The Synergy between Deep Learning and Organs-on-Chips for High-Throughput Drug Screening: A Review
Abstract
1. Introduction
- Show that deep learning has begun to be explored in OoCs for higher-throughput drug screening.
- Highlight the critical deep learning tasks in OoCs and the successful use cases that solve or improve the efficiency of drug screening in the real world.
- Describe the potential applications and future challenges between deep learning and OoCs.
2. Overview of Deep Learning Methods
2.1. NN and DNN
2.2. CNN
2.3. RNN
2.4. GAN
2.5. AE
3. Deep Learning Methods Potentially Useful for OoCs
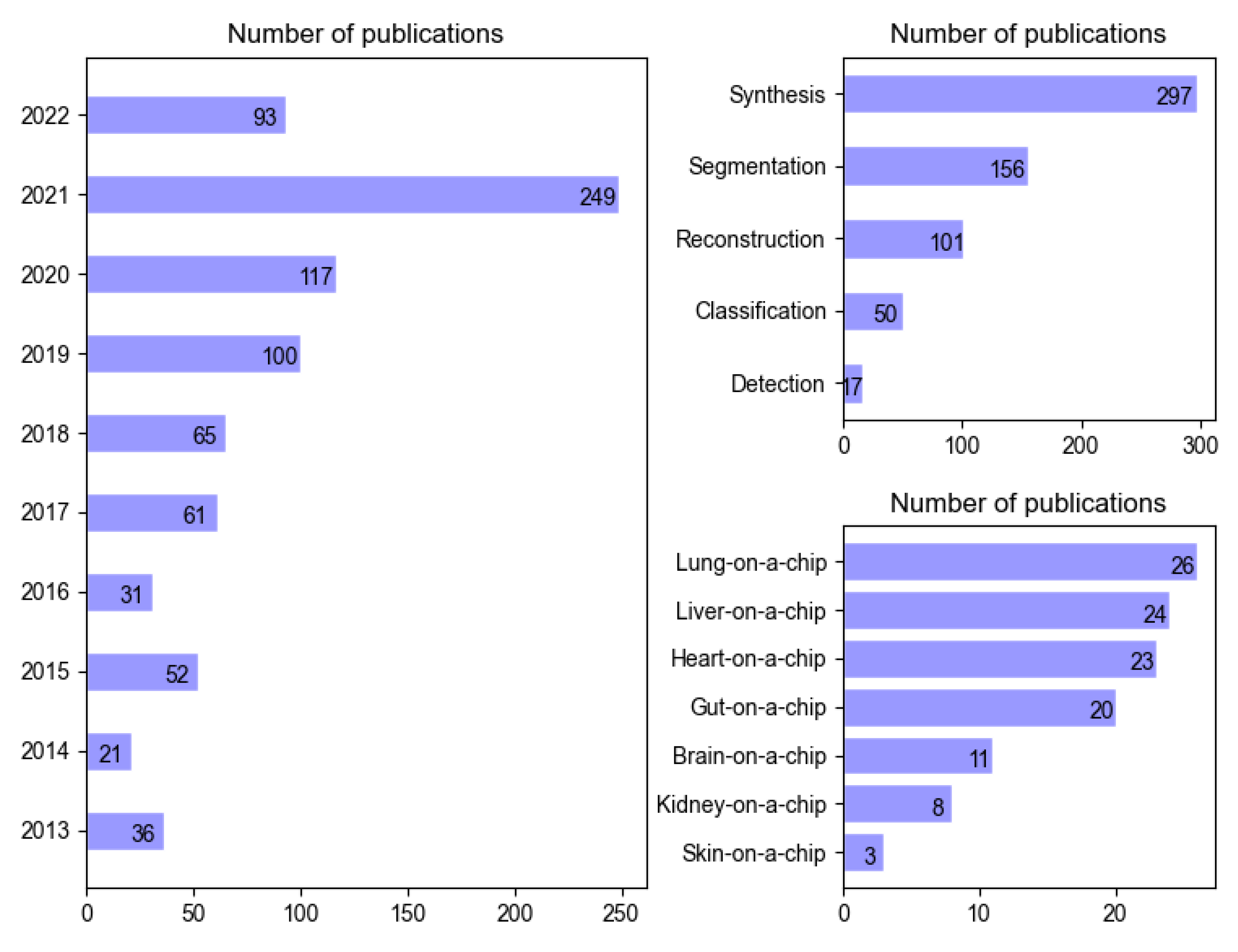
3.1. Image Synthesis (Super-Resolution, Data Augmentation)
3.2. Image Segmentation
3.3. Image Reconstruction
3.4. Image Classification
3.5. Image Detection
4. Case Studies in OoC Applications
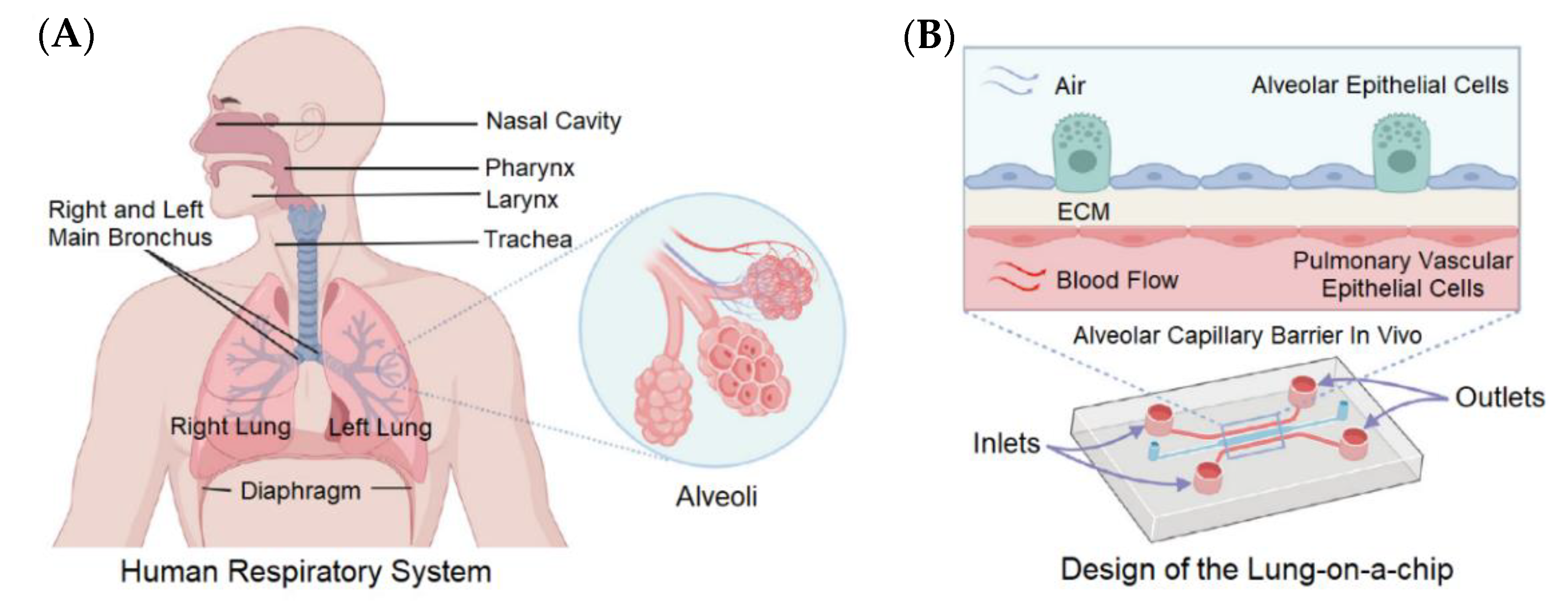
4.1. Lung-on-a-Chip
4.2. Liver-on-a-Chip
4.3. Heart-on-a-Chip
4.4. Gut-on-a-Chip
4.5. Brain-on-a-Chip and Brain Organoid-on-a-Chip
4.6. Kidney-on-a-Chip
4.7. Skin-on-a-Chip
5. Discussion
5.1. Upcoming Technical Challenges
5.2. Promising Applications
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, J.; Koo, B.K.; Knoblich, J.A. Human Organoids: Model Systems for Human Biology and Medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef]
- Li, J.; Chen, J.; Bai, H.; Wang, H.; Hao, S.; Ding, Y.; Peng, B.; Zhang, J.; Li, L.; Huang, W. An Overview of Organs-on-Chips Based on Deep Learning. Research 2022, 2022, 9869518. [Google Scholar] [CrossRef]
- Ma, C.; Peng, Y.; Li, H.; Chen, W. Organ-on-a-Chip: A New Paradigm for Drug Development. Trends Pharmacol. Sci. 2021, 42, 119–133. [Google Scholar] [CrossRef]
- Fontana, F.; Figueiredo, P.; Martins, J.P.; Santos, H.A. Requirements for Animal Experiments: Problems and Challenges. Small 2021, 17, 2004182. [Google Scholar] [CrossRef]
- Armenia, I.; Cuestas Ayllón, C.; Torres Herrero, B.; Bussolari, F.; Alfranca, G.; Grazú, V.; Martínez de la Fuente, J. Photonic and Magnetic Materials for on-Demand Local Drug Delivery. Adv. Drug Deliv. Rev. 2022, 191, 114584. [Google Scholar] [CrossRef]
- Leung, C.M.; de Haan, P.; Ronaldson-Bouchard, K.; Kim, G.-A.; Ko, J.; Rho, H.S.; Chen, Z.; Habibovic, P.; Jeon, N.L.; Takayama, S.; et al. A Guide to the Organ-on-a-Chip. Nat. Rev. Methods Prim. 2022, 2, 33. [Google Scholar] [CrossRef]
- Trapecar, M.; Wogram, E.; Svoboda, D.; Communal, C.; Omer, A.; Lungjangwa, T.; Sphabmixay, P.; Velazquez, J.; Schneider, K.; Wright, C.W.; et al. Human Physiomimetic Model Integrating Microphysiological Systems of the Gut, Liver, and Brain for Studies of Neurodegenerative Diseases. Sci. Adv. 2021, 7, eabd1707. [Google Scholar] [CrossRef]
- Ingber, D.E. Human Organs-on-Chips for Disease Modelling, Drug Development and Personalized Medicine. Nat. Rev. Genet. 2022, 23, 467–491. [Google Scholar] [CrossRef]
- Polini, A.; Moroni, L. The Convergence of High-Tech Emerging Technologies into the next Stage of Organ-on-a-Chips. Biomater. Biosyst. 2021, 1, 100012. [Google Scholar] [CrossRef] [PubMed]
- O’Mahony, N.; Campbell, S.; Carvalho, A.; Harapanahalli, S.; Hernandez, G.V.; Krpalkova, L.; Riordan, D.; Walsh, J. Deep Learning vs. Traditional Computer Vision. Adv. Intell. Syst. Comput. 2020, 943, 128–144. [Google Scholar]
- Chen, X.; Jin, L.; Zhu, Y.; Luo, C.; Wang, T. Text Recognition in the Wild: A survey. ACM Comput. Surv. (CSUR) 2021, 54, 42. [Google Scholar] [CrossRef]
- Akbik, A.; Bergmann, T.; Blythe, D.; Rasul, K.; Schweter, S.; Vollgraf, R. FLAIR: An Easy-to-Use Framework for State-of-the-Art NLP. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics (Demonstrations), Minneapolis, MN, USA, 2–7 June 2019; Association for Computational Linguistics: Minneapolis, MN, USA, 2019; pp. 54–59. [Google Scholar]
- Lundervold, A.S.; Lundervold, A. An Overview of Deep Learning in Medical Imaging Focusing on MRI. Z. Med. Phys. 2019, 29, 102–127. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, S.J.; Hauptmann, A. Deep D-Bar: Real-Time Electrical Impedance Tomography Imaging with Deep Neural Networks. IEEE Trans. Med. Imaging 2018, 37, 2367–2377. [Google Scholar] [CrossRef]
- Khatami, A.; Nazari, A.; Khosravi, A.; Lim, C.P.; Nahavandi, S. A Weight Perturbation-Based Regularisation Technique for Convolutional Neural Networks and the Application in Medical Imaging. Expert Syst. Appl. 2020, 149, 113196. [Google Scholar] [CrossRef]
- Lyu, Q.; Shan, H.; Xie, Y.; Kwan, A.C.; Otaki, Y.; Kuronuma, K.; Li, D.; Wang, G. Cine Cardiac MRI Motion Artifact Reduction Using a Recurrent Neural Network. IEEE Trans. Med. Imaging 2021, 40, 2170–2181. [Google Scholar] [CrossRef]
- Fernandes, F.E.; Yen, G.G. Pruning of Generative Adversarial Neural Networks for Medical Imaging Diagnostics with Evolution Strategy. Inf. Sci. 2021, 558, 91–102. [Google Scholar] [CrossRef]
- Öztürk, Ş. Stacked Auto-Encoder Based Tagging with Deep Features for Content-Based Medical Image Retrieval. Expert Syst. Appl. 2020, 161, 113693. [Google Scholar] [CrossRef]
- Mallows Ranking Models: Maximum Likelihood Estimate and Regeneration. Available online: https://proceedings.mlr.press/v97/tang19a.html (accessed on 21 June 2022).
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A Survey on Deep Learning in Medical Image Analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Novikov, A.A.; Major, D.; Wimmer, M.; Lenis, D.; Buhler, K. Deep Sequential Segmentation of Organs in Volumetric Medical Scans. IEEE Trans. Med. Imaging 2019, 38, 1207–1215. [Google Scholar] [CrossRef]
- Tuttle, J.F.; Blackburn, L.D.; Andersson, K.; Powell, K.M. A Systematic Comparison of Machine Learning Methods for Modeling of Dynamic Processes Applied to Combustion Emission Rate Modeling. Appl. Energy 2021, 292, 116886. [Google Scholar] [CrossRef]
- He, J.; Zhu, Q.; Zhang, K.; Yu, P.; Tang, J. An Evolvable Adversarial Network with Gradient Penalty for COVID-19 Infection Segmentation. Appl. Soft Comput. 2021, 113, 107947. [Google Scholar] [CrossRef] [PubMed]
- 3D Self-Supervised Methods for Medical Imaging. Available online: https://proceedings.neurips.cc/paper/2020/hash/d2dc6368837861b42020ee72b0896182-Abstract.html (accessed on 5 June 2022).
- Li, M.; Zhang, T.; Chen, Y.; Smola, A.J. Efficient Mini-Batch Training for Stochastic Optimization. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; pp. 661–670. [Google Scholar]
- Stapor, P.; Schmiester, L.; Wierling, C.; Merkt, S.; Pathirana, D.; Lange, B.M.H.; Weindl, D.; Hasenauer, J. Mini-Batch Optimization Enables Training of ODE Models on Large-Scale Datasets. Nat. Commun. 2022, 13, 34. [Google Scholar] [CrossRef] [PubMed]
- Generalization Bounds of Stochastic Gradient Descent for Wide and Deep Neural Networks. Available online: https://proceedings.neurips.cc/paper/2019/hash/cf9dc5e4e194fc21f397b4cac9cc3ae9-Abstract.html (accessed on 12 June 2022).
- Ilboudo, W.E.L.; Kobayashi, T.; Sugimoto, K. Robust Stochastic Gradient Descent with Student-t Distribution Based First-Order Momentum. IEEE Trans. Neural Netw. Learn. Syst. 2022, 33, 1324–1337. [Google Scholar] [CrossRef]
- Sexton, R.S.; Dorsey, R.E.; Johnson, J.D. Optimization of Neural Networks: A Comparative Analysis of the Genetic Algorithm and Simulated Annealing. Eur. J. Oper. Res. 1999, 114, 589–601. [Google Scholar] [CrossRef]
- Amine, K. Multiobjective Simulated Annealing: Principles and Algorithm Variants. Adv. Oper. Res. 2019, 2019, 8134674. [Google Scholar] [CrossRef]
- Qiao, J.; Li, S.; Li, W. Mutual Information Based Weight Initialization Method for Sigmoidal Feedforward Neural Networks. Neurocomputing 2016, 207, 676–683. [Google Scholar] [CrossRef]
- Zhu, D.; Lu, S.; Wang, M.; Lin, J.; Wang, Z. Efficient Precision-Adjustable Architecture for Softmax Function in Deep Learning. IEEE Trans. Circuits Syst. II Express Briefs 2020, 67, 3382–3386. [Google Scholar] [CrossRef]
- Liu, Y.; Gong, C.; Yang, L.; Chen, Y. DSTP-RNN: A Dual-Stage Two-Phase Attention-Based Recurrent Neural Network for Long-Term and Multivariate Time Series Prediction. Expert Syst. Appl. 2020, 143, 113082. [Google Scholar] [CrossRef]
- Gao, R.; Tang, Y.; Xu, K.; Huo, Y.; Bao, S.; Antic, S.L.; Epstein, E.S.; Deppen, S.; Paulson, A.B.; Sandler, K.L.; et al. Time-Distanced Gates in Long Short-Term Memory Networks. Med. Image Anal. 2020, 65, 101785. [Google Scholar] [CrossRef]
- Tan, Q.; Ye, M.; Yang, B.; Liu, S.Q.; Ma, A.J.; Yip, T.C.F.; Wong, G.L.H.; Yuen, P.C. DATA-GRU: Dual-Attention Time-Aware Gated Recurrent Unit for Irregular Multivariate Time Series. In Proceedings of the 36th AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; AAAI Press: Palo Alto, CA, USA, 2020; Volume 34, pp. 930–937. [Google Scholar]
- Nemeth, C.; Fearnhead, P. Stochastic Gradient Markov Chain Monte Carlo. J. Am. Stat. Assoc. 2021, 116, 433–450. [Google Scholar] [CrossRef]
- Lugmayr, A.; Danelljan, M.; Timofte, R. Unsupervised Learning for Real-World Super-Resolution. In Proceedings of the 2019 International Conference on Computer Vision Workshop, ICCVW 2019, Seoul, Republic of Korea, 27–28 October 2019; pp. 3408–3416. [Google Scholar]
- Karunasingha, D.S.K. Root Mean Square Error or Mean Absolute Error? Use Their Ratio as Well. Inf. Sci. 2022, 585, 609–629. [Google Scholar] [CrossRef]
- Polini, A.; Prodanov, L.; Bhise, N.S.; Manoharan, V.; Dokmeci, M.R.; Khademhosseini, A. Organs-on-a-Chip: A New Tool for Drug Discovery. Expert Opin. Drug Discov. 2014, 9, 335–352. [Google Scholar] [CrossRef]
- Dai, M.; Xiao, G.; Fiondella, L.; Shao, M.; Zhang, Y.S. Deep Learning-Enabled Resolution-Enhancement in Mini- and Regular Microscopy for Biomedical Imaging. Sens. Actuators A Phys. 2021, 331, 112928. [Google Scholar] [CrossRef] [PubMed]
- Cascarano, P.; Comes, M.C.; Mencattini, A.; Parrini, M.C.; Piccolomini, E.L.; Martinelli, E. Recursive Deep Prior Video: A Super Resolution Algorithm for Time-Lapse Microscopy of Organ-on-Chip Experiments. Med. Image Anal. 2021, 72, 102124. [Google Scholar] [CrossRef] [PubMed]
- Ulyanov, D.; Vedaldi, A.; Lempitsky, V. Deep Image Prior. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; IEEE: Salt Lake City, UT, USA, 2018. [Google Scholar]
- Comes, M.C.; Filippi, J.; Mencattini, A.; Casti, P.; Cerrato, G.; Sauvat, A.; Vacchelli, E.; de Ninno, A.; di Giuseppe, D.; D’Orazio, M.; et al. Multi-Scale Generative Adversarial Network for Improved Evaluation of Cell–Cell Interactions Observed in Organ-on-Chip Experiments. Neural Comput. Appl. 2020, 33, 3671–3689. [Google Scholar] [CrossRef]
- Stoecklein, D.; Lore, K.G.; Davies, M.; Sarkar, S.; Ganapathysubramanian, B. Deep Learning for Flow Sculpting: Insights into Efficient Learning Using Scientific Simulation Data. Sci. Rep. 2017, 7, 46368. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, Q. Research and Improvement of Content-Based Image Retrieval Framework. Int. J. Pattern Recognit. Artif. Intell. 2018, 32, 1850043. [Google Scholar] [CrossRef]
- Siddique, N.; Paheding, S.; Elkin, C.P.; Devabhaktuni, V. U-Net and Its Variants for Medical Image Segmentation: A Review of Theory and Applications. IEEE Access 2021, 9, 82031–82057. [Google Scholar] [CrossRef]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A Nested U-Net Architecture for Medical Image Segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; University of Toronto: Tornoto, ON, Canada, 2018; pp. 3–11. [Google Scholar]
- Schönfeld, E.; Schiele, B.; Khoreva, A. A U-Net Based Discriminator for Generative Adversarial Networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 8207–8216. [Google Scholar]
- Falk, T.; Mai, D.; Bensch, R. U-Net: Deep Learning for Cell Counting, Detection, and Morphometry. Nat. Methods 2019, 16, 67–70. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Lim, J.; Ayoub, A.B.; Psaltis, D. Three-Dimensional Tomography of Red Blood Cells Using Deep Learning. Adv. Photonics 2020, 2, 026001. [Google Scholar] [CrossRef]
- Pretini, V.; Koenen, M.H.; Kaestner, L.; Fens, M.H.A.M.; Schiffelers, R.M.; Bartels, M.; van Wijk, R. Red Blood Cells: Chasing Interactions. Front. Physiol. 2019, 10, 945. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.; Borges, B.-H.V.; Martins, E.R.; Liang, H.; Zhou, J.; Li, J.; Krauss, T.F. High Performance Metalenses: Numerical Aperture, Aberrations, Chromaticity, and Trade-Offs. Optica 2019, 6, 1461–1470. [Google Scholar]
- Mencattini, A.; di Giuseppe, D.; Comes, M.C.; Casti, P.; Corsi, F.; Bertani, F.R.; Ghibelli, L.; Businaro, L.; di Natale, C.; Parrini, M.C.; et al. Discovering the Hidden Messages within Cell Trajectories Using a Deep Learning Approach for in Vitro Evaluation of Cancer Drug Treatments. Sci. Rep. 2020, 10, 7653. [Google Scholar] [CrossRef]
- Lu, S.; Lu, Z.; Zhang, Y.D. Pathological Brain Detection Based on AlexNet and Transfer Learning. J. Comput. Sci. 2019, 30, 41–47. [Google Scholar] [CrossRef]
- Ditadi, A.; Sturgeon, C.M.; Keller, G. A View of Human Haematopoietic Development from the Petri Dish. Nat. Rev. Mol. Cell Biol. 2016, 18, 56–67. [Google Scholar] [CrossRef]
- Jing, Y.; Yang, Y.; Feng, Z.; Ye, J.; Yu, Y.; Song, M. Neural Style Transfer: A Review. IEEE Trans. Vis. Comput. Graph. 2020, 26, 3365–3385. [Google Scholar] [CrossRef] [PubMed]
- Heo, Y.J.; Lee, D.; Kang, J.; Lee, K.; Chung, W.K. Real-Time Image Processing for Microscopy-Based Label-Free Imaging Flow Cytometry in a Microfluidic Chip. Sci. Rep. 2017, 7, 11651. [Google Scholar] [CrossRef]
- Becht, E.; Tolstrup, D.; Dutertre, C.A.; Morawski, P.A.; Campbell, D.J.; Ginhoux, F.; Newell, E.W.; Gottardo, R.; Headley, M.B. High-Throughput Single-Cell Quantification of Hundreds of Proteins Using Conventional Flow Cytometry and Machine Learning. Sci. Adv. 2021, 7, 505–527. [Google Scholar] [CrossRef] [PubMed]
- Kieninger, J.; Weltin, A.; Flamm, H.; Urban, G.A. Microsensor Systems for Cell Metabolism–from 2D Culture to Organ-on-Chip. Lab Chip 2018, 18, 1274–1291. [Google Scholar] [CrossRef]
- Meijering, E.; Dzyubachyk, O.; Smal, I. Methods for Cell and Particle Tracking. Methods Enzymol. 2012, 504, 183–200. [Google Scholar] [PubMed]
- Li, S.; Li, A.; Molina Lara, D.A.; Gómez Marín, J.E.; Juhas, M.; Zhang, Y. Transfer Learning for Toxoplasma Gondii Recognition. mSystems 2020, 5, e00445-19. [Google Scholar] [CrossRef] [PubMed]
- Askari, S. Fuzzy C-Means Clustering Algorithm for Data with Unequal Cluster Sizes and Contaminated with Noise and Outliers: Review and Development. Expert Syst. Appl. 2021, 165, 113856. [Google Scholar] [CrossRef]
- Kwon, Y.-H.; Park, M.-G. Predicting Future Frames Using Retrospective Cycle GAN. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–19 June 2019; pp. 1811–1820. [Google Scholar]
- Riordon, J.; Sovilj, D.; Sanner, S.; Sinton, D.; Young, E.W.K. Deep Learning with Microfluidics for Biotechnology. Trends Biotechnol. 2019, 37, 310–324. [Google Scholar] [CrossRef] [PubMed]
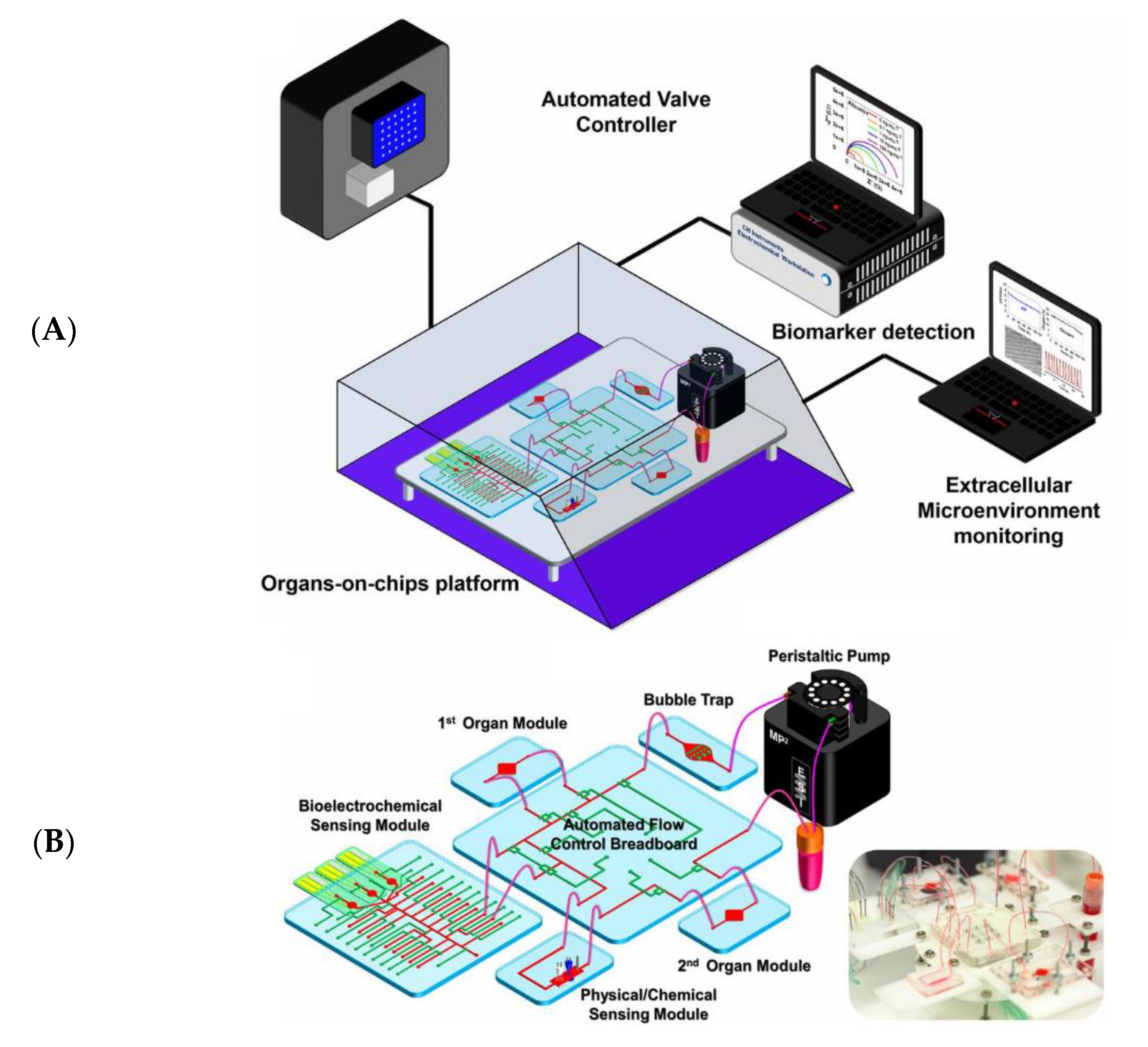
- Zhang, Y.S.; Aleman, J.; Shin, S.R.; Kilic, T.; Kim, D.; Shaegh, S.A.M.; Massa, S.; Riahi, R.; Chae, S.; Hu, N.; et al. Multisensor-Integrated Organs-on-Chips Platform for Automated and Continual in Situ Monitoring of Organoid Behaviors. Proc. Natl. Acad. Sci. USA 2017, 114, E2293–E2302. [Google Scholar] [CrossRef]
- Sun, A.M.; Hoffman, T.; Luu, B.Q.; Ashammakhi, N.; Li, S. Application of Lung Microphysiological Systems to COVID-19 Modeling and Drug Discovery: A Review. Bio-Des. Manuf. 2021, 4, 757–777. [Google Scholar] [CrossRef]
- Wang, L.; Han, J.; Su, W.; Li, A.; Zhang, W.; Li, H.; Hu, H.; Song, W.; Xu, C.; Chen, J. Gut-on-a-Chip for Exploring the Transport Mechanism of Hg(II). Microsyst. Nanoeng. 2023, 9, 2. [Google Scholar] [CrossRef]
- Sarker, I.H. Deep Learning: A Comprehensive Overview on Techniques, Taxonomy, Applications and Research Directions. SN Comput. Sci. 2021, 2, 420. [Google Scholar] [CrossRef]
- Su, R.; Li, Y.; Zink, D.; Loo, L.H. Supervised Prediction of Drug-Induced Nephrotoxicity Based on Interleukin-6 and -8 Expression Levels. BMC Bioinform. 2014, 15, S16. [Google Scholar] [CrossRef]
- Qu, C.; Gao, L.; Yu, X.Q.; Wei, M.; Fang, G.Q.; He, J.; Cao, L.X.; Ke, L.; Tong, Z.H.; Li, W.Q. Machine Learning Models of Acute Kidney Injury Prediction in Acute Pancreatitis Patients. Gastroenterol. Res. Pract. 2020, 2020, 3431290. [Google Scholar] [CrossRef]
- Kandasamy, K.; Chuah, J.K.C.; Su, R.; Huang, P.; Eng, K.G.; Xiong, S.; Li, Y.; Chia, C.S.; Loo, L.H.; Zink, D. Prediction of Drug-Induced Nephrotoxicity and Injury Mechanisms with Human Induced Pluripotent Stem Cell-Derived Cells and Machine Learning Methods. Sci. Rep. 2015, 5, 12337. [Google Scholar] [CrossRef]
- Wilmer, M.J.; Ng, C.P.; Lanz, H.L.; Vulto, P.; Suter-Dick, L.; Masereeuw, R. Kidney-on-a-Chip Technology for Drug-Induced Nephrotoxicity Screening. Trends Biotechnol. 2016, 34, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Sutterby, E.; Thurgood, P.; Baratchi, S.; Khoshmanesh, K.; Pirogova, E. Microfluidic Skin-on-a-Chip Models: Toward Biomimetic Artificial Skin. Small 2020, 16, 2002515. [Google Scholar] [CrossRef] [PubMed]
- Legrand, S.; Scheinberg, A.; Tillack, A.F.; Thavappiragasam, M.; Vermaas, J.V.; Agarwal, R.; Larkin, J.; Poole, D.; Santos-Martins, D.; Solis-Vasquez, L.; et al. GPU-Accelerated Drug Discovery with Docking on the Summit Supercomputer: Porting, Optimization, and Application to COVID-19 Research. In Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, Online, 21–24 September 2020; Association for Computing Machinery: New York, NY, USA, 2020; pp. 1–10. [Google Scholar]
- McDonald, K.A.; Holtz, R.B. From Farm to Finger Prick—A Perspective on How Plants Can Help in the Fight Against COVID-19. Front. Bioeng. Biotechnol. 2020, 8, 782. [Google Scholar] [CrossRef]
- Mazza, M.G.; de Lorenzo, R.; Conte, C.; Poletti, S.; Vai, B.; Bollettini, I.; Melloni, E.M.T.; Furlan, R.; Ciceri, F.; Rovere-Querini, P.; et al. Anxiety and Depression in COVID-19 Survivors: Role of Inflammatory and Clinical Predictors. Brain Behav. Immun. 2020, 89, 594–600. [Google Scholar] [CrossRef]
- Francis, I.; Shrestha, J.; Paudel, K.R.; Hansbro, P.M.; Warkiani, M.E.; Saha, S.C. Recent Advances in Lung-on-a-Chip Models. Drug Discov. Today 2022, 27, 2593–2602. [Google Scholar] [CrossRef]
- Novac, O.; Silva, R.; Young, L.M.; Lachani, K.; Hughes, D.; Kostrzewski, T. Human Liver Microphysiological System for Assessing Drug-Induced Liver Toxicity in Vitro. J. Vis. Exp. Jove 2022, 179, preprint. [Google Scholar]
- Liu, M.; Xiang, Y.; Yang, Y.; Long, X.; Xiao, Z.; Nan, Y.; Jiang, Y.; Qiu, Y.; Huang, Q.; Ai, K. State-of-the-Art Advancements in Liver-on-a-Chip (LOC): Integrated Biosensors for LOC. Biosens. Bioelectron. 2022, 218, 114758. [Google Scholar] [CrossRef]
- Gazaryan, A.; Shkurnikov, I.; Nikulin, M.; Drapkina, S.; Baranova, O.; Tonevitsky, A. In Vitro and in Silico Liver Models: Current Trends, Challenges and in Vitro and in Silico Liver Models: Current Trends, Challenges and Opportunities Opportunities. ALTEX 2018, 35, 397. [Google Scholar]
- Vanella, R.; Kovacevic, G.; Doffini, V.; Fernández De Santaella, J.; Nash, M.A. High-Throughput Screening, next Generation Sequencing and Machine Learning: Advanced Methods in Enzyme Engineering. Chem. Commun. 2022, 58, 2455–2467. [Google Scholar] [CrossRef]
- Capuzzi, S.J.; Politi, R.; Isayev, O.; Farag, S.; Tropsha, A. QSAR Modeling of Tox21 Challenge Stress Response and Nuclear Receptor Signaling Toxicity Assays. Front. Environ. Sci. 2016, 4, 3. [Google Scholar] [CrossRef]
- Ignacz, G.; Szekely, G. Deep Learning Meets Quantitative Structure–Activity Relationship (QSAR) for Leveraging Structure-Based Prediction of Solute Rejection in Organic Solvent Nanofiltration. J. Memb. Sci. 2022, 646, 120268. [Google Scholar] [CrossRef]
- Bai, J.; Li, Y.; Li, J.; Yang, X.; Jiang, Y.; Xia, S.T. Multinomial Random Forest. Pattern Recognit. 2022, 122, 108331. [Google Scholar] [CrossRef]
- Long-Term Impact of Johnson & Johnson’s Health & Wellness Program on Health Care Utilization and Expenditures on JSTOR. Available online: https://www.jstor.org/stable/44995849 (accessed on 10 July 2022).
- Zhang, C.; Lu, Y. Study on Artificial Intelligence: The State of the Art and Future Prospects. J. Ind. Inf. Integr. 2021, 23, 100224. [Google Scholar] [CrossRef]
- Matschinske, J.; Alcaraz, N.; Benis, A.; Golebiewski, M.; Grimm, D.G.; Heumos, L.; Kacprowski, T.; Lazareva, O.; List, M.; Louadi, Z.; et al. The AIMe Registry for Artificial Intelligence in Biomedical Research. Nat. Methods 2021, 18, 1128–1131. [Google Scholar] [CrossRef]
- Agarwal, A.; Goss, J.A.; Cho, A.; McCain, M.L.; Parker, K.K. Microfluidic Heart on a Chip for Higher Throughput Pharmacological Studies. Lab Chip 2013, 13, 3599–3608. [Google Scholar] [CrossRef]
- Jastrzebska, E.; Tomecka, E.; Jesion, I. Heart-on-a-Chip Based on Stem Cell Biology. Biosens. Bioelectron. 2016, 75, 67–81. [Google Scholar] [CrossRef]
- Yang, Q.; Xiao, Z.; Lv, X.; Zhang, T.; Liu, H. Fabrication and Biomedical Applications of Heart-on-a-Chip. Int. J. Bioprint. 2021, 7, 370. [Google Scholar] [CrossRef]
- Cho, K.W.; Lee, W.H.; Kim, B.S.; Kim, D.H. Sensors in Heart-on-a-Chip: A Review on Recent Progress. Talanta 2020, 219, 121269. [Google Scholar] [CrossRef]
- Fetah, K.L.; DiPardo, B.J.; Kongadzem, E.M.; Tomlinson, J.S.; Elzagheid, A.; Elmusrati, M.; Khademhosseini, A.; Ashammakhi, N. Cancer Modeling-on-a-Chip with Future Artificial Intelligence Integration. Small 2019, 15, 1901985. [Google Scholar] [CrossRef]
- Mencattini, A.; Mattei, F.; Schiavoni, G.; Gerardino, A.; Businaro, L.; di Natale, C.; Martinelli, E. From Petri Dishes to Organ on Chip Platform: The Increasing Importance of Machine Learning and Image Analysis. Front. Pharmacol. 2019, 10, 100. [Google Scholar] [CrossRef]
- Marrero, D.; Pujol-Vila, F.; Vera, D.; Gabriel, G.; Illa, X.; Elizalde-Torrent, A.; Alvarez, M.; Villa, R. Gut-on-a-Chip: Mimicking and Monitoring the Human Intestine. Biosens. Bioelectron. 2021, 181, 113156. [Google Scholar] [CrossRef]
- Hewes, S.A.; Wilson, R.L.; Estes, M.K.; Shroyer, N.F.; Blutt, S.E.; Grande-Allen, K.J. In Vitro Models of the Small Intestine: Engineering Challenges and Engineering Solutions. Tissue Eng. Part B Rev. 2020, 26, 313–326. [Google Scholar] [CrossRef]
- Park, H.; Na, M.; Kim, B.; Park, S.; Kim, K.H.; Chang, S.; Ye, J.C. Deep Learning Enables Reference-Free Isotropic Super-Resolution for Volumetric Fluorescence Microscopy. Nat. Commun. 2022, 13, 3297. [Google Scholar] [CrossRef]
- Tian, C.; Xu, Y.; Zuo, W.; Zhang, B.; Fei, L.; Lin, C.W. Coarse-to-Fine CNN for Image Super-Resolution. IEEE Trans. Multimed. 2021, 23, 1489–1502. [Google Scholar] [CrossRef]
- Shin, W.; Kim, H.J. 3D in Vitro Morphogenesis of Human Intestinal Epithelium in a Gut-on-a-Chip or a Hybrid Chip with a Cell Culture Insert. Nat. Protocols. 2022, 17, 910–939. [Google Scholar] [CrossRef]
- Trietsch, S.J.; Naumovska, E.; Kurek, D.; Setyawati, M.C.; Vormann, M.K.; Wilschut, K.J.; Lanz, H.L.; Nicolas, A.; Ng, C.P.; Joore, J.; et al. Membrane-Free Culture and Real-Time Barrier Integrity Assessment of Perfused Intestinal Epithelium Tubes. Nat. Commun. 2017, 8, 262. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Zhang, X.; Chen, X.; Fang, Y. Distributed Optimization of Visual Sensor Networks for Coverage of a Large-Scale 3-D Scene. IEEE/ASME Trans. Mechatron. 2020, 25, 2777–2788. [Google Scholar] [CrossRef]
- al Hayani, B.; Ilhan, H. Image Transmission Over Decode and Forward Based Cooperative Wireless Multimedia Sensor Networks for Rayleigh Fading Channels in Medical Internet of Things (MIoT) for Remote Health-Care and Health Communication Monitoring. J. Med. Imaging Health Inform. 2019, 10, 160–168. [Google Scholar] [CrossRef]
- Atat, O.E.; Farzaneh, Z.; Pourhamzeh, M.; Taki, F.; Abi-Habib, R.; Vosough, M.; El-Sibai, M. 3D Modeling in Cancer Studies. Hum. Cell 2022, 35, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Bang, S.; Choi, N.; Kim, H.N. Brain Organoid-on-a-Chip: A next-Generation Human Brain Avatar for Recapitulating Human Brain Physiology and Pathology. Biomicrofluidics 2022, 16, 061301. [Google Scholar] [CrossRef] [PubMed]
- Cakir, B.; Xiang, Y.; Tanaka, Y.; Kural, M.H.; Parent, M.; Kang, Y.J.; Chapeton, K.; Patterson, B.; Yuan, Y.; He, C.S.; et al. Engineering of Human Brain Organoids with a Functional Vascular-like System. Nat. Methods 2019, 16, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Ryu, H.; Chung, M.; Kim, Y.; Blum, Y.; Lee, S.S.; Pertz, O.; Jeon, N.L. Microfluidic Platform for Single Cell Analysis under Dynamic Spatial and Temporal Stimulation. Biosens. Bioelectron. 2018, 104, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Krauss, J.K.; Lipsman, N.; Aziz, T.; Boutet, A.; Brown, P.; Chang, J.W.; Davidson, B.; Grill, W.M.; Hariz, M.I.; Horn, A.; et al. Technology of Deep Brain Stimulation: Current Status and Future Directions. Nat. Rev. Neurol. 2020, 17, 75–87. [Google Scholar] [CrossRef]
- Blauwendraat, C.; Nalls, M.A.; Singleton, A.B. The Genetic Architecture of Parkinson’s Disease. Lancet Neurol. 2020, 19, 170–178. [Google Scholar] [CrossRef]
- Arber, S.; Costa, R.M. Networking Brainstem and Basal Ganglia Circuits for Movement. Nat. Rev. Neurosci. 2022, 23, 342–360. [Google Scholar] [CrossRef]
- Gao, Q.; Naumann, M.; Jovanov, I.; Lesi, V.; Kamaravelu, K.; Grill, W.M.; Pajic, M. Model-Based Design of Closed Loop Deep Brain Stimulation Controller Using Reinforcement Learning. In Proceedings of the 2020 ACM/IEEE 11th International Conference on Cyber-Physical Systems, Sydney, NSW, Australia, 21–25 April 2020; pp. 108–118. [Google Scholar]
- Eppe, M.; Gumbsch, C.; Kerzel, M.; Nguyen, P.D.H.; Butz, M.V.; Wermter, S. Intelligent Problem-Solving as Integrated Hierarchical Reinforcement Learning. Nat. Mach. Intell. 2022, 4, 11–20. [Google Scholar] [CrossRef]
- Kim, H.; Kim, Y.; Ji, H.; Park, H.; An, J.; Song, H.; Kim, Y.T.; Lee, H.S.; Kim, K. A Single-Chip FPGA Holographic Video Processor. IEEE Trans. Ind. Electron. 2019, 66, 2066–2073. [Google Scholar] [CrossRef]
- Milardi, D.; Quartarone, A.; Bramanti, A.; Anastasi, G.; Bertino, S.; Basile, G.A.; Buonasera, P.; Pilone, G.; Celeste, G.; Rizzo, G.; et al. The Cortico-Basal Ganglia-Cerebellar Network: Past, Present and Future Perspectives. Front. Syst. Neurosci. 2019, 13, 61. [Google Scholar] [CrossRef]
- Lake, M.; Lake, M.; Narciso, C.; Cowdrick, K.; Storey, T.; Zhang, S.; Zartman, J.; Hoelzle, D. Microfluidic Device Design, Fabrication, and Testing Protocols. Protoc. Exch. 2015. [Google Scholar] [CrossRef]
- Eve-Mary Leikeki, K. Machine Learning Application: Organs-on-a-Chip in Parellel. 2018. Available online: https://osuva.uwasa.fi/handle/10024/9314 (accessed on 16 December 2022).
- Hwang, S.H.; Lee, S.; Park, J.Y.; Jeon, J.S.; Cho, Y.J.; Kim, S. Potential of Drug Efficacy Evaluation in Lung and Kidney Cancer Models Using Organ-on-a-Chip Technology. Micromachines 2021, 12, 215. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, P. Prediction of Drug-Induced Kidney Injury in Drug Discovery. Drug Metab. Rev. 2021, 53, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hui, J.; Yang, P.; Mao, H. Microfluidic Organ-on-a-Chip System for Disease Modeling and Drug Development. Biosensors 2022, 12, 370. [Google Scholar] [CrossRef] [PubMed]
- Varga-Medveczky, Z.; Kocsis, D.; Naszlady, M.B.; Fónagy, K.; Erdő, F. Skin-on-a-Chip Technology for Testing Transdermal Drug Delivery—Starting Points and Recent Developments. Pharmaceutics 2021, 13, 1852. [Google Scholar] [CrossRef]
- Alicia Boos, J.; Mark Misun, P.; Michlmayr, A.; Hierlemann, A.; Frey, O.; Boos, J.A.; Misun, P.M.; Michlmayr, A.; Hierlemann, A.; Frey, O. Microfluidic Multitissue Platform for Advanced Embryotoxicity Testing in Vitro. Adv. Sci. 2019, 6, 1900294. [Google Scholar] [CrossRef]
- Wikswo, J.P.; Curtis, E.L.; Eagleton, Z.E.; Evans, B.C.; Kole, A.; Hofmeister, L.H.; Matloff, W.J. Scaling and Systems Biology for Integrating Multiple Organs-on-a-Chip. Lab Chip 2013, 13, 3496–3511. [Google Scholar] [CrossRef]
- Ke, X.; Zou, J.; Niu, Y. End-to-End Automatic Image Annotation Based on Deep CNN and Multi-Label Data Augmentation. IEEE Trans. Multimed. 2019, 21, 2093–2106. [Google Scholar] [CrossRef]
- He, X.; Zhao, K.; Chu, X. AutoML: A Survey of the State-of-the-Art. Knowl. Based Syst. 2021, 212, 106622. [Google Scholar] [CrossRef]
- Mok, J.; Na, B.; Choe, H.; Yoon, S. AdvRush: Searching for Adversarially Robust Neural Architectures. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 12322–12332. [Google Scholar]
- Hutter, F.; Kotthoff, L.; Vanschoren, J. The Springer Series on Challenges in Machine Learning Automated Machine Learning Methods, Systems, Challenges; Springer Nature: Cham, Switzerland, 2022. [Google Scholar]
- Sriram, A.; Jun, H.; Satheesh, S.; Coates, A. Cold Fusion: Training Seq2Seq Models Together with Language Models. arXiv 2017, arXiv:1708.06426. [Google Scholar]
- Lin, J.C.W.; Shao, Y.; Djenouri, Y.; Yun, U. ASRNN: A Recurrent Neural Network with an Attention Model for Sequence Labeling. Knowl. Based Syst. 2021, 212, 106548. [Google Scholar] [CrossRef]
- Chen, L.; Lu, K.; Rajeswaran, A.; Lee, K.; Grover, A.; Laskin, M.; Abbeel, P.; Srinivas, A.; Mordatch, I. Decision Transformer: Reinforcement Learning via Sequence Modeling. Adv. Neural. Inf. Process. Syst. 2021, 34, 15084–15097. [Google Scholar]
- Wu, Z.; Pan, S.; Long, G.; Jiang, J.; Chang, X.; Zhang, C. Connecting the Dots: Multivariate Time Series Forecasting with Graph Neural Networks. In Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Online, 6–10 July 2020; Association for Computing Machinery: New York, NY, USA, 2020; pp. 753–763. [Google Scholar]
- Shih, S.Y.; Sun, F.K.; Lee, H.Y. Temporal Pattern Attention for Multivariate Time Series Forecasting. Mach. Learn. 2019, 108, 1421–1441. [Google Scholar] [CrossRef]
- Wen, Q.; Zhou, T.; Zhang, C.; Chen, W.; Ma, Z.; Yan, J.; Sun, L. Transformers in Time Series: A Survey. arXiv 2022, arXiv:2202.07125. [Google Scholar]
- Ngiam, J.; Khosla, A.; Kim, M.; Nam, J.; Lee, H.; Ng, A.Y. Multimodal Deep Learning. In Proceedings of the 28th International Conference on Machine Learning, Bellevue, WA, USA, 28 June–2 July 2011; Omnipress: Madison, WI, USA, 2011. [Google Scholar]
- Boehm, K.M.; Khosravi, P.; Vanguri, R.; Gao, J.; Shah, S.P. Harnessing Multimodal Data Integration to Advance Precision Oncology. Nat. Rev. Cancer 2021, 22, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Low, L.A.; Mummery, C.; Berridge, B.R.; Austin, C.P.; Tagle, D.A. Organs-on-Chips: Into the next Decade. Nat. Rev. Drug Discov. 2020, 20, 345–361. [Google Scholar] [CrossRef]
- Gawehn, E.; Hiss, J.A.; Schneider, G. Deep Learning in Drug Discovery. Mol. Inform. 2016, 35, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Lane, T.R.; Foil, D.H.; Minerali, E.; Urbina, F.; Zorn, K.M.; Ekins, S.B.C. Bioactivity Comparison across Multiple Machine Learning Algorithms Using over 5000 Datasets for Drug Discovery. Mol. Pharm. 2020, 18, 403–415. [Google Scholar] [CrossRef] [PubMed]


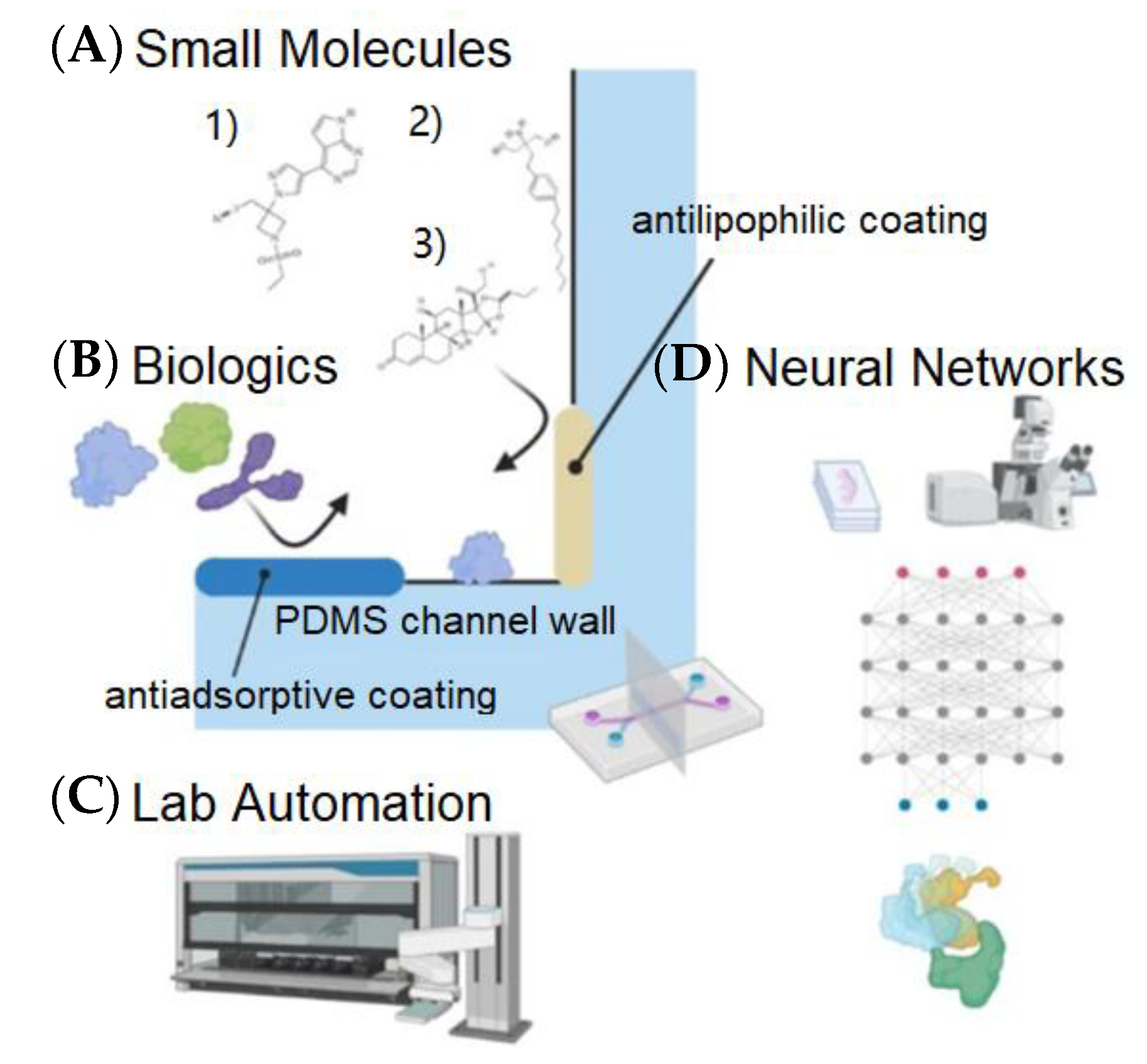

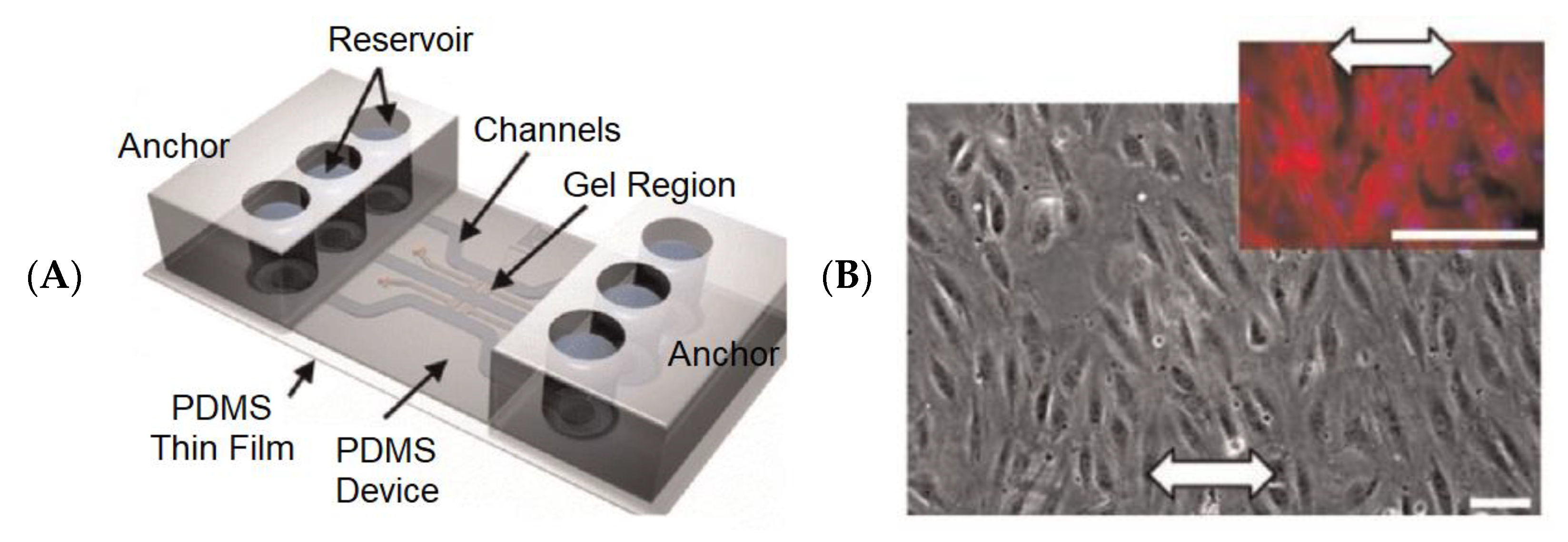



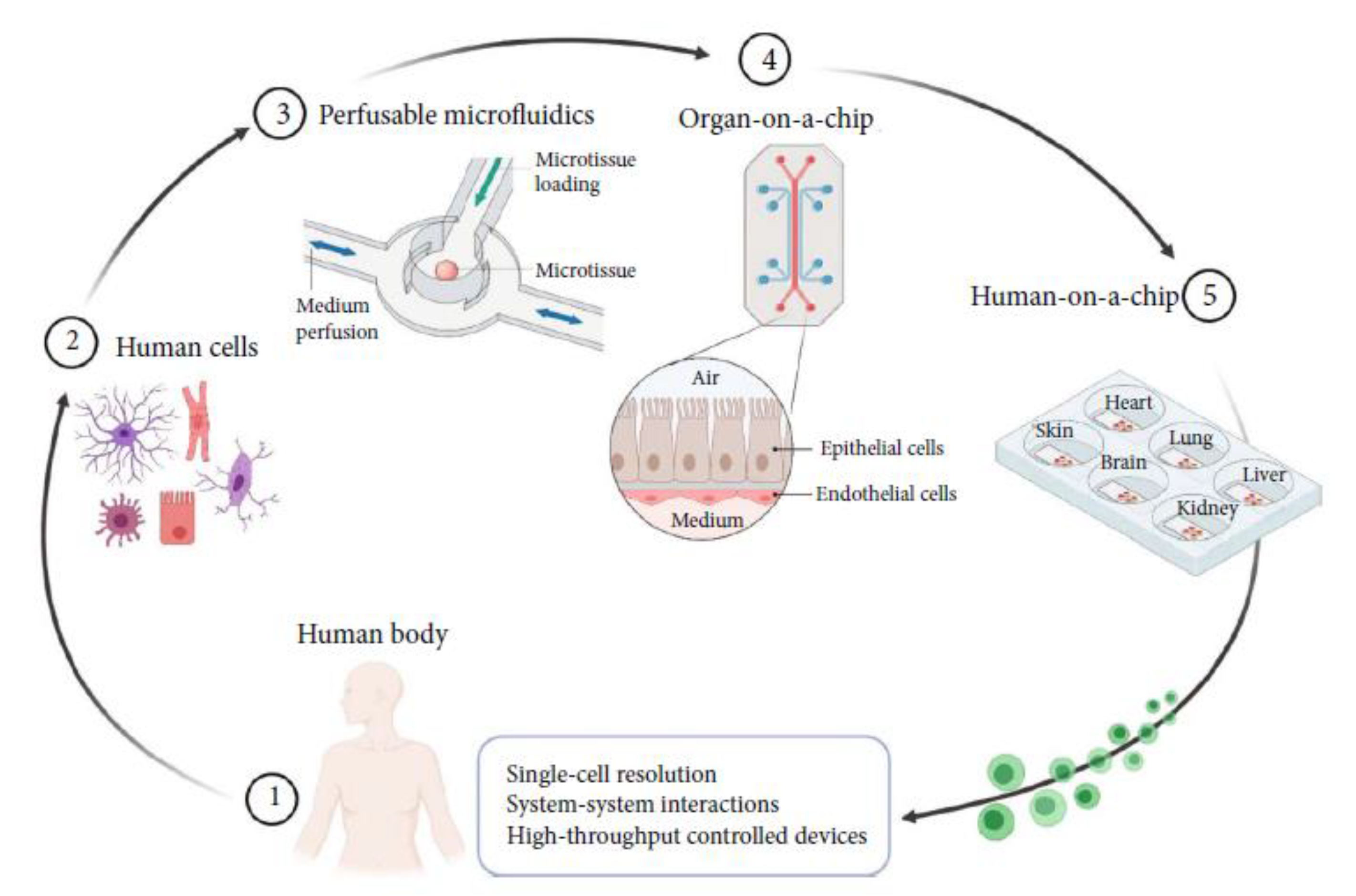
| Network | Platform | Function | Refs |
|---|---|---|---|
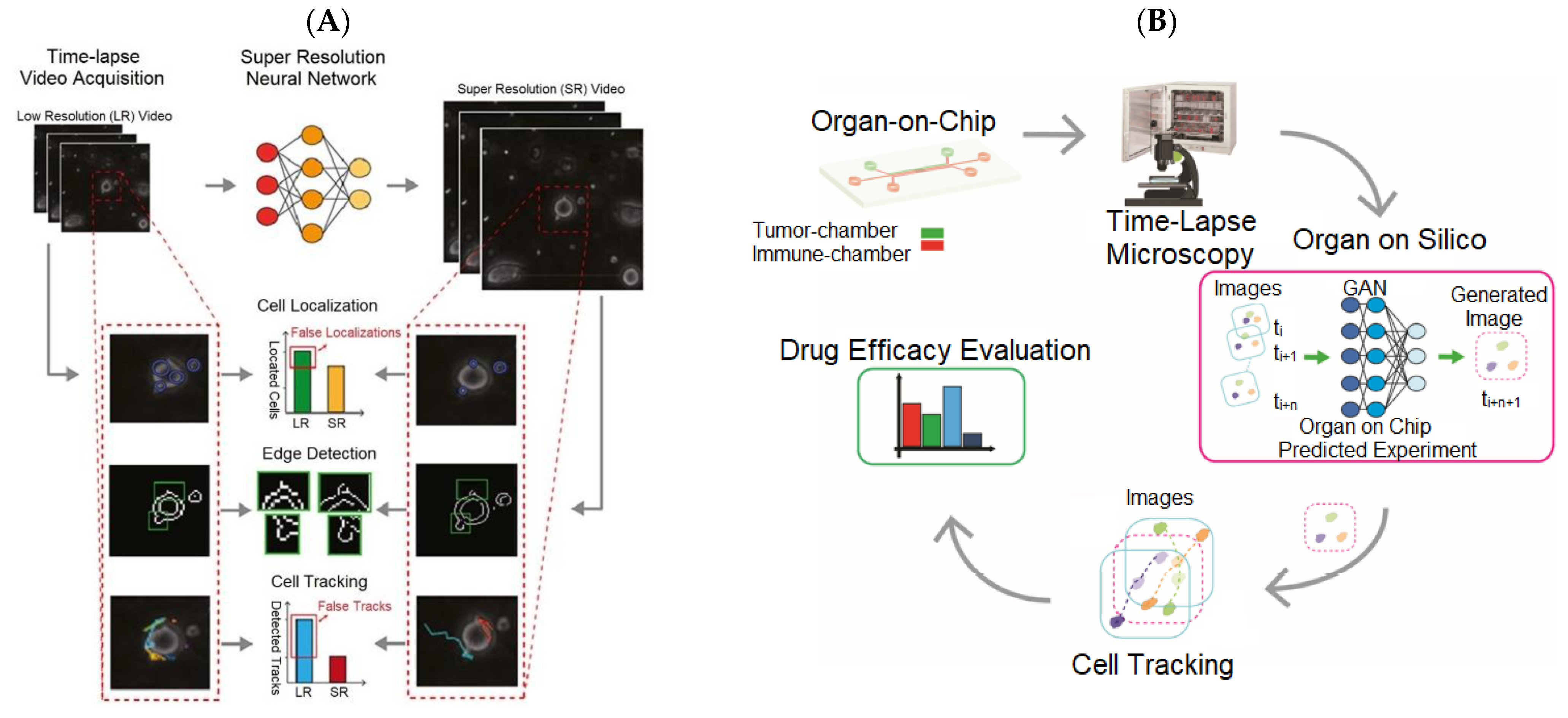
| CNN | OoC | Improve the spatial resolution of TLM videos for observing cell dynamics and interactions. | [41] |
| GAN | OoC | Providing high-throughput videos with more cell content for accurately reconstructing cell-interaction dynamics. | [43] |
| CNN | OoC | Segment nerve cell images into axons, myelins, and background. | [44] |
| AlexNET | OoC | Classify the treated cancer cells and untreated cancer cells according to their trajectories. | [54] |
| NN | Lung-on-a-chip | Predict the toxicity for drug discovery via image analysis. | [67] |
| GAN, CNN | Gut-on-a-chip | Enhance the resolution of confocal fluorescence photographs and conduct a better analysis of protein expression. | [68] |
| CNN, RNN | Brain-on-a-chip, Brain organoid-on-a-chip | Read the data for analysis in both HCS and HTS via deep learning rather than in a labor-intensive manner. | [69] |
| CNN | Kidney-on-a-chip | Improve early prediction of DIKI. | [70,71,72,73] |
| CNN | Skin-on-a-chip | Classify the skin cells as healthy or unhealthy based on metabolic parameters acquired from sensors. | [74] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, M.; Xiao, G.; Shao, M.; Zhang, Y.S. The Synergy between Deep Learning and Organs-on-Chips for High-Throughput Drug Screening: A Review. Biosensors 2023, 13, 389. https://doi.org/10.3390/bios13030389
Dai M, Xiao G, Shao M, Zhang YS. The Synergy between Deep Learning and Organs-on-Chips for High-Throughput Drug Screening: A Review. Biosensors. 2023; 13(3):389. https://doi.org/10.3390/bios13030389
Chicago/Turabian StyleDai, Manna, Gao Xiao, Ming Shao, and Yu Shrike Zhang. 2023. "The Synergy between Deep Learning and Organs-on-Chips for High-Throughput Drug Screening: A Review" Biosensors 13, no. 3: 389. https://doi.org/10.3390/bios13030389
APA StyleDai, M., Xiao, G., Shao, M., & Zhang, Y. S. (2023). The Synergy between Deep Learning and Organs-on-Chips for High-Throughput Drug Screening: A Review. Biosensors, 13(3), 389. https://doi.org/10.3390/bios13030389







