Surface Plasmon Resonance (SPR) Sensor for Cancer Biomarker Detection
Abstract
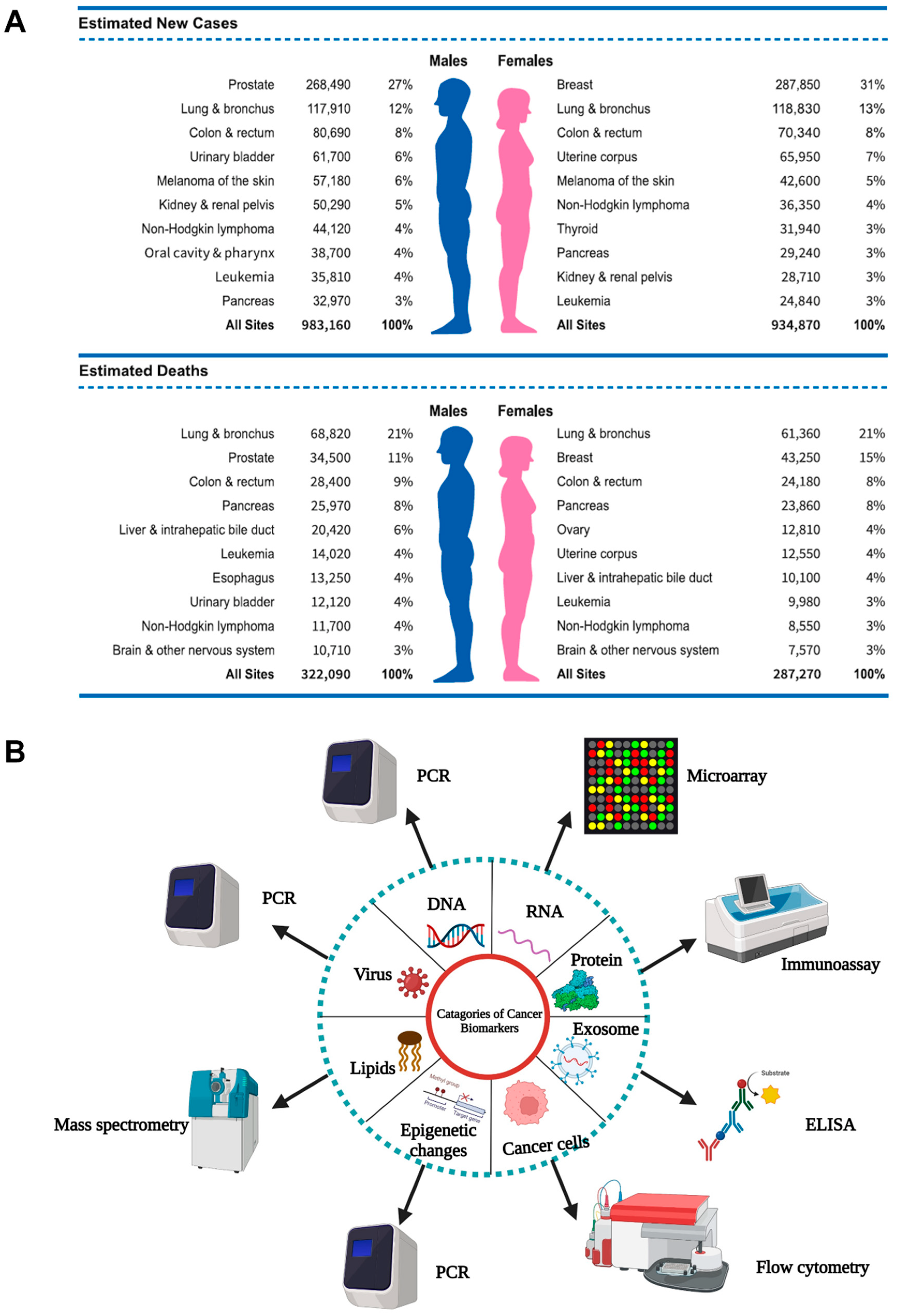
1. Introduction
2. Biomarkers in Cancer Detection, Diagnosis, and Prognosis
3. Biosensors: Diagnostic Devices to Detect Biomarkers


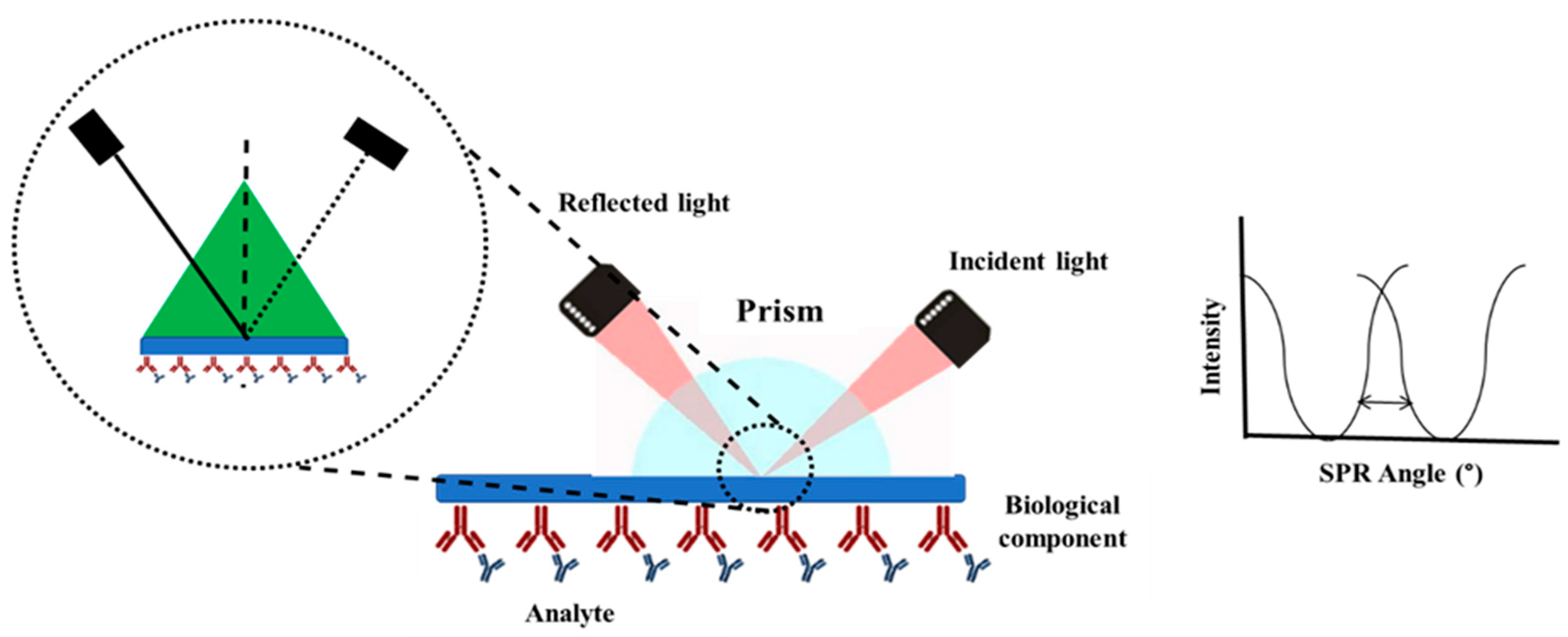
4. Surface Plasmon Resonance
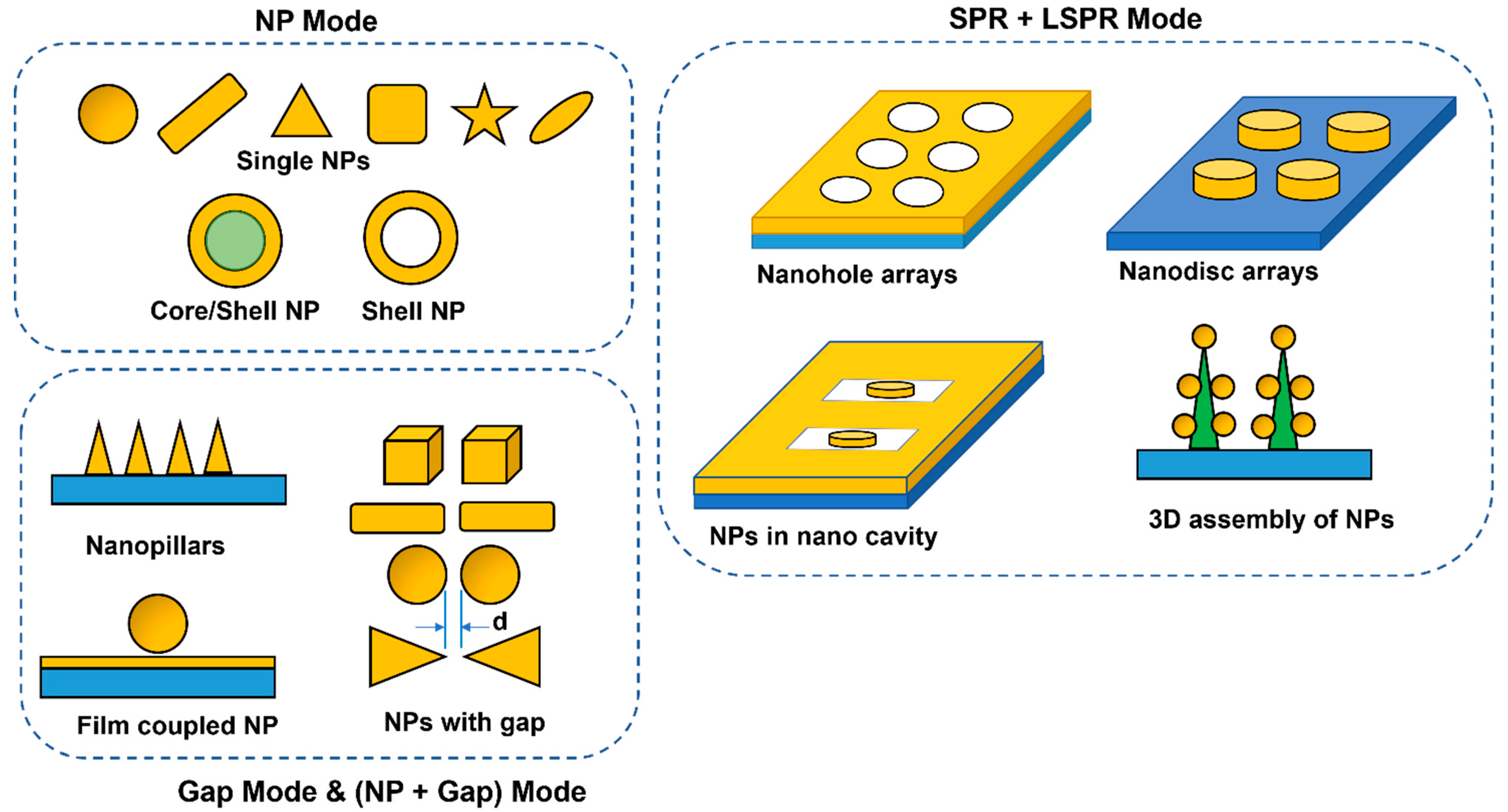
4.1. Role of the Placement of Molecules on the Plasmonic Sensor
4.2. Role of Geometry of the Plasmonic Sensor
5. SPR in Cancer Detection
5.1. SPR for Detecting CTC and ctDNA
5.2. SPR for Detection of miRNA
5.3. SPR for Detecting Cancer Stem Cells
5.4. SPR for Detection of Protein
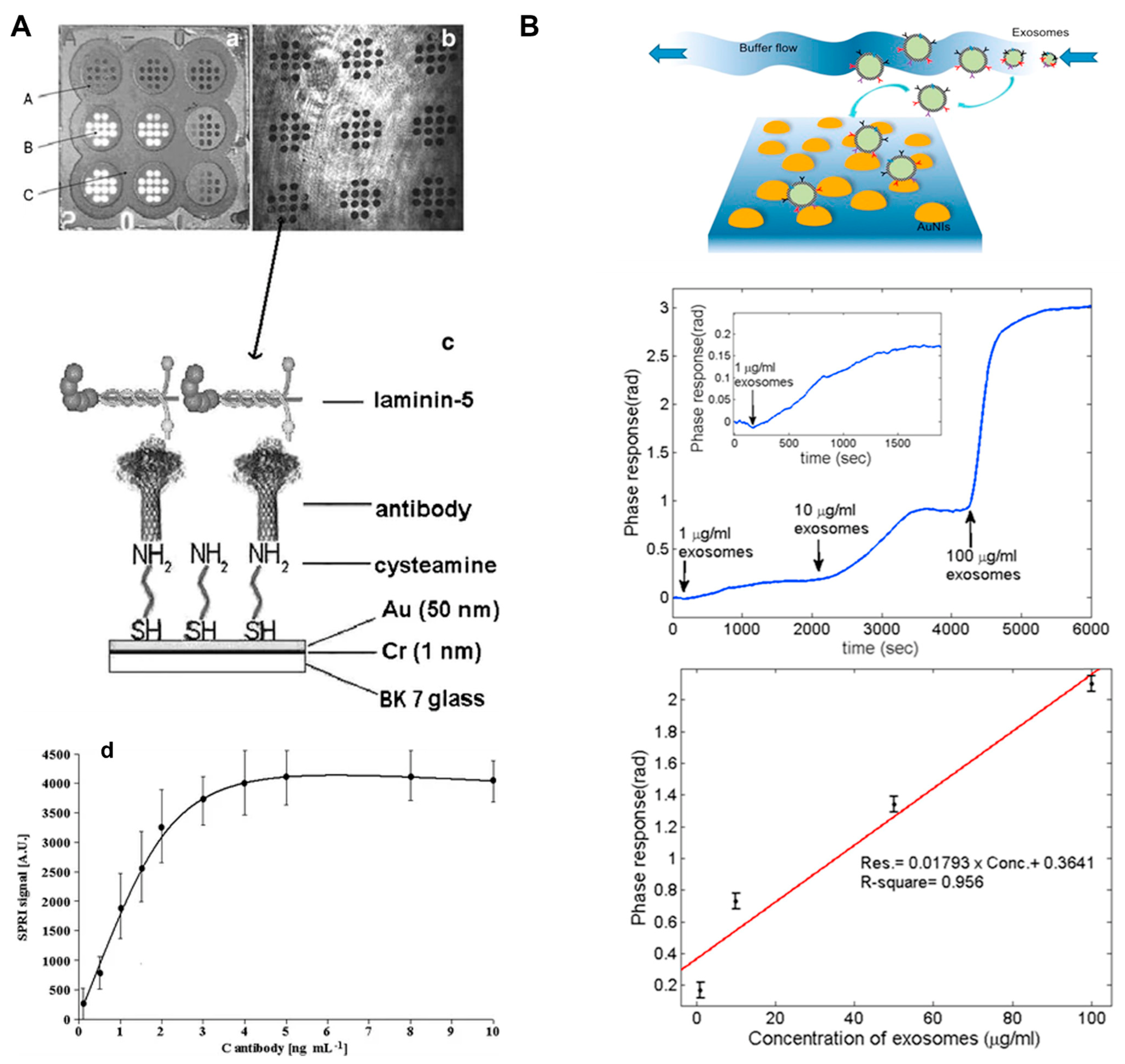
5.5. SPR for Detection of Exosomes

5.6. SPR for Detection of Lipids
6. Other Applications
6.1. SPR for High-Throughput Anti-Cancer Drug Screening
6.2. SPR for Anti-Cancer Antibody Development
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aronson, J.K.; Ferner, R.E. Biomarkers—A general review. Curr. Protoc. Pharmacol. 2017, 2017, 9.23.1–9.23.17. [Google Scholar] [CrossRef] [PubMed]
- Blackadar, C.B. Historical review of the causes of cancer. World J. Clin. Oncol. 2016, 7, 54–86. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Mordente, A.; Meucci, E.; Martorana, G.E.; Silvestrini, A. Cancer biomarkers discovery and validation: State of the art, problems and future perspectives. Adv. Exp. Med. Biol. 2015, 867, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Gorgannezhad, L.; Umer, M.; Islam, M.N.; Nguyen, N.T.; Shiddiky, M.J.A. Circulating tumor DNA and liquid biopsy: Opportunities, challenges, and recent advances in detection technologies. Lab Chip 2018, 18, 1174–1196. [Google Scholar] [CrossRef] [PubMed]
- Khanmohammadi, A.; Aghaie, A.; Vahedi, E.; Qazvini, A.; Ghanei, M.; Afkhami, A.; Hajian, A.; Bagheri, H. Electrochemical biosensors for the detection of lung cancer biomarkers: A review. Talanta 2020, 206, 120251. [Google Scholar] [CrossRef]
- Xiao, L.; Zhu, A.; Xu, Q.; Chen, Y.; Xu, J.; Weng, J. Colorimetric Biosensor for Detection of Cancer Biomarker by Au Nanoparticle-Decorated Bi2Se3 Nanosheets. ACS Appl. Mater. Interfaces 2017, 9, 6931–6940. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, Q.; Duan, X.; Wu, W.; Zeng, G. Phosphofructokinase-M inhibits cell growth via modulating the FOXO3 pathway in renal cell carcinoma cells. Biochem. Biophys. Res. Commun. 2020, 530, 67–74. [Google Scholar] [CrossRef]
- Cai, X.; Zhang, H.; Yu, X.; Wang, W. A microfluidic paper-based laser-induced fluorescence sensor based on duplex-specific nuclease amplification for selective and sensitive detection of miRNAs in cancer cells. Talanta 2020, 216, 120996. [Google Scholar] [CrossRef]
- Qu, J.H.; Dillen, A.; Saeys, W.; Lammertyn, J.; Spasic, D. Advancements in SPR biosensing technology: An overview of recent trends in smart layers design, multiplexing concepts, continuous monitoring and in vivo sensing. Anal. Chim. Acta 2020, 1104, 10–27. [Google Scholar] [CrossRef]
- Mahmoudpour, M.; Ezzati Nazhad Dolatabadi, J.; Torbati, M.; Pirpour Tazehkand, A.; Homayouni-Rad, A.; de la Guardia, M. Nanomaterials and new biorecognition molecules based surface plasmon resonance biosensors for mycotoxin detection. Biosens. Bioelectron. 2019, 143, 111603. [Google Scholar] [CrossRef]
- Masson, J.F. Surface Plasmon Resonance Clinical Biosensors for Medical Diagnostics. ACS Sens. 2017, 2, 16–30. [Google Scholar] [CrossRef]
- Jebelli, A.; Oroojalian, F.; Fathi, F.; Mokhtarzadeh, A.; de la Guardia, M. Recent advances in surface plasmon resonance biosensors for microRNAs detection. Biosens. Bioelectron. 2020, 169, 112599. [Google Scholar] [CrossRef]
- Homola, J.; Yee, S.S.; Gauglitz, G. Surface plasmon resonance sensors: Review. Sens. Actuators B Chem. 1999, 54, 3–15. [Google Scholar] [CrossRef]
- He, L.; Pagneux, Q.; Larroulet, I.; Serrano, A.Y.; Pesquera, A.; Zurutuza, A.; Mandler, D.; Boukherroub, R.; Szunerits, S. Label-free femtomolar cancer biomarker detection in human serum using graphene-coated surface plasmon resonance chips. Biosens. Bioelectron. 2017, 89, 606–611. [Google Scholar] [CrossRef]
- Patil, P.O.; Pandey, G.R.; Patil, A.G.; Borse, V.B.; Deshmukh, P.K.; Patil, D.R.; Tade, R.S.; Nangare, S.N.; Khan, Z.G.; Patil, A.M.; et al. Graphene-based nanocomposites for sensitivity enhancement of surface plasmon resonance sensor for biological and chemical sensing: A review. Biosens. Bioelectron. 2019, 139, 111324. [Google Scholar] [CrossRef]
- Shao, B.; Xiao, Z. Recent achievements in exosomal biomarkers detection by nanomaterials-based optical biosensors—A review. Anal. Chim. Acta 2020, 1114, 74–84. [Google Scholar] [CrossRef]
- Mesri, E.A.; Feitelson, M.A.; Munger, K. Human viral oncogenesis: A cancer hallmarks analysis. Cell Host Microbe 2014, 15, 266–282. [Google Scholar] [CrossRef]
- Weissleder, R.; Ntziachristos, V. Shedding light onto live molecular targets. Nat. Med. 2003, 9, 123–128. [Google Scholar] [CrossRef]
- Sidransky, D. Emerging molecular markers of cancer. Nat. Rev. Cancer 2002, 2, 210–219. [Google Scholar] [CrossRef]
- Vogelstein, B.; Kinzler, K.W. Cancer genes and the pathways they control. Nat. Med. 2004, 10, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.B.; Ohm, J.E. Epigenetic gene silencing in cancer—A mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer 2006, 6, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Jaenisch, R.; Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.C.S. Contribution of oncoproteomics to cancer biomarker discovery. Mol. Cancer 2007, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Ludwig, J.A.; Weinstein, J.N. Biomarkers in cancer staging, prognosis and treatment selection. Nat. Rev. Cancer 2005, 5, 845–856. [Google Scholar] [CrossRef]
- Bhatt, A.N.; Mathur, R.; Farooque, A.; Verma, A.; Dwarakanath, B.S. Cancer biomarkers—Current perspectives. Indian J. Med. Res. 2010, 132, 129–149. [Google Scholar]
- Sulzyc-Bielicka, V.; Bielicki, D.; Binczak-Kuleta, A.; Kaczmarczyk, M.; Pioch, W.; Machoy-Mokrzynska, A.; Ciechanowicz, A.; Gołȩbiewska, M.; Drozdzik, M. Thymidylate synthase gene polymorphism and survival of colorectal cancer patients receiving adjuvant 5-fluorouracil. Genet. Test. Mol. Biomarkers 2013, 17, 799–806. [Google Scholar] [CrossRef]
- Henry, N.L.; Hayes, D.F. Cancer biomarkers. Mol. Oncol. 2012, 6, 140–146. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, J.; Luo, F. Serum tumor markers for detection of hepatocellular carcinoma. World J. Gastroenterol. 2006, 12, 1175–1181. [Google Scholar] [CrossRef]
- Sarosdy, F.; Hudson, M.L.A.; Ellis, W.J.; Soloway, M.S.; White, R.D.; Sheinfeld, J.; Jarowenko, M.V.; Schellhammer, P.F.; Schervish, E.D.W.; Patel, J.A.Y.V.; et al. Using the Bard Bta Stat Test Cancer. Urology 1995, 4295, 349–353. [Google Scholar]
- Vaz, F.H.; Machado, P.M.; Brandão, R.D.; Laranjeira, C.T.; Eugénio, J.S.; Fernandes, A.H.; André, S.P. Familial breast/ovarian cancer and BRCA1/2 genetic screening: The role of immunohistochemistry as an additional method in the selection of patients. J. Histochem. Cytochem. 2007, 55, 1105–1113. [Google Scholar] [CrossRef] [PubMed]
- Ballehaninna, U.K.; Chamberlain, R.S. Serum CA 19-9 as a Biomarker for Pancreatic Cancer-A Comprehensive Review. Indian J. Surg. Oncol. 2011, 2, 88–100. [Google Scholar] [CrossRef]
- Safi, F.; Kohler, I.; Rottinger, E.; Suhr, P.; Beger, H.G. Comparison of CA 15-3 and CEA in diagnosis and monitoring of breast cancer. Int. J. Biol. Markers 1989, 4, 207–214. [Google Scholar] [CrossRef]
- Scholler, N.; MUrban, N. CA125 in ovarian cancer. Biomark. Med. 2007, 1, 513–523. [Google Scholar] [CrossRef]
- Frenette, P.S.; Thirlwell, M.P.; Trudeau, M.; Thomson, D.M.P.; Joseph, L.; Shuster, J.S. The diagnostic value of CA 27-29, CA 15-3, mucin-like carcinoma antigen, carcinoembryonic antigen and CA 19-9 in breast and gastrointestinal malignancies. Tumor Biol. 1994, 15, 247–254. [Google Scholar] [CrossRef]
- Uygur, M.M.; Gümüş, M. The utility of serum tumor markers CEA and CA 15–3 for breast cancer prognosis and their association with clinicopathological parameters. Cancer Treat. Res. Commun. 2021, 28, 100402. [Google Scholar] [CrossRef]
- Tachezy, M.; Zander, H.; Marx, A.H.; Stahl, P.R.; Gebauer, F.; Izbicki, J.R.; Bockhorn, M. ALCAM (CD166) expression and serum levels in pancreatic cancer. PLoS ONE 2012, 7, e39018. [Google Scholar] [CrossRef] [PubMed]
- Lindgren, M.; Jansson, M.; Tavelin, B.; Dirix, L.; Vermeulen, P.; Nyström, H. Type IV collagen as a potential biomarker of metastatic breast cancer. Clin. Exp. Metastasis 2021, 38, 175–185. [Google Scholar] [CrossRef]
- Banerjee, J.; Pradhan, R.; Gupta, A.; Kumar, R.; Sahu, V.; Upadhyay, A.D.; Chaterjee, P.; Dwivedi, S.; Dey, S.; Dey, A.B. CDK4 in lung, and head and neck cancers in old age: Evaluation as a biomarker. Clin. Transl. Oncol. 2016, 19, 571–578. [Google Scholar] [CrossRef]
- Gao, J.; Lv, F.; Li, J.; Wu, Z.; Qi, J. Serum cytokeratin 19 fragment, CK19-2G2, as a newly identified biomarker for lung cancer. PLoS ONE 2014, 9, e101979. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Sun, X.Y.; Xu, L.C.; Fu, R.Z. Abnormal expression of serum soluble e-cadherin is correlated with clinicopathological features and prognosis of breast cancer. Med. Sci. Monit. 2014, 20, 2776–2782. [Google Scholar] [CrossRef] [PubMed]
- Dekker, T.J.A.; ter Borg, S.; Hooijer, G.K.J.; Meijer, S.L.; Wesseling, J.; Boers, J.E.; Schuuring, E.; Bart, J.; van Gorp, J.; Bult, P.; et al. Quality assessment of estrogen receptor and progesterone receptor testing in breast cancer using a tissue microarray-based approach. Breast Cancer Res. Treat. 2015, 152, 247–252, Erratum in Breast Cancer Res. Treat. 2015, 152, 253. https://doi.org/10.1007/s10549-015-3478-0. [Google Scholar] [CrossRef] [PubMed]
- Schmetter, B.S.; Habicht, K.K.; Lamm, D.L.; Morales, A.; Bander, N.H.; Grossman, H.B.; Hanna, M.G.; Silberman, S.R.; Butman, B.T. A multicenter trial evaluation of the fibrin/fibrinogen degradation products test for detection and monitoring of bladder cancer. J. Urol. 1997, 158, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Dong, Y.; Sun, X.; Sui, X.; Zhu, H.; Zhao, Y.; Zhang, Y.; Mason, C.; Zhu, Q.; Han, S. High levels of serum glypican-1 indicate poor prognosis in pancreatic ductal adenocarcinoma. Cancer Med. 2018, 7, 5525–5533. [Google Scholar] [CrossRef]
- Lu, J.; Wang, Y.; Yan, M.; Feng, P.; Yuan, L.; Cai, Y.; Xia, X.; Liu, M.; Luo, J.; Li, L. High serum haptoglobin level is associated with tumor progression and predicts poor prognosis in non-small cell lung cancer. Oncotarget 2016, 7, 41758–41766. [Google Scholar] [CrossRef]
- Dekker, T.J.A.; Borg, S.T.; Hooijer, G.K.J.; Meijer, S.L.; Wesseling, J.; Boers, J.E.; Schuuring, E.; Bart, J.; van Gorp, J.; Mesker, W.E.; et al. Determining sensitivity and specificity of HER2 testing in breast cancer using a tissue micro-array approach. Breast Cancer Res. 2012, 14, R93. [Google Scholar] [CrossRef]
- Lenhard, M.; Tsvilina, A.; Schumacher, L.; Kupka, M.; Ditsch, N.; Mayr, D.; Friese, K.; Jeschke, U. Human chorionic gonadotropin and its relation to grade, stage and patient survival in ovarian cancer. BMC Cancer 2012, 12, 2. [Google Scholar] [CrossRef]
- Moore, R.G.; Brown, A.K.; Miller, M.C.; Badgwell, D.; Lu, Z.; Allard, W.J.; Granai, C.O.; Bast, R.C.; Lu, K. Utility of a novel serum tumor biomarker HE4 in patients with endometrioid adenocarcinoma of the uterus. Gynecol. Oncol. 2008, 110, 196–201. [Google Scholar] [CrossRef]
- Xu, Y.; Shen, Z.; Wiper, D.W.; Wu, M.; Morton, R.E.; Elson, P.; Kennedy, A.W.; Belinson, J.; Markman, M.; Casey, G. Lysophosphatidic acid as a potential biomarker for ovarian and other gynecologic cancers. J. Am. Med. Assoc. 1998, 280, 719–723. [Google Scholar] [CrossRef]
- Li, L.; Wang, H.; Lu, Y.; Pan, Z. The expression and clinical significance of serum miR-205 for breast cancer and its role in detection of human cancers. Int. J. Clin. Exp. Med. 2015, 8, 3034–3043. [Google Scholar]
- Shariat, S.F.; Marberger, M.J.; Lotan, Y.; Sanchez-Carbayo, M.; Zippe, C.; Lüdecke, G.; Boman, H.; Sawczuk, I.; Friedrich, M.G.; Casella, R.; et al. Variability in the Performance of Nuclear Matrix Protein 22 for the Detection of Bladder Cancer. J. Urol. 2006, 176, 919–926. [Google Scholar] [CrossRef]
- Khan, A.S.; Ahmad, S.; Ullah, Z.; Sadiq, N.; Haq, M.; Sheikh, A.K. Predictive value of tissue p53 protein expression and serum p53 antibodies in oral potentially malignant disorders: Relative to oral squamous cell carcinoma. J. Taibah Univ. Med. Sci. 2022, 17, 415–423. [Google Scholar] [CrossRef]
- Al Saidi, S.S.; Al Riyami, N.B.; Al Marhoon, M.S.; Al Saraf, M.S.; Al Busaidi, S.S.; Bayoumi, R.; Mula-Abed, W.A.S. Validity of prostate health index and percentage of [-2] pro-prostate-specific antigen as novel biomarkers in the diagnosis of prostate cancer: Omani tertiary hospitals experience. Oman Med. J. 2017, 32, 275–283. [Google Scholar] [CrossRef]
- Der Lin, J.; Huang, M.J.; Hsu, B.R.S.; Chao, T.C.; Hsueh, C.; Liu, F.H.; Liou, M.J.; Weng, H.F. Significance of postoperative serum thyroglobulin levels in patients with papillary and follicular thyroid carcinomas. J. Surg. Oncol. 2002, 80, 45–51. [Google Scholar] [CrossRef]
- Shehata, F.; Monem, N.A.; Sakr, M.; Kasem, S.; Balbaa, M. Epidermal growth factor, its receptor and transforming growth factor-β1 in the diagnosis of HCV-induced hepatocellular carcinoma. Med. Oncol. 2013, 30, 673. [Google Scholar] [CrossRef]
- Santin, A.D.; Bellone, S.; Ravaggi, A.; Roman, J.; Smith, C.V.; Pecorelli, S.; Cannon, M.J.; Parham, G.P. Increased levels of interleukin-10 and transforming growth factor-β in the plasma and ascitic fluid of patients with advanced ovarian cancer. Br. J. Obstet. Gynaecol. 2001, 108, 804–808. [Google Scholar] [CrossRef]
- Kut, C.; Mac Gabhann, F.; Popel, A.S. Where is VEGF in the body? A meta-analysis of VEGF distribution in cancer. Br. J. Cancer 2007, 97, 978–985. [Google Scholar] [CrossRef]
- Kim, D.M.; Noh, H.B.; Park, D.S.; Ryu, S.H.; Koo, J.S.; Shim, Y.B. Immunosensors for detection of Annexin II and MUC5AC for early diagnosis of lung cancer. Biosens. Bioelectron. 2009, 25, 456–462. [Google Scholar] [CrossRef]
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical biosensors—Sensor principles and architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef]
- Jia, Y.; Qin, M.; Zhang, H.; Niu, W.; Li, X.; Wang, L.; Li, X.; Bai, Y.; Cao, Y.; Feng, X. Label-free biosensor: A novel phage-modified Light Addressable Potentiometric Sensor system for cancer cell monitoring. Biosens. Bioelectron. 2007, 22, 3261–3266. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhao, X.; Khimji, I.; Akbas, R.; Qiu, W.; Edwards, D.; Cramer, D.W.; Ye, B.; Demirci, U. Integration of cell phone imaging with microchip ELISA to detect ovarian cancer HE4 biomarker in urine at the point-of-care. Lab Chip 2011, 11, 3411–3418. [Google Scholar] [CrossRef] [PubMed]
- Underwood, J.J.; Quadri, R.S.; Kalva, S.P.; Shah, H.; Sanjeeviah, A.R.; Beg, M.S.; Sutphin, P.D. Liquid biopsy for cancer: Review and implications for the radiologist. Radiology 2020, 294, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.; Zhu, Z.; Jia, H.; Lu, C.; Liu, X.; Mao, D.; Chen, G. Colorimetric detection of cancer biomarker based on enzyme enrichment and pH sensing. Sens. Actuators B Chem. 2020, 320, 128435. [Google Scholar] [CrossRef]
- Won, H.J.; Robby, A.I.; Jhon, H.S.; In, I.; Ryu, J.H.; Park, S.Y. Wireless label-free electrochemical detection of cancer cells by MnO2-Decorated polymer dots. Sens. Actuators B Chem. 2020, 320, 128391. [Google Scholar] [CrossRef]
- Loyez, M.; Lobry, M.; Hassan, E.M.; DeRosa, M.C.; Caucheteur, C.; Wattiez, R. HER2 breast cancer biomarker detection using a sandwich optical fiber assay. Talanta 2021, 221, 121452. [Google Scholar] [CrossRef]
- Hahn, J.; Kim, E.; You, Y.; Choi, Y.J. Colorimetric switchable linker-based bioassay for ultrasensitive detection of prostate-specific antigen as a cancer biomarker. Analyst 2019, 144, 4439–4446. [Google Scholar] [CrossRef]
- Prasad, A.; Chaichi, A.; Mahigir, A.; Sahu, S.P.; Ganta, D.; Veronis, G.; Gartia, M.R. Ripple mediated surface enhanced Raman spectroscopy on graphene. Carbon. 2020, 157, 525–536. [Google Scholar] [CrossRef]
- Kretschmann, E.; Raether, H. Radiative Decay of Non Radiative Surface Plasmons Excited by Light. Z. Fur Naturforsch.-Sect. A J. Phys. Sci. 1968, 23, 2135–2136. [Google Scholar] [CrossRef]
- Otto, A. Excitation of nonradiative surface plasma waves in silver by the method of frustrated total reflection. Z. Für Phys. 1968, 216, 398–410. [Google Scholar] [CrossRef]
- Wood, R.W. On a remarkable case of uneven distribution of light in a diffraction grating spectrum. Proc. Phys. Soc. Lond. 1901, 18, 269–275. [Google Scholar] [CrossRef]
- Homola, J. Present and future of surface plasmon resonance biosensors. Anal. Bioanal. Chem. 2003, 377, 528–539. [Google Scholar] [CrossRef]
- Willets, K.A.; Van Duyne, R.P. Localized surface plasmon resonance spectroscopy and sensing. Annu. Rev. Phys. Chem. 2007, 58, 267–297. [Google Scholar] [CrossRef]
- Ferhan, A.R.; Jackman, J.A.; Cho, N.J. Integration of quartz crystal microbalance-dissipation and reflection-mode localized surface plasmon resonance sensors for biomacromolecular interaction analysis. Anal. Chem. 2016, 88, 12524–12531. [Google Scholar] [CrossRef]
- Stewart, M.E.; Anderton, C.R.; Thompson, L.B.; Maria, J.; Gray, S.K.; Rogers, J.A.; Nuzzo, R.G. Nanostructured plasmonic sensors. Chem. Rev. 2008, 108, 494–521. [Google Scholar] [CrossRef]
- Yang, J.; Giessen, H.; Lalanne, P. Simple analytical expression for the peak-frequency shifts of plasmonic resonances for sensing. Nano Lett. 2015, 15, 3439–3444. [Google Scholar] [CrossRef]
- Rezabakhsh, A.; Rahbarghazi, R.; Fathi, F. Surface plasmon resonance biosensors for detection of Alzheimer’s biomarkers; an effective step in early and accurate diagnosis. Biosens. Bioelectron. 2020, 167, 112511. [Google Scholar] [CrossRef]
- Achen, M.G.; Roufail, S.; Domagala, T.; Catimel, B.; Nice, E.C.; Geleick, D.M.; Murphy, R.; Scott, A.M.; Caesar, C.; Makinen, T.; et al. Monoclonal antibodies to vascular endothelial growth factor-D block its interactions with both VEGF receptor-2 and VEGF receptor-3. Eur. J. Biochem. 2000, 267, 2505–2515. [Google Scholar] [CrossRef]
- Vogel, M.; Miescher, S.; Kuhn, S.; Zürcher, A.W.; Stadler, M.B.; Ruf, C.; Effenberger, F.; Kricek, F.; Stadler, B.M. Mimicry of human IgE epitopes by anti-idiotypic antibodies. J. Mol. Biol. 2000, 298, 729–735. [Google Scholar] [CrossRef]
- Namgung, S.; Mohr, D.A.; Yoo, D.; Bharadwaj, P.; Koester, S.J.; Oh, S.H. Ultrasmall Plasmonic Single Nanoparticle Light Source Driven by a Graphene Tunnel Junction. ACS Nano 2018, 12, 2780–2788. [Google Scholar] [CrossRef]
- Lee, Y.; Kamal, A.S.A.; Abasaki, M.; Ho, Y.L.; Takakura, Y.; Delaunay, J.J. Gap Plasmons Multiple Mirroring from Spheres in Pyramids for Surface-Enhanced Raman Scattering. ACS Photonics 2016, 3, 2405–2412. [Google Scholar] [CrossRef]
- Chen, H.; Shao, L.; Li, Q.; Wang, J. Gold nanorods and their plasmonic properties. Chem. Soc. Rev. 2013, 42, 2679–2724. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.Y.; Yi, J.; Li, J.F.; Ren, B.; Wu, D.Y.; Panneerselvam, R.; Tian, Z.Q. Nanostructure-based plasmon-enhanced Raman spectroscopy for surface analysis of materials. Nat. Rev. Mater. 2016, 1, 16021. [Google Scholar] [CrossRef]
- Kennedy, B.J.; Spaeth, S.; Dickey, M.; Carron, K.T. Substrates Based on Self-Assembled Monolayers Formed Using Alkanethiols. J. Phys. Chem. B 1999, 103, 3640–3646. [Google Scholar] [CrossRef]
- Masango, S.S.; Hackler, R.A.; Large, N.; Henry, A.I.; McAnally, M.O.; Schatz, G.C.; Stair, P.C.; Van Duyne, R.P. High-Resolution Distance Dependence Study of Surface-Enhanced Raman Scattering Enabled by Atomic Layer Deposition. Nano Lett. 2016, 16, 4251–4259. [Google Scholar] [CrossRef]
- Devaraj, V.; Lee, J.M.; Kim, Y.J.; Jeong, H.; Oh, J.W. Engineering efficient self-assembled plasmonic nanostructures by configuring metallic nanoparticle’s morphology. Int. J. Mol. Sci. 2021, 22, 10595. [Google Scholar] [CrossRef]
- Zong, C.; Xu, M.; Xu, L.J.; Wei, T.; Ma, X.; Zheng, X.S.; Hu, R.; Ren, B. Surface-Enhanced Raman Spectroscopy for Bioanalysis: Reliability and Challenges. Chem. Rev. 2018, 118, 4946–4980. [Google Scholar] [CrossRef]
- Devaraj, V.; Lee, J.M.; Oh, J.W. Distinguishable plasmonic nanoparticle and gap mode properties in a silver nanoparticle on a gold film system using three-dimensional FDTD simulations. Nanomaterials 2018, 8, 582. [Google Scholar] [CrossRef]
- Helmerhorst, E.; Chandler, D.J.; Nussio, M.; Mamotte, C.D. Real-time and label-free bio-sensing of molecular interactions by surface plasmon resonance: A laboratory medicine perspective. Clin. Biochem. Rev. 2012, 33, 161–173. [Google Scholar]
- Kotlarek, D.; Fossati, S.; Venugopalan, P.; Gisbert Quilis, N.; Slabý, J.; Homola, J.; Lequeux, M.; Amiard, F.; Lamy De La Chapelle, M.; Jonas, U.; et al. Actuated plasmonic nanohole arrays for sensing and optical spectroscopy applications. Nanoscale 2020, 12, 9756–9768. [Google Scholar] [CrossRef]
- Corso, A.J.; Zuccon, S.; Zuppella, P.; Pelizzo, M.G. Flexible SPR system able to switch between Kretschmann and SPRi. Opt. Sens. 2015, 9506, 261–265. [Google Scholar] [CrossRef]
- Shao, Y.; Li, Y.; Gu, D.; Zhang, K.; Qu, J.; He, J.; Li, X.; Wu, S.-Y.; Ho, H.-P.; Somekh, M.G.; et al. Wavelength-multiplexing phase-sensitive surface plasmon imaging sensor. Opt. Lett. 2013, 38, 1370. [Google Scholar] [CrossRef]
- Bottazzi, B.; Fornasari, L.; Frangolho, A.; Giudicatti, S.; Mantovani, A.; Marabelli, F.; Marchesini, G.; Pellacani, P.; Therisod, R.; Valsesia, A. Multiplexed label-free optical biosensor for medical diagnostics. J. Biomed. Opt. 2014, 19, 017006. [Google Scholar] [CrossRef]
- Guner, H.; Ozgur, E.; Kokturk, G.; Celik, M.; Esen, E.; Topal, A.E.; Ayas, S.; Uludag, Y.; Elbuken, C.; Dana, A. A smartphone based surface plasmon resonance imaging (SPRi) platform for on-site biodetection. Sens. Actuators B Chem. 2017, 239, 571–577. [Google Scholar] [CrossRef]
- Cappi, G.; Spiga, F.M.; Moncada, Y.; Ferretti, A.; Beyeler, M.; Bianchessi, M.; Decosterd, L.; Buclin, T.; Guiducci, C. Label-Free detection of tobramycin in serum by transmission-localized surface plasmon resonance. Anal. Chem. 2015, 87, 5278–5285. [Google Scholar] [CrossRef]
- Zhang, J.; Dai, S.; Ma, C.; Di, J.; Zhao, J. Compact surface plasmon holographic microscopy for near-field film mapping. Opt. Lett. 2017, 42, 3462. [Google Scholar] [CrossRef]
- Zhang, Y.; Lai, J.; Yin, C.; Li, Z. Determination of effective complex refractive index of a turbid liquid with surface plasmon resonance phase detection. Appl. Opt. 2009, 48, 1262–1267. [Google Scholar] [CrossRef]
- Karlsson, R. SPR for molecular interaction analysis: A review of emerging application areas. J. Mol. Recognit. 2004, 17, 151–161. [Google Scholar] [CrossRef]
- Lu, J.; Spasic, D.; Delport, F.; Van Stappen, T.; Detrez, I.; Daems, D.; Vermeire, S.; Gils, A.; Lammertyn, J. Immunoassay for Detection of Infliximab in Whole Blood Using a Fiber-Optic Surface Plasmon Resonance Biosensor. Anal. Chem. 2017, 89, 3664–3671. [Google Scholar] [CrossRef]
- Vashist, S.K.; Vashist, S.K.; Schneider, E.M.; Luong, J.H.T. Surface plasmon resonance-based immunoassay for human fetuin A. Analyst 2014, 139, 2237–2242. [Google Scholar] [CrossRef]
- Vashist, S.K.; Schneider, E.M.; Luong, J.H.T. Surface plasmon resonance-based immunoassay for human C-reactive protein. Analyst 2015, 140, 4445–4452. [Google Scholar] [CrossRef] [PubMed]
- Piliarik, M.; Bocková, M.; Homola, J. Surface plasmon resonance biosensor for parallelized detection of protein biomarkers in diluted blood plasma. Biosens. Bioelectron. 2010, 26, 1656–1661. [Google Scholar] [CrossRef] [PubMed]
- Vance, S.A.; Sandros, M.G. Zeptomole detection of C-reactive protein in serum by a nanoparticle amplified surface plasmon resonance imaging aptasensor. Sci. Rep. 2014, 4, 5129. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Gupta, A.; Vaishnavi, T.V.; Walia, S.; Bhatia, D.; Chakraborty, B. Aptamers functionalized biomolecular nano-vehicles for applications in cancer diagnostics & therapeutics. Appl. NanoMedicine 2022, 22, 360. [Google Scholar]
- Hassan, E.M.; DeRosa, M.C. Recent advances in cancer early detection and diagnosis: Role of nucleic acid based aptasensors. TrAC-Trends Anal. Chem. 2020, 124, 115806. [Google Scholar] [CrossRef]
- Bellassai, N.; D’Agata, R.; Jungbluth, V.; Spoto, G. Surface Plasmon Resonance for Biomarker Detection: Advances in Non-invasive Cancer Diagnosis. Front. Chem. 2019, 7, 570. [Google Scholar] [CrossRef]
- Kumar, V.; Kukkar, D.; Hashemi, B.; Kim, K.H.; Deep, A. Advanced Functional Structure-Based Sensing and Imaging Strategies for Cancer Detection: Possibilities, Opportunities, Challenges, and Prospects. Adv. Funct. Mater. 2019, 29, 1807859. [Google Scholar] [CrossRef]
- Špringer, T.; Homola, J. Biofunctionalized gold nanoparticles for SPR-biosensor-based detection of CEA in blood plasma. Anal. Bioanal. Chem. 2012, 404, 2869–2875. [Google Scholar] [CrossRef]
- Špringer, T.; Chadtová Song, X.; Ermini, M.L.; Lamačová, J.; Homola, J. Functional gold nanoparticles for optical affinity biosensing. Anal. Bioanal. Chem. 2017, 409, 4087–4097. [Google Scholar] [CrossRef]
- Suwansa-ard, S.; Kanatharana, P.; Asawatreratanakul, P.; Wongkittisuksa, B.; Limsakul, C.; Thavarungkul, P. Comparison of surface plasmon resonance and capacitive immunosensors for cancer antigen 125 detection in human serum samples. Biosens. Bioelectron. 2009, 24, 3436–3441. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.; Wang, J.; Fu, W.; Yao, C. A SPR biosensor based on signal amplification using antibody-QD conjugates for quantitative determination of multiple tumor markers OPEN. Nat. Publ. Gr. 2016, 6, 33140. [Google Scholar] [CrossRef]
- Yi, X.; Ma, J.; Guan, Y.; Chen, R.; Yang, L.; Xia, X. The feasibility of using mutation detection in ctDNA to assess tumor dynamics. Int. J. Cancer 2017, 140, 2642–2647. [Google Scholar] [CrossRef]
- Chen, H.; Hou, Y.; Ye, Z.; Wang, H.; Koh, K.; Shen, Z.; Shu, Y. Label-free surface plasmon resonance cytosensor for breast cancer cell detection based on nano-conjugation of monodisperse magnetic nanoparticle and folic acid. Sens. Actuators B Chem. 2014, 201, 433–438. [Google Scholar] [CrossRef]
- Zhu, S.; Li, H.; Yang, M.; Pang, S.W. Label-free detection of live cancer cells and DNA hybridization using 3D multilayered plasmonic biosensor. Nanotechnology 2018, 29, 365503. [Google Scholar] [CrossRef]
- Wang, S.-S.; Zhao, X.-P.; Liu, F.-F.; Younis, M.R.; Xia, X.-H.; Wang, C. Direct Plasmon-Enhanced Electrochemistry for Enabling Ultrasensitive and Label-Free Detection of Circulating Tumor Cells in Blood. Anal. Chem. 2019, 91, 4413–4420. [Google Scholar] [CrossRef]
- Huang, X.; Hu, X.; Song, S.; Mao, D.; Lee, J.; Koh, K.; Zhu, Z.; Chen, H. Triple-enhanced surface plasmon resonance spectroscopy based on cell membrane and folic acid functionalized gold nanoparticles for dual-selective circulating tumor cell sensing. Sens. Actuators B Chem. 2020, 305, 127543. [Google Scholar] [CrossRef]
- Tadimety, A.; Zhang, Y.; Kready, K.M.; Palinski, T.J.; Tsongalis, G.J.; Zhang, J.X. Design of peptide nucleic acid probes on plasmonic gold nanorods for detection of circulating tumor DNA point mutations. Biosens. Bioelectron. 2019, 130, 236–244. [Google Scholar] [CrossRef]
- Adams, B.D.; Anastasiadou, E.; Esteller, M.; He, L.; Slack, F.J. The Inescapable Influence of Noncoding RNAs in Cancer. Cancer Res 2015, 75, 5206–5210. [Google Scholar] [CrossRef]
- Van der Ven, C.F.T.; Hogewoning, B.C.R.; van Mil, A.; Sluijter, J.P.G. Non-coding RNAs in Cardiac Regeneration. Adv. Exp. Med. Biol. 2020, 1229, 163–180. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, J.; Zhang, J.; Liu, Y.; Wu, L.; Shen, J.; Zhang, Y.; Hu, Y.; Fan, Q.; Huang, W.; et al. Individual Au-Nanocube Based Plasmonic Nanoprobe for Cancer Relevant MicroRNA Biomarker Detection. ACS Sensors 2017, 2, 1435–1440. [Google Scholar] [CrossRef]
- Ki, J.S.; Lee, H.Y.; Son, H.Y.; Huh, Y.-M.; Haam, S. Sensitive Plasmonic Detection of miR-10b in Biological Samples Using Enzyme-Assisted Target Recycling and Developed LSPR Probe. ACS Appl. Mater. Interfaces 2019, 11, 18923–18929. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, Q.; Yang, X.; Wang, K.; Zhang, H.; Nie, W. High sensitivity surface plasmon resonance biosensor for detection of microRNA and small molecule based on graphene oxide-gold nanoparticles composites. Talanta 2017, 174, 521–526. [Google Scholar] [CrossRef]
- Xue, T.; Liang, W.; Li, Y.; Sun, Y.; Xiang, Y.; Zhang, Y.; Dai, Z.; Duo, Y.; Wu, L.; Qi, K.; et al. Ultrasensitive detection of miRNA with an antimonene-based surface plasmon resonance sensor. Nat. Commun. 2019, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Mujica, M.L.; Zhang, Y.; Bédioui, F.; Gutiérrez, F.; Rivas, G. Label-free graphene oxide–based SPR genosensor for the quantification of microRNA21. Anal. Bioanal. Chem. 2020, 412, 3539–3546. [Google Scholar] [CrossRef] [PubMed]
- Fathi, F.; Rahbarghazi, R.; Movassaghpour, A.A.; Rashidi, M.-R. Detection of CD133-marked cancer stem cells by surface plasmon resonance: Its application in leukemia patients. Biochim. Biophys. Acta-Gen. Subj. 2019, 1863, 1575–1582. [Google Scholar] [CrossRef]
- Yavas, O.; Aćimović, S.S.; Guirado, J.G.; Berthelot, J.; Dobosz, P.; Sanz, V.; Quidant, R. Self-Calibrating On-Chip Localized Surface Plasmon Resonance Sensing for Quantitative and Multiplexed Detection of Cancer Markers in Human Serum. ACS Sens. 2018, 3, 1376–1384. [Google Scholar] [CrossRef]
- Szymańska, B.; Lukaszewski, Z.; Hermanowicz-Szamatowicz, K.; Gorodkiewicz, E. A biosensor for determination of the circulating biomarker CA125/MUC16 by Surface Plasmon Resonance Imaging. Talanta 2020, 206, 120187. [Google Scholar] [CrossRef]
- Chiu, N.-F.; Lin, T.-L.; Kuo, C.-T. Highly sensitive carboxyl-graphene oxide-based surface plasmon resonance immunosensor for the detection of lung cancer for cytokeratin 19 biomarker in human plasma. Sens. Actuators B Chem. 2018, 265, 264–272. [Google Scholar] [CrossRef]
- Karami, P.; Khoshsafar, H.; Johari-Ahar, M.; Arduini, F.; Afkhami, A.; Bagheri, H. Colorimetric immunosensor for determination of prostate specific antigen using surface plasmon resonance band of colloidal triangular shape gold nanoparticles. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 222, 117218. [Google Scholar] [CrossRef]
- Sankiewicz, A.; Romanowicz, L.; Laudanski, P.; Zelazowska-Rutkowska, B.; Puzan, B.; Cylwik, B.; Gorodkiewicz, E. SPR imaging biosensor for determination of laminin-5 as a potential cancer marker in biological material. Anal. Bioanal. Chem. 2016, 408, 5269–5276. [Google Scholar] [CrossRef]
- Wu, W.; Wu, Q.; Ren, S.-N.; Liu, Z.; Chen, F.-F. Ti3C2-MXene-assisted signal amplification for sensitive and selective surface plasmon resonance biosensing of biomarker. Chin. J. Anal. Chem. 2021, 50, 13–18. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef]
- El Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J.A. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- Azmi, A.S.; Bao, B.; Sarkar, F.H. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Rev. 2013, 32, 623–642. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, J.-Q.; Liu, J.-L.; Tian, L. Exosomes in tumor microenvironment: Novel transporters and biomarkers. J. Transl. Med. 2016, 14, 297. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, X.; Shi, H.; Wu, L.; Qian, H.; Xu, W. Exosomes in cancer: Small particle, big player. J. Hematol. Oncol. 2015, 8, 83. [Google Scholar] [CrossRef]
- Wang, Q.; Zou, L.; Yang, X.; Liu, X.; Nie, W.; Zheng, Y.; Cheng, Q.; Wang, K. Direct quantification of cancerous exosomes via surface plasmon resonance with dual gold nanoparticle-assisted signal amplification. Biosens. Bioelectron. 2019, 135, 129–136. [Google Scholar] [CrossRef]
- Mao, Z.; Zhao, J.; Chen, J.; Hu, X.; Koh, K.; Chen, H. A simple and direct SPR platform combining three-in-one multifunctional peptides for ultra-sensitive detection of PD-L1 exosomes. Sens. Actuators B Chem. 2021, 346, 130496. [Google Scholar] [CrossRef]
- Thakur, A.; Qiu, G.; Ng, S.-P.; Guan, J.; Yue, J.; Lee, Y.; Wu, C.-M.L. Direct detection of two different tumor-derived extracellular vesicles by SAM-AuNIs LSPR biosensor. Biosens. Bioelectron. 2017, 94, 400–407. [Google Scholar] [CrossRef]
- Ibn Sina, A.A.; Vaidyanathan, R.; Wuethrich, A.; Carrascosa, L.G.; Trau, M. Label-free detection of exosomes using a surface plasmon resonance biosensor. Anal. Bioanal. Chem. 2019, 411, 1311–1318. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ji, C.; Lü, B.; Rodin, M.; Paradies, J.H.H.; Yin, M.; Kuckling, D. Dually Crosslinked Supramolecular Hydrogel for Cancer Biomarker Sensing. ACS Appl. Mater. Interfaces 2020, 12, 36873–36881. [Google Scholar] [CrossRef] [PubMed]
- Thurmond, R.; Wadsworth, S.A.; Schafer, P.H.; Zivin, R.A.; Siekierka, J.J. Kinetics of small molecule inhibitor binding to p38 kinase. JBIC J. Biol. Inorg. Chem. 2001, 268, 5747–5754. [Google Scholar] [CrossRef] [PubMed]
- Adamczyk, M.; Moore, J.A.; Yu, Z. Application of Surface Plasmon Resonance toward Studies of Low-Molecular-Weight Antigen–Antibody Binding Interactions. Methods 2000, 20, 319–328. [Google Scholar] [CrossRef]
- Loo, J.F.-C.; Yang, C.; Tsang, H.L.; Lau, P.M.; Yong, K.-T.; Ho, H.P.; Kong, S.K. An Aptamer Bio-barCode (ABC) assay using SPR, RNase H, and probes with RNA and gold-nanorods for anti-cancer drug screening. Analyst 2017, 142, 3579–3587. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Lu, F.; Liu, L.; Ji, Q.; Song, K.; Yin, Q.; Lerner, R.A.; Yang, G.; Xu, H.; et al. A DNA-encoded library for the identification of natural product binders that modulate poly (ADP-ribose) polymerase 1, a validated anti-cancer target. Biochem. Biophys. Res. Commun. 2020, 533, 241–248. [Google Scholar] [CrossRef]
- Huang, R.; Jing, X.; Huang, X.; Pan, Y.; Fang, Y.; Liang, G.; Liao, Z.-X.; Wang, H.-S.; Chen, Z.-F.; Zhang, Y. Bifunctional Naphthoquinone Aromatic Amide-Oxime Derivatives Exert Combined Immunotherapeutic and Antitumor Effects through Simultaneous Targeting of Indoleamine-2,3-dioxygenase and Signal Transducer and Activator of Transcription 3. J. Med. Chem. 2020, 63, 1544–1563. [Google Scholar] [CrossRef]
- Wen, M.; Deng, Z.-K.; Jiang, S.-L.; Guan, Y.-D.; Wu, H.-Z.; Wang, X.-L.; Xiao, S.-S.; Zhang, Y.; Yang, J.-M.; Cao, D.-S.; et al. Identification of a Novel Bcl-2 Inhibitor by Ligand-Based Screening and Investigation of Its Anti-cancer Effect on Human Breast Cancer Cells. Front. Pharmacol. 2019, 10, 391. [Google Scholar] [CrossRef]
- Gassner, C.; Lipsmeier, F.; Metzger, P.; Beck, H.; Schnueriger, A.; Regula, J.T.; Moelleken, J. Development and validation of a novel SPR-based assay principle for bispecific molecules. J. Pharm. Biomed. Anal. 2014, 102, 144–149. [Google Scholar] [CrossRef]
- Ritter, G.; Cohen, L.S.; Williams, C.J.; Richards, E.C.; Old, L.J.; Welt, S. Serological Analysis of Human Anti-Human Antibody Responses in Colon Cancer Patients Treated with Repeated Doses of Humanized Monoclonal Antibody A33. Cancer Res. 2001, 61, 6851–6859. [Google Scholar]
- Rogozińska-Szczepka, J.; Utracka-Hutka, B.; Grzybowska, E.; Mąka, B.; Nowicka, E.; Smok-Ragankiewicz, A.; Zientek, H.; Steffen, J.; Wojciechowska-Łącka, A. BRCA1 and BRCA2 mutations as prognostic factors in bilateral breast cancer patients. Ann. Oncol. 2004, 15, 1373–1376. [Google Scholar] [CrossRef]
- Iovine, R.; Loscrí, V.; Pizzi, S.; Tarparelli, R.; Vegni, A.M. Model of Multi-Source Nanonetworks for the Detection of BRCA1 DNA Alterations Based on LSPR Phenomenon. Adv. Nanoparticles 2013, 2, 301–312. [Google Scholar] [CrossRef][Green Version]
- Meenakshi, A.; Kumar, R.S.; Kumar, N.S. Elisa for Quantitation of Serum C-erbB-2 Oncoprotein in Breast Cancer Patients. J. Immunoass. Immunochem. 2002, 23, 293–305. [Google Scholar] [CrossRef]
- Eletxigerra, U.; Martinez-Perdiguero, J.; Barderas, R.; Pingarrón, J.M.; Campuzano, S.; Merino, S. Surface plasmon resonance immunosensor for ErbB2 breast cancer biomarker determination in human serum and raw cancer cell lysates. Anal. Chim. Acta 2016, 905, 156–162. [Google Scholar] [CrossRef]
- Shabani, A.; Tabrizian, M. Design of a universal biointerface for sensitive, selective, and multiplex detection of biomarkers using surface plasmon resonance imaging. Analyst 2013, 138, 6052–6062. [Google Scholar] [CrossRef]
- Li, R.; Feng, F.; Chen, Z.-Z.; Bai, Y.-F.; Guo, F.-F.; Wu, F.-Y.; Zhou, G. Sensitive detection of carcinoembryonic antigen using surface plasmon resonance biosensor with gold nanoparticles signal amplification. Talanta 2015, 140, 143–149. [Google Scholar] [CrossRef]
- Lee, J.U.; Nguyen, A.H.; Sim, S.J. A nanoplasmonic biosensor for label-free multiplex detection of cancer biomarkers. Biosens. Bioelectron. 2015, 74, 341–346. [Google Scholar] [CrossRef]
- Su, F.; Xu, C.; Taya, M.; Murayama, K.; Shinohara, Y.; Nishimura, S.-I. Detection of Carcinoembryonic Antigens Using a Surface Plasmon Resonance Biosensor. Sensors 2008, 8, 4282–4295. [Google Scholar] [CrossRef]
- Altintas, Z.; Uludag, Y.; Gurbuz, Y.; Tothill, I.E. Surface plasmon resonance based immunosensor for the detection of the cancer biomarker carcinoembryonic antigen. Talanta 2011, 86, 377–383. [Google Scholar] [CrossRef]
- Duan, F.; Zhang, S.; Yang, L.; Zhang, Z.; He, L.; Wang, M. Bifunctional aptasensor based on novel two-dimensional nanocomposite of MoS2 quantum dots and g-C3N4 nanosheets decorated with chitosan-stabilized Au nanoparticles for selectively detecting prostate specific antigen. Anal. Chim. Acta 2018, 1036, 121–132. [Google Scholar] [CrossRef]
- Malic, L.; Sandros, M.G.; Tabrizian, M. Designed Biointerface Using Near-Infrared Quantum Dots for Ultrasensitive Surface Plasmon Resonance Imaging Biosensors. Anal. Chem. 2011, 83, 5222–5229. [Google Scholar] [CrossRef] [PubMed]
- Law, W.-C.; Yong, K.-T.; Baev, A.; Prasad, P.N. Sensitivity Improved Surface Plasmon Resonance Biosensor for Cancer Biomarker Detection Based on Plasmonic Enhancement. ACS Nano 2011, 5, 4858–4864. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Na, K.; Lee, J.; Kim, K.-W.; Hyun, J. Enhanced surface plasmon resonance by Au nanoparticles immobilized on a dielectric SiO2 layer on a gold surface. Anal. Chim. Acta 2009, 651, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Reekmans, G.; Saerens, D.; Friedt, J.-M.; Frederix, F.; Francis, L.; Muyldermans, S.; Campitelli, A.; Van Hoof, C. Prostate-specific antigen immunosensing based on mixed self-assembled monolayers, camel antibodies and colloidal gold enhanced sandwich assays. Biosens. Bioelectron. 2005, 21, 483–490. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.-H.; Li, S.-M.; Cho, Y.-H. Enhanced detection sensitivity of pegylated CdSe/ZnS quantum dots-based prostate cancer biomarkers by surface plasmon-coupled emission. Biosens. Bioelectron. 2012, 33, 284–287. [Google Scholar] [CrossRef]
- Acimovic, S.S.; Ortega, M.; Sanz, V.; Berthelot, J.; Garcia-Cordero, J.L.; Renger, J.; Maerkl, S.J.; Kreuzer, M.P.; Quidant, R. LSPR Chip for Parallel, Rapid, and Sensitive Detection of Cancer Markers in Serum. Nano Lett. 2014, 14, 2636–2641. [Google Scholar] [CrossRef]
- Vaisocherová, H.; Šípová, H.; Víšová, I.; Bocková, M.; Špringer, T.; Ermini, M.L.; Song, X.; Krejčík, Z.; Chrastinová, L.; Pastva, O.; et al. Rapid and sensitive detection of multiple microRNAs in cell lysate by low-fouling surface plasmon resonance biosensor. Biosens. Bioelectron. 2015, 70, 226–231. [Google Scholar] [CrossRef]
- Chiu, N.-F.; Kuo, C.-T.; Lin, T.-L.; Chang, C.-C.; Chen, C.-Y. Ultra-high sensitivity of the non-immunological affinity of graphene oxide-peptide-based surface plasmon resonance biosensors to detect human chorionic gonadotropin. Biosens. Bioelectron. 2017, 94, 351–357. [Google Scholar] [CrossRef]
- Chung, J.; Bernhardt, R.; Pyun, J. Sequential analysis of multiple analytes using a surface plasmon resonance (SPR) biosensor. J. Immunol. Methods 2006, 311, 178–188. [Google Scholar] [CrossRef]
- Piliarik, M.; Vaisocherová, H.; Homola, J. A new surface plasmon resonance sensor for high-throughput screening applications. Biosens. Bioelectron. 2005, 20, 2104–2110. [Google Scholar] [CrossRef]
- Boozer, C.; Ladd, J.; Chen, S.; Jiang, S. DNA-Directed Protein Immobilization for Simultaneous Detection of Multiple Analytes by Surface Plasmon Resonance Biosensor. Anal. Chem. 2006, 78, 1515–1519. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Bernhardt, R.; Pyun, J. Additive assay of cancer marker CA 19-9 by SPR biosensor. Sens. Actuators B Chem. 2006, 118, 28–32. [Google Scholar] [CrossRef]
- Chang, C.-C.; Chiu, N.-F.; Lin, D.S.; Chu-Su, Y.; Liang, Y.-H.; Lin, C.-W. High-Sensitivity Detection of Carbohydrate Antigen 15-3 Using a Gold/Zinc Oxide Thin Film Surface Plasmon Resonance-Based Biosensor. Anal. Chem. 2010, 82, 1207–1212. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.-H.; Chang, C.-C.; Chen, C.-C.; Chu-Su, Y.; Lin, C.-W. Development of an Au/ZnO thin film surface plasmon resonance-based biosensor immunoassay for the detection of carbohydrate antigen 15-3 in human saliva. Clin. Biochem. 2012, 45, 1689–1693. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-M.; Jeong, D.H.; Lee, H.-Y.; Park, J.-H.; Lee, S.-K. Design and validation of fiber optic localized surface plasmon resonance sensor for thyroglobulin immunoassay with high sensitivity and rapid detection. Sci. Rep. 2021, 11, 15985. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef]
- Xiong, H.; Huang, Z.; Lin, Q.; Yang, B.; Yan, F.; Liu, B.; Chen, H.; Kong, J. Surface Plasmon Coupling Electrochemiluminescence Immunosensor Based on Polymer Dots and AuNPs for Ultrasensitive Detection of Pancreatic Cancer Exosomes. Anal. Chem. 2022, 94, 837–846. [Google Scholar] [CrossRef]
- Raghu, D.; Christodoulides, J.A.; Christophersen, M.; Liu, J.L.; Anderson, G.P.; Robitaille, M.; Byers, J.M.; Raphael, M.P. Nanoplasmonic pillars engineered for single exosome detection. PLoS ONE 2018, 13, e0202773. [Google Scholar] [CrossRef]
- Ahmed, N.; Barker, G.; Oliva, K.T.; Hoffmann, P.; Riley, C.; Reeve, S.; Smith, I.; E Kemp, B.; A Quinn, M.; E Rice, G. Proteomic-based identification of haptoglobin-1 precursor as a novel circulating biomarker of ovarian cancer. Br. J. Cancer 2004, 91, 129–140. [Google Scholar] [CrossRef]
- Bharti, A.; Ma, P.C.; Maulik, G.; Singh, R.; Khan, E.; Skarin, A.T.; Salgia, R. Haptoglobin α-Subunit and Hepatocyte Growth Factor Can Potentially Serve as Serum Tumor Biomarkers in Small Cell Lung Cancer. Anticancer Res. 2004, 24, 1031–1038. [Google Scholar]
- Xing, P.X.; Young, G.; Ho, D.; Sinatra, M.A.; Hoj, P.B.; McKenzie, I.F.C. A new approach to fecal occult blood testing based on the detection of haptoglobin. Cancer 1996, 78, 48–56. [Google Scholar] [CrossRef]
- Bresalier, R.S.; Byrd, J.C.; Tessler, D.; Lebel, J.; Koomen, J.; Hawke, D.; Half, E.; Liu, K.-F.; Mazurek, N. A circulating ligand for galectin-3 is a haptoglobin-related glycoprotein elevated in individuals with colon cancer. Gastroenterology 2004, 127, 741–748. [Google Scholar] [CrossRef]
- Kazuno, S.; Fujimura, T.; Arai, T.; Ueno, T.; Nagao, K.; Fujime, M.; Murayama, K. Multi-sequential surface plasmon resonance analysis of haptoglobin–lectin complex in sera of patients with malignant and benign prostate diseases. Anal. Biochem. 2011, 419, 241–249. [Google Scholar] [CrossRef]
- Lleonart, M.E.; Cajal, S.R.Y.; Groopman, J.D.; Friesen, M.D. Sensitive and specific detection of K-ras mutations in colon tumors by short oligonucleotide mass analysis. Nucleic Acids Res. 2004, 32, e53. [Google Scholar] [CrossRef][Green Version]
- D’Agata, R.; Bellassai, N.; Allegretti, M.; Rozzi, A.; Korom, S.; Manicardi, A.; Melucci, E.; Pescarmona, E.; Corradini, R.; Giacomini, P.; et al. Direct plasmonic detection of circulating RAS mutated DNA in colorectal cancer patients. Biosens. Bioelectron. 2020, 170, 112648. [Google Scholar] [CrossRef]
- Sutphen, R.; Xu, Y.; Wilbanks, G.D.; Fiorica, J.; Grendys, J.E.C.; LaPolla, J.P.; Arango, H.; Hoffman, M.S.; Martino, M.; Wakeley, K.; et al. Lysophospholipids Are Potential Biomarkers of Ovarian Cancer. Cancer Epidemiol. Biomark. Prev. 2004, 13, 1185–1191. [Google Scholar] [CrossRef]
- Yuan, J.; Duan, R.; Yang, H.; Luo, X.; Xi, M. Detection of serum human epididymis secretory protein 4 in patients with ovarian cancer using a label-free biosensor based on localized surface plasmon resonance. Int. J. Nanomed. 2012, 7, 2921–2928. [Google Scholar] [CrossRef]
- Wu, A.; Wu, B.; Guo, J.; Luo, W.; Wu, D.; Yang, H.; Zhen, Y.; Yu, X.; Wang, H.; Zhou, Y.; et al. Elevated expression of CDK4 in lung cancer. J. Transl. Med. 2011, 9, 38. [Google Scholar] [CrossRef]
- Sankiewicz, A.; Lukaszewski, Z.; Trojanowska, K.; Gorodkiewicz, E. Determination of collagen type IV by Surface Plasmon Resonance Imaging using a specific biosensor. Anal. Biochem. 2016, 515, 40–46. [Google Scholar] [CrossRef]
- Zhou, Y.; Liao, Q.; Han, Y.; Chen, J.; Liu, Z.; Ling, H.; Zhang, J.; Yang, W.; Oyang, L.; Xia, L.; et al. Rac1 overexpression is correlated with epithelial mesenchymal transition and predicts poor prognosis in non-small cell lung cancer. J. Cancer 2016, 7, 2100–2109. [Google Scholar] [CrossRef]
- Sahu, V.; Gupta, A.; Kumar, R.; Gupta, T.; Mohan, A.; Dey, S. Quantification of Rac1 and Rac1b in serum of non small cell lung cancer by label free real time assay. Clin. Chim. Acta 2016, 460, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Cennamo, N.; Pesavento, M.; Lunelli, L.; Vanzetti, L.; Pederzolli, C.; Zeni, L.; Pasquardini, L. An easy way to realize SPR aptasensor: A multimode plastic optical fiber platform for cancer biomarkers detection. Talanta 2015, 140, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Yeh, E.-C.; Sinha, R.; Laurence, T.A.; Bearinger, J.P.; Lee, L.P. Single-Step Nanoplasmonic VEGF165Aptasensor for Early Cancer Diagnosis. ACS Nano 2012, 6, 7607–7614. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-Y.; Brooks, E.; Li, Y.; Denny, P.; Ho, C.-M.; Qi, F.; Shi, W.; Wolinsky, L.; Wu, B.; Wong, D.T.W.; et al. Detection of picomolar levels of interleukin-8 in human saliva by SPR. Lab Chip 2005, 5, 1017–1023. [Google Scholar] [CrossRef]
- Lin, Y.-T.; Darvishi, S.; Preet, A.; Huang, T.-Y.; Lin, S.-H.; Girault, H.H.; Wang, L.; Lin, T.-E. A Review: Electrochemical Biosensors for Oral Cancer. Chemosensors 2020, 8, 54. [Google Scholar] [CrossRef]
- Vergara, D.; Bianco, M.; Pagano, R.; Priore, P.; Lunetti, P.; Guerra, F.; Bettini, S.; Carallo, S.; Zizzari, A.; Pitotti, E.; et al. An SPR based immunoassay for the sensitive detection of the soluble epithelial marker E-cadherin. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 1963–1971. [Google Scholar] [CrossRef]
- Zhou, W.; Ma, Y.; Yang, H.; Ding, Y.; Luo, X. A label-free biosensor based on silver nanoparticles array for clinical detection of serum p53 in head and neck squamous cell carcinoma. Int. J. Nanomed. 2011, 6, 381–386. [Google Scholar] [CrossRef]
- Myers, M.J.; Smith, E.R.; Turfle, P.G. Biomarkers in Veterinary Medicine. Annu. Rev. Anim. Biosci. 2017, 5, 65–87. [Google Scholar] [CrossRef]
- Loyez, M.; Albert, J.; Caucheteur, C.; Wattiez, R. Cytokeratins Biosensing Using Tilted Fiber Gratings. Biosensors 2018, 8, 74. [Google Scholar] [CrossRef]
- Aubé, A.; Charbonneau, D.M.; Pelletier, J.N.; Masson, J.-F. Response Monitoring of Acute Lymphoblastic Leukemia Patients Undergoing l-Asparaginase Therapy: Successes and Challenges Associated with Clinical Sample Analysis in Plasmonic Sensing. ACS Sens. 2016, 1, 1358–1365. [Google Scholar] [CrossRef]
- Krupin, O.; Wang, C.; Berini, P. Detection of leukemia markers using long-range surface plasmon waveguides functionalized with Protein G. Lab Chip 2015, 15, 4156–4165. [Google Scholar] [CrossRef]
- Zhao, Y.; Tong, L.; Li, Y.; Pan, H.; Zhang, W.; Guan, M.; Li, W.; Chen, Y.; Li, Q.; Li, Z.; et al. Lactose-Functionalized Gold Nanorods for Sensitive and Rapid Serological Diagnosis of Cancer. ACS Appl. Mater. Interfaces 2016, 8, 5813–5820. [Google Scholar] [CrossRef]
- Lee, K.-A.; Ahn, J.-Y.; Lee, S.-H.; Sekhon, S.S.; Kim, D.-G.; Min, J.; Kim, Y.-H. Aptamer-based Sandwich Assay and its Clinical Outlooks for Detecting Lipocalin-2 in Hepatocellular Carcinoma (HCC). Sci. Rep. 2015, 5, 10897. [Google Scholar] [CrossRef]
- Gorodkiewicz, E.; Charkiewicz, R.; Rakowska, A.; Bajko, P.; Chyczewski, L.; Niklinski, J. SPR imaging biosensor for podoplanin: Sensor development and application to biological materials. Microchim. Acta 2012, 176, 337–343. [Google Scholar] [CrossRef]
- Gill, K.; Mohanti, B.K.; Ashraf, S.; Singh, A.K.; Dey, S. Quantification of p38αMAP kinase: A prognostic marker in HNSCC with respect to radiation therapy. Clin. Chim. Acta 2012, 413, 219–225. [Google Scholar] [CrossRef]
- Gorodkiewicz, E.; Sieńczyk, M.; Regulska, E.; Grzywa, R.; Pietrusewicz, E.; Lesner, A.; Łukaszewski, Z. Surface plasmon resonance imaging biosensor for cathepsin G based on a potent inhibitor: Development and applications. Anal. Biochem. 2012, 423, 218–223. [Google Scholar] [CrossRef]
- Hsieh, H.-Y.; Chang, R.; Huang, Y.-Y.; Juan, P.-H.; Tahara, H.; Lee, K.-Y.; Vo, D.N.K.; Tsai, M.-H.; Wei, P.-K.; Sheen, H.-J.; et al. Continuous polymerase chain reaction microfluidics integrated with a gold-capped nanoslit sensing chip for Epstein-Barr virus detection. Biosens. Bioelectron. 2022, 195, 113672. [Google Scholar] [CrossRef]


| Biomarker | Biomarker Type | Sensitivity/Specificity and Predictive Value | Cancer Type | Source | Biological Concentration | Clinical Use | Conventional Technique | Sample No. (n) | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Alpha-fetoprotein (AFP) | Protein | Sensitivity: 65%/Specificity: 89% | Hepatocellular carcinomas | Serum | >400 ng/mL | Diagnostic and prognostic | Immunoassay | ----- | [30] |
| Bladder tumor antigen (BTA) | Protein | Sensitivity: 83%/Specificity: 92% | Bladder cancer | Urine | ----- | Monitoring | Immunoassay | 220 | [31] |
| BRCA-1 and BRCA-2 mutations | Genomic | Sensitivity: 80%/Specificity: 100% | Breast cancer | Blood | ---- | Prognosis | DNA sequencing | ----- | [32] |
| Cancer antigen 19-9 (CA 19-9) | Protein | Sensitivity: 81%/Specificity: 90% | Pancreatic cancer | Serum | s100 U/mL | Diagnostic and prognostic | ELISA | 1040 | [33] |
| Cancer antigen 15-3 (CA 15-3) | Protein | Sensitivity: 31%/Specificity: 86% | Breast cancer | Serum | 25 U/mL | Monitoring | Immunoassay | 1342 | [34] |
| Cancer antigen 125 (CA 125) | Glycoprotein | Sensitivity: 80%/Specificity: 99.6% | Ovarian cancer | Serum | 35 units/mL | Detection, diagnosis, and prognosis | Immunoassay | ----- | [35] |
| Carbohydrate antigen 27.29 (CA 27.29) | Protein | Sensitivity: 65%/Specificity: 100% | Breast cancer | Serum | >35 U/mL | Monitoring | Immunoassay | 213 | [36] |
| Carcinoembryonic antigen (CEA) | Protein | Sensitivity: 88.3%/Specificity: 46.2% | Lung cancer | Serum | 8.2 ng/mL | Detection, diagnosis, and prognosis | Immunoassay | ----- | [37] |
| CD133 | Protein (cancer stem cell marker) | ------ | Acute myeloid leukemia | Cells | ------- | Diagnostic, prognostic, and therapeutic | Flow cytometry | ------- | |
| Cluster of differentiation 9 (CD9) | Exosomal protein | ----- | Breast cancer | Cells | ----- | Diagnostic | ELISA | ----- | |
| Cluster of differentiation 147 (CD147) | Exosomal protein | ----- | Colorectal cancer | Serum | 103.59 pg/mL | Diagnostic | ELISA | 108 | |
| CD166 | Protein | Sensitivity: 58.6%/Specificity: 78.9% | Pancreatic cancer | Serum | 22 ng/mL | Prognosis | ELISA | 600 | [38] |
| Collagen IV | Protein | ----- | Breast cancer | Serum | 103 ng/mL | Diagnostic | ELISA | 41 | [39] |
| Cyclin-dependent kinase 4 (CDK4) | Protein | Sensitivity: 70%/Specificity: 69% | Lung, head, and neck cancers | Serum | >29.6 ng/µL | Diagnostic | Immunochemistry | ----- | [40] |
| Cytokeratin 19 | Protein | ---- | Non-small cell lung carcinoma | Serum | 8.92 ± 9.95 mU/mL | Prognosis | ELISA (0.5 ng/mL) | ----- | [41] |
| Cytokeratin 19 fragments (CYFRA 21-1) | Protein | ----- | Non-small cell lung cancer | Serum | ----- | Prognostic and predictive | ELISA | ----- | |
| Cytochrome P450 mutations | Genomic | ----- | Prostate and breast cancer | Blood | ----- | Risk and assessment and prognosis | DNA sequencing | ----- | |
| E-cadherin | Protein | Sensitivity: 74.3%/Specificity: 97.1% | Breast cancer | Serum | 2218.9 ± 319.6 ng/mL | Diagnostic | ELISA | 35 | [42] |
| Estrogen receptor (ER) | Protein | Sensitivity: 99.7%/Specificity: 95.4% | Breast cancer | Tissue | ----- | Prognosis and prediction | Immunohistochemistry | 1569 | [43] |
| Fibrin/fibrinogen degradation products (FDPs) | Protein | Sensitivity: 100%/Specificity: 80% | Bladder cancer | Serum | ----- | Monitoring | Immunoassay | 192 | [44] |
| Glutathione S-transferase (GSTP1) polymorphisms | Genomic | ----- | Breast and prostate cancer | Blood | ----- | Risk assessment, prognosis, and treatment | PCR restriction fragment-length polymorphism assay (PCR-RFLP assay) | ----- | |
| Glypican-1 (GPC-1) | Exosomal protein | Sensitivity: 76.92%/Specificity: 70.85% | Pancreatic cancer | Serum | 8.75 ng/mL | Diagnosis | Mass spectrometry | 595 | [45] |
| Haptoglobin | Protein | Sensitivity: 63.9%/Specificity: 88.1% | Lung cancer | Serum | 1.985 mg/mL | Diagnosis, therapy response | Immunoassay | 205 | [46] |
| HER2 | Protein | Sensitivity: 98.7%/Specificity: 99.3% | Breast cancer | Serum | 65.38 ± 37.92 ng/mL (serum) | Prognosis and treatment | FISH, PCR | 1210 | [47] |
| Human chorionic gonadotrophin (hCG) | Protein | ----- | Ovarian and testicular cancer | Serum | 1000–10,000 IU/L | Diagnostic | ELISA | ------ | [38,48] |
| Human epidermidis protein 4 (HE4) | Protein | Sensitivity: 76%/Specificity: 92% | Ovarian cancer | Serum | ----- | Diagnostic | Chemiluminescent immunoassay | 986 | [49] |
| Interleukin 8 (IL-8) | Protein | ---- | Multiple cancers | Serum | ----- | Diagnostic and prognostic | ELISA (1–3 pg mL−1) | ----- | |
| Laminin 5 | Protein | ----- | Bladder cancer | Serum | ----- | Diagnostic | ELISA | ----- | |
| Lysophosphatidic acid (LPA) | Lipid | ----- | Ovarian cancer | Serum | 8.6 µ mol/L | Detection, diagnosis, and prognosis | Mass spectrometry | 48 | [50] |
| MCF-7 cells | Cells | ------ | Breast cancer | Tumor sample | ------- | Diagnostic and prognostic | Immunocytometry | ------ | |
| Melanoma-associated antigen 3/6 (MAGE 3/6) | Protein | ----- | Ovarian cancer | Plasma | ----- | Prognostic and therapy monitoring | Western blots | ----- | |
| miR-16, miR-181, miR-34a, and miR-125b | RNA | ---- | Malignant tumors | Serum | ----- | Diagnosis | RT-qPCR | ----- | |
| miR-205 | RNA | Sensitivity: 78%/Specificity: 69% | Lung cancer | Serum | ---- | Diagnosis | RT-qPCR | ----- | [36,51] |
| Nuclear matrix protein 22 (NMP-22) | Protein | Sensitivity: 83%/Specificity: 71% | Bladder cancer | Urine | 10 U/mL | Screening and monitoring | Immunoassay | 2951 | [52] |
| Osteopontin | Genomic | ----- | Ovarian Cancer | Blood | ----- | Detection, diagnosis, and prognosis | Microarray | ----- | |
| p53 | Protein | Sensitivity: 81.1%/Specificity: 83.3% | Head and neck cancer | Serum | 401 pg/mL | Prognosis | ELISA | ------ | [53] |
| Progesterone receptor (PR) | Protein | Sensitivity: 94.8%/Specificity: 92.6%; | Breast cancer | Tissue | ----- | Prognosis and prediction | Immunohistochemistry | 1347 | [43] |
| Prostate-specific antigen (PSA) | Protein | Sensitivity: 82.1%/Specificity: 80.6% | Prostate | Serum | 2.6–4.0 ng/mL | Diagnostic and prognostic | Immunoassay | 136 | [54] |
| Ras-related C3 botulinum (Rac1) | Protein | ----- | Non-small cell lung cancer | Tumor Tissue | ----- | Prognostic | Immunohistochemistry | ----- | |
| Ras mutations | Genomic | ----- | Colon and lung cancer | Blood | ----- | Risk assessment | Short oligonucleotide mass analysis (SOMA) | ----- | |
| Thyroglobulin (Tg) | Protein | ------ | Papillary and follicular thyroid cancer | Serum | >10 ng/mL | Diagnostic and prognostic | ELISA | 72 | [41,55] |
| Transforming growth factor β (TGFβ) | Protein | Sensitivity: 86.7%/Specificity: 100% | Malignant tumors | Serum | 370 pg/mL | Diagnostic and prognostic | ELISA | 180 | [37,56] |
| Transforming growth factor beta 1 (TGFβ1) | Protein | ----- | Ovarian cancer | Plasma | 31.2–2000 pg/mL | Prognostic and therapy monitoring | Western blots | 28 | [57] |
| Vascular endothelial growth factor (VEGF) | Protein | Sensitivity: 50%/Specificity: >90% | Multiple cancers | Serum | 92–390 pg/mL | Prognosis | ELISA | ----- | [58] |
| v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (KIT) | Protein | ----- | Gastrointestinal | Tissue | ----- | Prediction, diagnosis, and selection of therapy | Immunohistochemistry | ----- |
| Biomarker | Type of Probe | Type of SPR | Clinical Sample | Linear Range | LOD | Reference |
|---|---|---|---|---|---|---|
| BRCA-1 and BRCA-2 Mutations | LSPR | Non-Validated | ---- | ---- | ---- | [151,152] |
| HER2 | SPR with Optical Fiber | Non-Validated | ---- | 10−12–10−6 g/mL | 9.3 × 10−9/mL | [66] |
| SPR Based on Extraordinary Optical Transmission (EOT) | Non-Validated | Human Serum | ---- | 3.0 ng/mL | [153] | |
| SPR Direct Method | Non-Validated | Human Serum | ---- | 3.8 ng/mL | [154] | |
| SPRi Direct Method | Non-Validated | Buffer | 1–200 ng/mL | 2.06 ng/mL | [155] | |
| Carcinoembryonic Antigen (CEA) | SPR Fluidic | Validated | Serum | 1–60 ng/mL | 1.0 ng/mL | [156] |
| LSPR | Non-Validated | Serum | 1–1 × 106 fM | 94 fM | [157] | |
| SPR with Antibody | Non-Validated | Serum | 25–800 ng/mL | 6.2 ng/mL | [158] | |
| SPR Sandwich Assay | Non-Validated | Buffer | 3–400 ng/mL | 3 ng/mL | [159] | |
| SPR with Antibody | Non-Validated | Buffer Spiked Human Serum | 25–800 ng/mL | 6.2 ng/mL | [158] | |
| SPR with Direct Detection | Non-Validated | Buffer | 8–1000 ng/mL | 8 ng/mL | [108] | |
| Prostate Specific Antigen (PSA) | LSPR | Non-Validated | Serum | ---- | 0.71 pg/mL | [160] |
| SPRi Signal Enhancement with Quantum Dots | Non-Validated | HBS Buffer | 1 ng/mL–100 pg/mL | 100 pg/mL | [161] | |
| LSPR Integrated with Microfluidics | Non-Validated | 50% Human Serum | 10–100 ng/mL | 1 ng/mL | [162] | |
| SPR Enhancement due to Resonant Coupling between Au Thin film and AuNP | Non-Validated | ---- | 0.1–100 ng/mL | 0.1 ng/mL | [163] | |
| SPR Direct Detection and Enhancement using Sandwich Assay | Non-Validated | PSA Spiked In Human Serum | 1 ng/mL–10 μg/mL | 10 ng/mL | [164] | |
| SPRi Signal Enhancement using Pegylated CdSe/ZnS Quantum Dots | Non-Validated | PBS | 100 μg/mL–10 fg/mL | 10 fg/mL | [165] | |
| Cancer Antigen 125 (CA 125) | Non-Fluidic SPRi | Validated | Serum | 2.2–150 units/mL | 0.66 units/mL | [127] |
| SPR Capacitive System | Non-Validated | Human Serum | 0.1–40 units/mL | 0.1 units/mL | [110] | |
| Alpha-Fetoprotein (AFP) | Fluidic SPR | Validated | Serum | ---- | 0.1 ng/mL | [111] |
| LSPR Integrated With Microfluidics | Non-Validated | 50 % Human Serum | 5–1000 ng/mL | 500 pg/mL | [166] | |
| LSPR | Non-Validated | Serum | 1 fM–1 × 106 fM | 91 fM | [157] | |
| SPR Signal Enhancement using Fe3O4@Au–antibody | Non-Validated | ---- | 1.0–200.0 ng/mL | 0.65 ng/mL | [66] | |
| Mir-205 | LSPR | Non-Validated | Serum | 10 pM–1 μM | 5 pM | [120] |
| mir-181, mir-125b, mir-34a, and mir-16 | SPRi | Non-Validated | Erythrocyte Lysate | 0.1–500 pM | 0.5 pM | [167] |
| Human Chorionic Gonadotrophin (hCG) | SPR | Non-Validated | Blood | 8.32–0.065 nM | 0.065 nM | [27,168] |
| SPR using Antibody–DNA Conjugated Array | Non-Validated | Buffer 10% Plasma | 10–100 ng/mL | 13 ng/mL | [102] | |
| SPR Signal Enhancement using Secondary Antibody | Non-Validated | Urine | 46–415 miu/mL (milli-international units per milliliter) | 46.4 miu/mL (milli-international units per milliliter) | [169] | |
| Combining SPRi with Polarization Contrast | Non-Validated | PBS | 0.5–10 μg/mL | 500 ng/mL | [170] | |
| SPR based on Single Strand/Oligo(Ethylene Glycol) Self-Assembled Monolayer | Non-Validated | ---- | 1 μg/mL | ---- | [171] | |
| Cancer Antigen 19-9 (CA 19-9) | Fluidic SPR | Non-Validated | ---- | 400–192,000 units/mL | 410.9 units/mL | [172] |
| Cancer Antigen 15-3 (CA 15-3) | SPR | Non-Validated | Pleural Fluid | ---- | 0.025 units/mL | [173] |
| SPR based on Au/ZnO Thin Film | Non-Validated | Saliva | 40–300 units/mL | ---- | [174] | |
| Cancer Antigen 125 (CA 125) | SPR and Capacitive System | Non-Validated | Human Serum | 0.1–40 units/mL | 0.1 units/mL | [110] |
| Thyroglobulin (Tg) | LSPR | Non-Validated | Serum | 0.001–100,000 pg/mL | 93.11 fg/mL | [175] |
| MCF-7 Cells | SPR | Non-Validated | Serum | 104–106 cells/mL | 500 cells/mL | [113] |
| Cd133 | SPR | Non-Validated | Blood | ---- | 1 × 105 cells/mL | [125] |
| Glypican-1 (GPC-1) | LSPR | Non-Validated | Serum | 103 to 106 particles/mL | 400 particles/mL | [176,177] |
| Cluster of Differentiation 9 (CD9) | LSPRi | Non-Validated | Cells | ---- | ---- | [178] |
| Activated Leukocyte Cell Adhesion Molecule (ALCAM) | SPR based on Antibody-DNA Conjugate Array | Non-Validated | 10% Blood Plasma | 10–1000 ng/mL | 45 ng/mL | [102] |
| SPRi based on Antibody–Oligo(Ethylene Glycol) Array | Non-Validated | 10% Human Serum | ---- | 6 ng/mL | [102] | |
| Haptoglobin | SPR | Non-Validated | Serum | ---- | ---- | [179,180,181,182,183] |
| Ras Mutations | SPRi | Non-Validated | Plasma | ---- | ---- | [184,185] |
| Lysophosphatidic Acid (LPA) | SPR | Non-Validated | Blood Plasma | 2 to 30 μM | 0.122 μM | [142,186] |
| Human Epidermidis Protein 4 (HE4) | Non-Fluidic LSPR | Validated | Serum | 10–10,000 pM | 4 pM | [187] |
| Laminin 5 | Non-Fluidic SPRi | Validated | Serum | 0.014–0.1 ng mL−1 | 4 pg mL−1 | [130] |
| Cyclin-Dependent Kinase 4 (CDK4) | Fluidic SPR | Validated | Serum | ---- | ---- | [188] |
| Collagen IV | Non-Fluidic SPRi | Validated | Serum | 10–300 ng/mM | 2.4 ng mL−1 | [189] |
| Ras-Related C3 Botulinum (Rac1) | Fluidic SPR | Validated | Serum | 1 to 5 mmol/L | ---- | [190,191] |
| CYFRA 21-1 | SPR Fluidic Chip | Validated | Serum | 10−1 to 103 ng/mL | 0.1 ng/mL | [111] |
| Vascular Endothelial Growth Factor (VEGF) | LSPR | Non-Validated | - | Nanomolar Range | Nanomolar Range | [192] |
| LSPR based on the Fluorophore-Conjugated Aptamer | Non-Validated | Diluted Serum and Saliva | 1.25 pM–1.25 μM | ---- | [193] | |
| Interleukin 8 (IL-8) | Fluidic SPR | Non-Validated | Saliva | 0–2 nM | 2.5 pM | [194,195] |
| Cytokeratin 19 | Fluidic SPR | Non-Validated | Serum | 1.6–128.3 ng/mL | 0.05 pg/mL | [128] |
| E-Cadherin | Non-Fluidic SPR | Non-Validated | Serum | 0–200 ng/mL | 16 ng/mL | [196] |
| P53 | LSPR | Non-Validated | Serum | ---- | ---- | [197] |
| Cd166 | Fluidic SPR | Non-Validated | Serum | ---- | <1 ng/mL | [198] |
| Cytokeratin | LSPR | Non-Validated | ---- | ---- | 14 pM | [199] |
| Antiasparaginase | SPR | Validated | Serum | ---- | 500 pM | [200] |
| Immunoglobulins Kappa and Lambda | SPR | Non-Validated | Serum | ---- | ---- | [201] |
| Galectin-1 | LSPR | Non-Validated | Serum | ---- | 10−13 M | [202] |
| Lipocalin-2 | SPR | Non-Validated | Serum | 2.5–500 ng/mL | 0.6 ng/mL | [203] |
| Podoplanin | SPRi | Non-Validated | Blood Plasma | 0.25–1 ng/mL | 15 ng/mL | [204] |
| p38αMAP kinase | SPR | Non-Validated | Serum | ---- | ---- | [205] |
| Cathepsin G | SPRi | Non-Validated | Blood | ---- | ---- | [206] |
| Epstein–Barr virus | SPR | Non-Validated | Serum | ---- | 10 pg/mL | [207] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Das, S.; Devireddy, R.; Gartia, M.R. Surface Plasmon Resonance (SPR) Sensor for Cancer Biomarker Detection. Biosensors 2023, 13, 396. https://doi.org/10.3390/bios13030396
Das S, Devireddy R, Gartia MR. Surface Plasmon Resonance (SPR) Sensor for Cancer Biomarker Detection. Biosensors. 2023; 13(3):396. https://doi.org/10.3390/bios13030396
Chicago/Turabian StyleDas, Sreyashi, Ram Devireddy, and Manas Ranjan Gartia. 2023. "Surface Plasmon Resonance (SPR) Sensor for Cancer Biomarker Detection" Biosensors 13, no. 3: 396. https://doi.org/10.3390/bios13030396
APA StyleDas, S., Devireddy, R., & Gartia, M. R. (2023). Surface Plasmon Resonance (SPR) Sensor for Cancer Biomarker Detection. Biosensors, 13(3), 396. https://doi.org/10.3390/bios13030396






