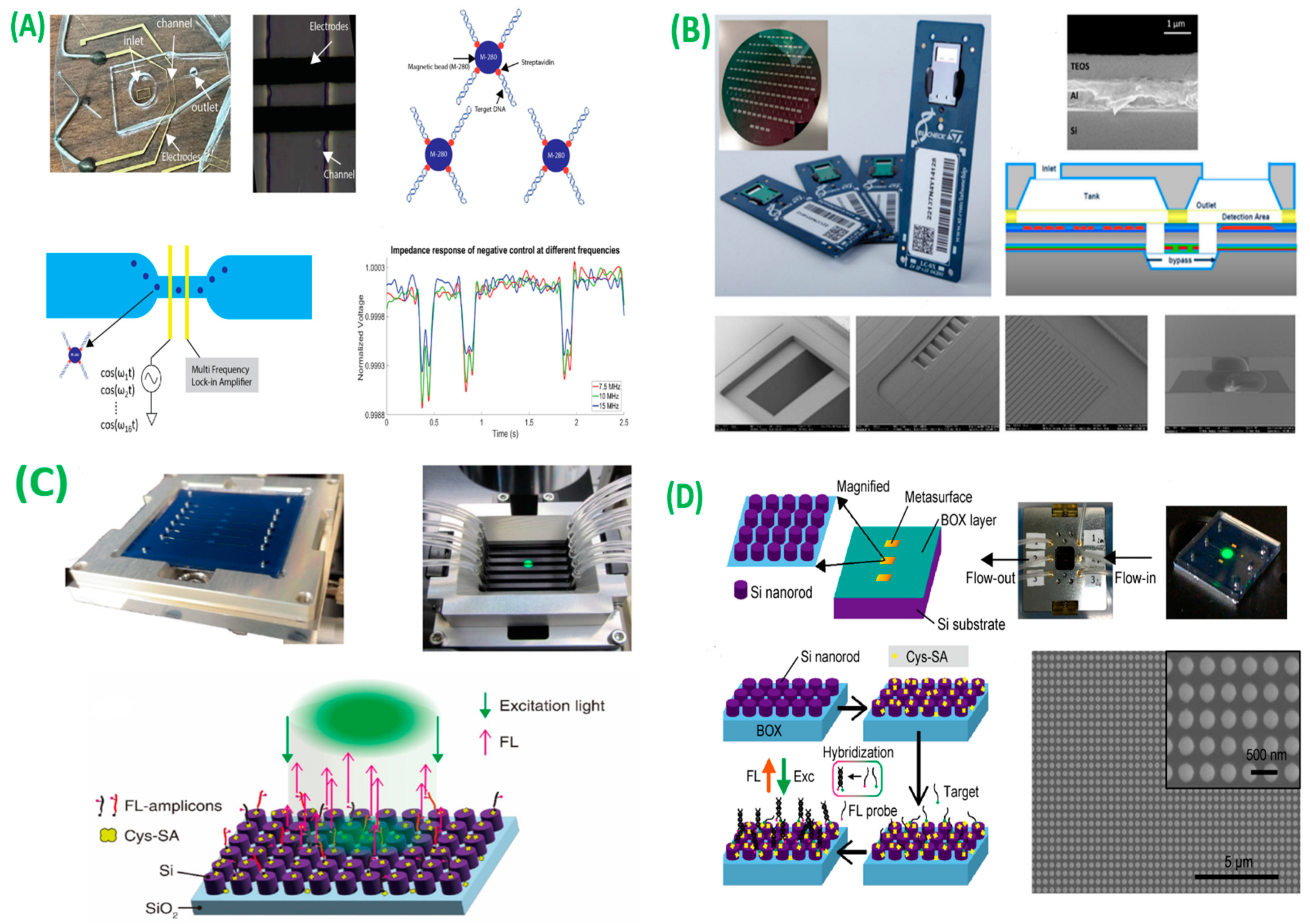
- Kokabi, M.; Sui, J.; Gandotra, N.; Pournadali Khamseh, A.; Scharfe, C.; Javanmard, M. Nucleic Acid Quantification by Multi-Frequency Impedance Cytometry and Machine Learning. Biosensors 2023, 13, 316. [Google Scholar] [CrossRef]
- Ventimiglia, G.; Pesaturo, M.; Malcolm, A.; Petralia, S. A Miniaturized Silicon Lab-on-Chip for Integrated PCR and Hybridization Microarray for High Multiplexing Nucleic Acids Analysis. Biosensors 2022, 12, 563. [Google Scholar] [CrossRef] [PubMed]
- Iwanaga, M. Rapid Detection of Attomolar SARS-CoV-2 Nucleic Acids in All-Dielectric Metasurface Biosensors. Biosensors 2022, 12, 987. [Google Scholar] [CrossRef] [PubMed]
- Iwanaga, M. High-Sensitivity High-Throughput Detection of Nucleic Acid Targets on Metasurface Fluorescence Biosensors. Biosensors 2021, 11, 33. [Google Scholar] [CrossRef] [PubMed]

| Company | Foundation Year | Targets | Cost (~$) | Ref. |
|---|---|---|---|---|
| Abbott | 1888 | Glucose biosensor | 48.85 | [51] |
| Affymetrix | 1992 | Pharmaceutical research | - | [52] |
| Applied Biosystems | 1981 | Affinity chip | 69.95 | [53] |
| ARKRAY, Inc. | 1960 | Creatinine biosensor | 21.25 | [54] |
| Bayer Diagnostics | 1958 | Glucose biosensor | 42.78 | [55] |
| Becton Dickinson | 1897 | Glucose biosensor | 45.37 | [56] |
| Biacore | 1984 | Affinity sensors for medical research | 56.32 | [57] |
| Cleome Innovations | 2021 | Medical POCT devices | 18.26 | [58] |
| Eppendorf Inc. | 1989 | Medical diagnostics | 19.37 | [59] |
| LifeScan | 1981 | Lactate | 14.31 | [60] |
| Molecular devices | 1983 | Pharmaceutical | - | [61] |
| Nanogen | 1993 | Glucose, urea, creatinine, and lactate biosensors | 20.35 | [62] |
| Roche Diagnostics | 1896 | Glucose biosensors | 38.21 | [63] |
| Renalyx Healthcare Systems | 2013 | Creatinine and albumin sensors | 19.21 | [64] |
| YSI Inc. | 1948 | Lactate | 13.24 | [65] |
| Biorecognition Component | Bioanalyte | Transducer | Limit of Detection | Application | Ref. |
|---|---|---|---|---|---|
| DNA | Nuclei acids | QCM | 450 fM | Eco-friendly biointerfaces | [98] |
| ssDNA | RNA | Fluorescence | 180 pM | Early disease diagnosis | [99] |
| PNA | ssDNA | GCE | 2.58 pM | NS | [100] |
| PNA | miRNA-492 suggested biomarker for PDCA | Graphite oxide with gold NPs | 8 nM | Early identification of PDCA | [101] |
| PNA | RNA | G-FET | 0.2 aM | NS | [102] |
| Aptamer | PDGF | CNT | - | Atherosclerosis, fibrosis, malevolent viruses | [103] |
| ssDNA | Synthetic DNA of E. faecalis | Electrode with Gold NPs | 3.4 amol L−1 | Early detection of pathogens in food | [104] |
| Aptamer | Synthetic DNA of Group B Streptococci | Gold NPs | 0.5 fM | Early detection of bacteria | [105] |
| Arched probeaptamer, Hairpin probe-1 and 2 | Salmonella typhimurium | Luminescence | 1.6 CFU mL−1 | Adulteration in milk | [106] |
| 3D-Printed PMMA chip | Salmonella enteritis and Staphylococcus aureus | Luminescence | 5 CFU mL−1 | Microfluidic-based biosensor for identification of virulent | [107] |
| Polyacrylamidehydrogel:aptamer strands | Microcystin-LR | Colorimetric | 3.5 ng L−1 | Detection of fresh fish | [108] |
| Aptamer hairpin | Salmonella typhimurium | Gold electrode | 0.98 fM | Genomic DNA from clinical anal/vaginal samples | [109] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kulkarni, M.B.; Ayachit, N.H.; Aminabhavi, T.M. A Short Review on Miniaturized Biosensors for the Detection of Nucleic Acid Biomarkers. Biosensors 2023, 13, 412. https://doi.org/10.3390/bios13030412
Kulkarni MB, Ayachit NH, Aminabhavi TM. A Short Review on Miniaturized Biosensors for the Detection of Nucleic Acid Biomarkers. Biosensors. 2023; 13(3):412. https://doi.org/10.3390/bios13030412
Chicago/Turabian StyleKulkarni, Madhusudan B., Narasimha H. Ayachit, and Tejraj M. Aminabhavi. 2023. "A Short Review on Miniaturized Biosensors for the Detection of Nucleic Acid Biomarkers" Biosensors 13, no. 3: 412. https://doi.org/10.3390/bios13030412
APA StyleKulkarni, M. B., Ayachit, N. H., & Aminabhavi, T. M. (2023). A Short Review on Miniaturized Biosensors for the Detection of Nucleic Acid Biomarkers. Biosensors, 13(3), 412. https://doi.org/10.3390/bios13030412







