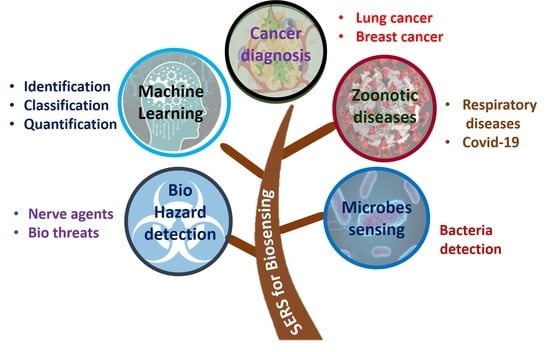
Recent Trends in SERS-Based Plasmonic Sensors for Disease Diagnostics, Biomolecules Detection, and Machine Learning Techniques
Abstract
1. Introduction
2. SERS for Disease Diagnosis
2.1. Cancer Diagnosis and Theranostics
2.1.1. Lung Cancer
2.1.2. Breast Cancer
2.1.3. Miscellaneous
2.2. SARS-CoV-2 and Other Respiratory Diseases
3. SERS-Based Detection of Microorganisms
3.1. Bacteria Sensing
3.2. Sensing of Biohazardous Molecules for Homeland Security
4. Machine Learning in SERS-Based Biosensing
4.1. Introduction to Machine Learning
4.2. Identification
4.3. Quantification
4.4. Classification
5. Conclusions and Scope
Author Contributions
Funding
Conflicts of Interest
References
- Stewart, M.E.; Anderton, C.R.; Thompson, L.B.; Maria, J.; Gray, S.K.; Rogers, J.A.; Nuzzo, R.G. Nanostructured Plasmonic Sensors. Chem. Rev. 2008, 108, 494–521. [Google Scholar] [CrossRef]
- Costanzo, H.; Gooch, J.; Frascione, N. Nanomaterials for Optical Biosensors in Forensic Analysis. Talanta 2023, 253, 123945. [Google Scholar] [CrossRef]
- Chen, G.; Chen, Y.; Huang, W.; Shi, Y. Plasmonic Nanobiosensors for Detection of Different Targets. In Proceedings of the Second International Conference on Medical Imaging and Additive Manufacturing (ICMIAM 2022), Xiamen, China, 25–27 February 2022. [Google Scholar] [CrossRef]
- Sadani, K.; Nag, P.; Thian, X.Y.; Mukherji, S. Enzymatic Optical Biosensors for Healthcare Applications. Biosens. Bioelectron. X 2022, 12, 100278. [Google Scholar] [CrossRef]
- Erkmen, C.; Selcuk, O.; Unal, D.N.; Kurbanoglu, S.; Uslu, B. Layer-by-Layer Modification Strategies for Electrochemical Detection of Biomarkers. Biosens. Bioelectron. X 2022, 12, 100270. [Google Scholar] [CrossRef]
- Spillman, W.B. Fiber Optic Biosensors; Elsevier: Amsterdam, The Netherlands, 2011; Volume 3, ISBN 9780470126844. [Google Scholar]
- Kazanskiy, N.L.; Khonina, S.N.; Butt, M.A.; Kaźmierczak, A.; Piramidowicz, R. State-of-the-Art Optical Devices for Biomedical Sensing Applications—A Review. Electronics 2021, 10, 973. [Google Scholar] [CrossRef]
- Ramirez, J.C.; Grajales García, D.; Maldonado, J.; Fernández-Gavela, A. Current Trends in Photonic Biosensors: Advances towards Multiplexed Integration. Chemosensors 2022, 10, 398. [Google Scholar] [CrossRef]
- Chadha, U.; Bhardwaj, P.; Agarwal, R.; Rawat, P.; Agarwal, R.; Gupta, I.; Panjwani, M.; Singh, S.; Ahuja, C.; Selvaraj, S.K.; et al. Recent Progress and Growth in Biosensors Technology: A Critical Review. J. Ind. Eng. Chem. 2022, 109, 21–51. [Google Scholar] [CrossRef]
- Dutta, G. Next-Generation Nanobiosensor Devices for Point-of-Care Diagnostics; Springer: Singapore, 2023; ISBN 9789811971303. [Google Scholar] [CrossRef]
- Ahangari, A.; Mahmoodi, P.; Mohammadzadeh, A. Advanced Nano Biosensors for Rapid Detection of Zoonotic Bacteria. Biotechnol. Bioeng. 2022, 120, 41–56. [Google Scholar] [CrossRef]
- Taha, B.A.; Al Mashhadany, Y.; Bachok, N.N.; Ashrif, A.; Bakar, A.; Hafiz Mokhtar, M.H.; Dzulkefly Bin Zan, M.S.; Arsad, N. Detection of COVID-19 Virus on Surfaces Using Photonics: Challenges and Perspectives. Diagnostics 2021, 11, 1119. [Google Scholar] [CrossRef]
- Soma, V.R.; Podagatlapalli, G.K.; Hamad, S.; Mechanics, F. Ultrafast Laser Ablation in Liquids for Nanomaterials and Applications. J. Nanosci. Nanotechnol. 2014, 14, 1364–1388. [Google Scholar] [CrossRef]
- Soler, M.; Lechuga, L.M. Principles, Technologies, and Applications of Plasmonic Biosensors. J. Appl. Phys. 2021, 129, 111102. [Google Scholar] [CrossRef]
- Barbillon, G. Plasmonics and Its Applications. Materials 2019, 12, 1502. [Google Scholar] [CrossRef] [PubMed]
- Michaels, A.M.; Jiang, J.; Brus, L. Ag Nanocrystal Junctions as the Site for Surface-Enhanced Raman Scattering of Single Rhodamine 6G Molecules. J. Phys. Chem. B 2000, 104, 11965–11971. [Google Scholar] [CrossRef]
- Golightly, R.S.; Doering, W.E.; Natan, M.J. Surface-Enhanced Raman Spectroscopy and Homeland Security: A Perfect Match? ACS Nano 2009, 3, 2859–2869. [Google Scholar] [CrossRef] [PubMed]
- Vendamani, V.S.; Beeram, R.; Nageswara Rao, S.V.S.; Pathak, A.P.; Soma, V.R. Trace Level Detection of Explosives and Pesticides Using Robust, Low-Cost, Free-Standing Silver Nanoparticles Decorated Porous Silicon. Opt. Express 2021, 29, 30045. [Google Scholar] [CrossRef]
- Liu, C.; Xu, D.; Dong, X.; Huang, Q. A Review: Research Progress of SERS-Based Sensors for Agricultural Applications. Trends Food Sci. Technol. 2022, 128, 90–101. [Google Scholar] [CrossRef]
- Zhang, D.; Pu, H.; Huang, L.; Sun, D.W. Advances in Flexible Surface-Enhanced Raman Scattering (SERS) Substrates for Nondestructive Food Detection: Fundamentals and Recent Applications. Trends Food Sci. Technol. 2021, 109, 690–701. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Lin, Z. Recent Developments and Applications of Surface Plasmon Resonance Biosensors for the Detection of Mycotoxins in Foodstuffs. Food Chem. 2012, 132, 1549–1554. [Google Scholar] [CrossRef]
- Dies, H.; Raveendran, J.; Escobedo, C.; Docoslis, A. Rapid Identification and Quantification of Illicit Drugs on Nanodendritic Surface-Enhanced Raman Scattering Substrates. Sens. Actuators B Chem. 2018, 257, 382–388. [Google Scholar] [CrossRef]
- Vendamani, V.S.; Beeram, R.; Rao, S.V.S.N.; Rao, S.V. Protocol for Designing AuNP-Capped Ag Dendrites as Surface-Enhanced Raman Scattering Sensors for Trace Molecular Detection Protocol for Designing AuNP-Capped Ag Dendrites as Surface-Enhanced Raman Scattering Sensors for Trace Molecular Detection. STAR Protoc. 2023, 4, 102068. [Google Scholar] [CrossRef]
- He, L.; Rodda, T.; Haynes, C.L.; Deschaines, T.; Strother, T.; Diez-Gonzalez, F.; Labuza, T.P. Detection of a Foreign Protein in Milk Using Surface-Enhanced Raman Spectroscopy Coupled with Antibody-Modified Silver Dendrites. Anal. Chem. 2011, 83, 1510–1513. [Google Scholar] [CrossRef]
- Jebakumari, K.A.E.; Murugasenapathi, N.K. Engineered Two-Dimensional Nanostructures as SERS Substrates for Biomolecule Sensing: A Review. Biosensors 2023, 13, 102. [Google Scholar] [CrossRef]
- Bantz, K.C.; Meyer, A.F.; Wittenberg, N.J.; Im, H.; Kurtuluş, Ö.; Lee, S.H.; Lindquist, N.C.; Oh, S.H.; Haynes, C.L. Recent Progress in SERS Biosensing. Phys. Chem. Chem. Phys. 2011, 13, 11551–11567. [Google Scholar] [CrossRef] [PubMed]
- Vendamani, V.S.; Nageswara Rao, S.V.S.; Venugopal Rao, S.; Kanjilal, D.; Pathak, A.P. Three-Dimensional Hybrid Silicon Nanostructures for Surface Enhanced Raman Spectroscopy Based Molecular Detection. J. Appl. Phys. 2018, 123, 014301. [Google Scholar] [CrossRef]
- Huang, Z.; Zhang, A.; Zhang, Q.; Cui, D. Nanomaterial-Based SERS Sensing Technology for Biomedical Application. J. Mater. Chem. B 2019, 7, 3755–3774. [Google Scholar] [CrossRef]
- Szaniawska, A.; Kudelski, A. Applications of Surface-Enhanced Raman Scattering in Biochemical and Medical Analysis. Front. Chem. 2021, 9, 664134. [Google Scholar] [CrossRef]
- Chen, Y.; An, Q.; Teng, K.; Liu, C.; Sun, F.; Li, G. Application of SERS in In-Vitro Biomedical Detection. Chem. Asian J. 2022, 18, e202201194. [Google Scholar] [CrossRef]
- Hegde, M.; Pai, P.; Gangadhar Shetty, M.; Sundara Babitha, K. Gold Nanoparticle Based Biosensors for Rapid Pathogen Detection: A Review. Environ. Nanotechnol. Monit. Manag. 2022, 18, 100756. [Google Scholar] [CrossRef]
- Fleischmann, M.; Hendra, P.J.; McQuillan, A.J. Raman Spectra of Pyridine Adsorbed at a Silver Electrode. Chem. Phys. Lett. 1974, 26, 163–166. [Google Scholar] [CrossRef]
- Jeanmaire, D.L.; Duyne, R.P.V.A.N. Surface Raman spectroelectrochemistry: Part I. Heterocyclic, aromatic, and aliphatic amines adsorbed on the anodized silver electrode. J. Electroanal. Chem. Interfacial Electrochem. 1977, 84, 1–20. [Google Scholar] [CrossRef]
- Albrecht, M.G.; Creighton, J.A. Anomalously Intense Raman Spectra of Pyridine at a Silver Electrode. J. Am. Chem. Soc. 1977, 99, 5215–5217. [Google Scholar] [CrossRef]
- Pilot, R.; Signorini, R.; Durante, C.; Orian, L.; Bhamidipati, M.; Fabris, L. A Review on Surface-Enhanced Raman Scattering. Biosensors 2019, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Le Ru, E.; Etchegoin, P. Principles of Surface Enhanced Raman Spectroscopy and Related Plasmonic Effects; Elseveir: Amsterdam, The Netherlands, 2008. [Google Scholar]
- Etchegoin, P.G.; Ru, E.C.L. Basic Electromagnetic Theory of SERS. In Surface Enhanced Raman Spectroscopy: Analytical, Biophysical and Life Science Applications; John Wiley & Sons: Hoboken, NJ, USA, 2011; pp. 1–37. ISBN 9783527325672. [Google Scholar]
- Le Ru, E.C.; Blackie, E.; Meyer, M.; Etchegoint, P.G. Surface Enhanced Raman Scattering Enhancement Factors: A Comprehensive Study. J. Phys. Chem. C 2007, 111, 13794–13803. [Google Scholar] [CrossRef]
- Sharma, B.; Frontiera, R.R.; Henry, A.I.; Ringe, E.; Van Duyne, R.P. SERS: Materials, Applications, and the Future. Mater. Today 2012, 15, 16–25. [Google Scholar] [CrossRef]
- Li, Q.; Huo, H.; Wu, Y.; Chen, L.; Su, L.; Zhang, X.; Song, J.; Yang, H. Design and Synthesis of SERS Materials for In Vivo Molecular Imaging and Biosensing. Adv. Sci. 2023, 2023, 2202051. [Google Scholar] [CrossRef]
- Israelsen, N.D.; Hanson, C.; Vargis, E. Nanoparticle Properties and Synthesis Effects on Surface-Enhanced Raman Scattering Enhancement Factor: An Introduction. Sci. World J. 2015, 2015, 124582. [Google Scholar] [CrossRef]
- Wang, A.X.; Kong, X. Review of Recent Progress of Plasmonic Materials and Nano-Structures for Surface-Enhanced Raman Scattering. Materials 2015, 8, 3024–3052. [Google Scholar] [CrossRef]
- Moram, S.S.B.; Byram, C.; Soma, V.R. Gold-Nanoparticle- and Nanostar-Loaded Paper-Based SERS Substrates for Sensing Nanogram-Level Picric Acid with a Portable Raman Spectrometer. Bull. Mater. Sci. 2020, 43, 53. [Google Scholar] [CrossRef]
- Zhang, Z.; Guan, R.; Li, J. Engineering Rational SERS Nanotags for Parallel Detection of Multiple Cancer Circulating Biomarkers. Chemosensors. 2023, 11, 110. [Google Scholar] [CrossRef]
- Pilot, R.; Massari, M. Silver Nanoparticle Aggregates: Wavelength Dependence of Their SERS Properties in the First Transparency Window of Biological Tissues. Chem. Phys. Impact 2021, 2, 100014. [Google Scholar] [CrossRef]
- Zhang, Y.; Mi, X.; Tan, X.; Xiang, R. Recent Progress on Liquid Biopsy Analysis Using Surface-Enhanced Raman Spectroscopy. Theranostics 2019, 9, 491–525. [Google Scholar] [CrossRef] [PubMed]
- Aitekenov, S.; Sultangaziyev, A.; Ilyas, A.; Dyussupova, A.; Boranova, A.; Gaipov, A.; Bukasov, R. Surface-Enhanced Raman Spectroscopy (SERS) for Protein Determination in Human Urine. Sens. Bio-Sens. Res. 2022, 38, 100535. [Google Scholar] [CrossRef]
- Akgönüllü, S.; Denizli, A. Recent Advances in Optical Biosensing Approaches for Biomarkers Detection. Biosens. Bioelectron. X 2022, 12, 100269. [Google Scholar] [CrossRef]
- Lussier, F.; Thibault, V.; Charron, B.; Wallace, G.Q.; Masson, J.F. Deep Learning and Artificial Intelligence Methods for Raman and Surface-Enhanced Raman Scattering. TrAC Trends Anal. Chem. 2020, 124, 115796. [Google Scholar] [CrossRef]
- Lin, X.; Lin, D.; Chen, Y.; Lin, J.; Weng, S.; Song, J.; Feng, S. High Throughput Blood Analysis Based on Deep Learning Algorithm and Self-Positioning Super-Hydrophobic SERS Platform for Non-Invasive Multi-Disease Screening. Adv. Funct. Mater. 2021, 31, 2103382. [Google Scholar] [CrossRef]
- Breuch, R.; Klein, D.; Siefke, E.; Hebel, M.; Herbert, U.; Wickleder, C.; Kaul, P. Differentiation of Meat-Related Microorganisms Using Paper-Based Surface-Enhanced Raman Spectroscopy Combined with Multivariate Statistical Analysis. Talanta 2020, 219, 121315. [Google Scholar] [CrossRef]
- Ilkhani, H.; Hughes, T.; Li, J.; Zhong, C.J.; Hepel, M. Nanostructured SERS-Electrochemical Biosensors for Testing of Anticancer Drug Interactions with DNA. Biosens. Bioelectron. 2016, 80, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Leong, S.X.; Leong, Y.X.; Tan, E.X.; Sim, H.Y.F.; Koh, C.S.L.; Lee, Y.H.; Chong, C.; Ng, L.S.; Chen, J.R.T.; Pang, D.W.C.; et al. Noninvasive and Point-of-Care Surface-Enhanced Raman Scattering (SERS)-Based Breathalyzer for Mass Screening of Coronavirus Disease 2019 (COVID-19) under 5 Min. ACS Nano 2022, 16, 2629–2639. [Google Scholar] [CrossRef]
- Bharati, M.S.S.; Soma, V.R. Flexible SERS Substrates for Hazardous Materials Detection: Recent Advances. Opto-Electron. Adv. 2021, 4, 210048. [Google Scholar] [CrossRef]
- Ali, A.; Nettey-Oppong, E.E.; Effah, E.; Yu, C.Y.; Muhammad, R.; Soomro, T.A.; Byun, K.M.; Choi, S.H. Miniaturized Raman Instruments for SERS-Based Point-of-Care Testing on Respiratory Viruses. Biosensors 2022, 12, 590. [Google Scholar] [CrossRef]
- Mejía-Salazar, J.R.; Oliveira, O.N. Plasmonic Biosensing. Chem. Rev. 2018, 118, 10617–10625. [Google Scholar] [CrossRef]
- Han, X.; Liu, K.; Sun, C. Plasmonics for Biosensing. Materials 2019, 12, 1411. [Google Scholar] [CrossRef] [PubMed]
- Shrivastav, A.M.; Cvelbar, U.; Abdulhalim, I. A Comprehensive Review on Plasmonic-Based Biosensors Used in Viral Diagnostics. Commun. Biol. 2021, 4, 70. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ren, Z.H.; Zhao, W.M.; Wang, L.; Yan, X.; Zhu, A.S.; Qiu, F.M.; Zhang, K.K. Research Advances on Surface Plasmon Resonance Biosensors. Nanoscale 2022, 14, 564–591. [Google Scholar] [CrossRef] [PubMed]
- Homola, J. Present and Future of Surface Plasmon Resonance Biosensors. Anal. Bioanal. Chem. 2003, 377, 528–539. [Google Scholar] [CrossRef]
- Piliarik, M.; Vaisocherová, H.; Homola, J. Surface Plasmon Resonance Biosensing. In Biosensors and Biodetection; Humana Press: Totowa, NJ, USA, 2009; pp. 65–88. [Google Scholar] [CrossRef]
- Hong, Y.; Huh, Y.M.; Yoon, D.S.; Yang, J. Nanobiosensors Based on Localized Surface Plasmon Resonance for Biomarker Detection. J. Nanomater. 2012, 2012, 759830. [Google Scholar] [CrossRef]
- Unser, S.; Bruzas, I.; He, J.; Sagle, L. Localized Surface Plasmon Resonance Biosensing: Current Challenges and Approaches. Sensors 2015, 15, 15684–15716. [Google Scholar] [CrossRef]
- Brolo, A.G. Plasmonics for Future Biosensors. Nat. Photonics 2012, 6, 709–713. [Google Scholar] [CrossRef]
- Liu, J.; Jalali, M.; Mahshid, S.; Wachsmann-Hogiu, S. Are Plasmonic Optical Biosensors Ready for Use in Point-of-Need Applications? Analyst 2020, 145, 364–384. [Google Scholar] [CrossRef]
- Moore, T.J.; Moody, A.S.; Payne, T.D.; Sarabia, G.M.; Daniel, A.R.; Sharma, B. In Vitro and in Vivo Sers Biosensing for Disease Diagnosis. Biosensors 2018, 8, 46. [Google Scholar] [CrossRef]
- Alvarez-Puebla, R.A.; Liz-Marzán, L.M. SERS-Based Diagnosis and Biodetection. Small 2010, 6, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, S.; Song, X.; Wang, H.; Wang, J.; Wang, Y.; Huang, J.; Yu, J. Robust and Universal SERS Sensing Platform for Multiplexed Detection of Alzheimer’s Disease Core Biomarkers Using PAapt-AuNPs Conjugates. ACS Sens. 2019, 4, 2140–2149. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Cho, S.; Kim, M.; Jung, Y.S. Carboxylic Acid-Functionalized, Graphitic Layer-Coated Three-Dimensional SERS Substrate for Label-Free Analysis of Alzheimer’s Disease Biomarkers. Nano Lett. 2020, 20, 2576–2584. [Google Scholar] [CrossRef]
- Dang, H.; Joung, Y.; Jeong, C.; Jeon, C.S.; Pyun, S.H.; Park, S.-G.; Choo, J. Nanoplasmonic Assay Platforms for Reproducible SERS Detection of Alzheimer’s Disease Biomarker. Bull. Korean Chem. Soc. 2023, 2023, 1. [Google Scholar] [CrossRef]
- Momenpour, A.; Lima, P.D.A.; Chen, Y.-A.; Tzeng, C.-R.; Tsang, B.K.; Anis, H. Surface-Enhanced Raman Scattering for the Detection of Polycystic Ovary Syndrome. Biomed. Opt. Express 2018, 9, 801. [Google Scholar] [CrossRef] [PubMed]
- Lyandres, O.; Shah, N.C.; Yonzon, C.R.; Walsh, J.T.; Glucksberg, M.R.; Van Duyne, R.P. Real-Time Glucose Sensing by Surface-Enhanced Raman Spectroscopy in Bovine Plasma Facilitated by a Mixed Decanethiol/Mercaptohexanol Partition Layer. Anal. Chem. 2005, 77, 6134–6139. [Google Scholar] [CrossRef]
- Qi, G.; Jia, K.; Fu, C.; Xu, S.; Xu, W. A Highly Sensitive SERS Sensor for Quantitative Analysis of Glucose Based on the Chemical Etching of Silver Nanoparticles. J. Opt. 2015, 17, 114020. [Google Scholar] [CrossRef]
- Rong, Z.; Xiao, R.; Xing, S.; Xiong, G.; Yu, Z.; Wang, L.; Jia, X.; Wang, K.; Cong, Y.; Wang, S. SERS-Based Lateral Flow Assay for Quantitative Detection of C-Reactive Protein as an Early Bio-Indicator of a Radiation-Induced Inflammatory Response in Nonhuman Primates. Analyst 2018, 143, 2115–2121. [Google Scholar] [CrossRef]
- Li, B.; Wu, Y.; Wang, Z.; Xing, M.; Xu, W.; Zhu, Y.; Du, P.; Wang, X.; Yang, H. Non-Invasive Diagnosis of Crohn’s Disease Based on SERS Combined with PCA-SVM. Anal. Methods 2021, 13, 5264–5273. [Google Scholar] [CrossRef]
- Xu, H.; Bjerneld, E.J.; Käll, M.; Börjesson, L. Spectroscopy of Single Hemoglobin Molecules by Surface Enhanced Raman Scattering. Phys. Rev. Lett. 1999, 83, 4357–4360. [Google Scholar] [CrossRef]
- World Health Organization Report. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 30 January 2023).
- Blanco-Formoso, M.; Alvarez-Puebla, R.A. Cancer Diagnosis through Sers and Other Related Techniques. Int. J. Mol. Sci. 2020, 21, 2253. [Google Scholar] [CrossRef]
- Guerrini, L.; Pazos-Perez, N.; Garcia-Rico, E.; Alvarez-Puebla, R. Cancer Characterization and Diagnosis with SERS-Encoded Particles. Cancer Nanotechnol. 2017, 8, 5. [Google Scholar] [CrossRef]
- Kaur, B.; Kumar, S.; Kaushik, B.K. Recent Advancements in Optical Biosensors for Cancer Detection. Biosens. Bioelectron. 2022, 197, 113805. [Google Scholar] [CrossRef]
- Thenrajan, T.; Wilson, J. Biosensors for Cancer Theranostics. Biosens. Bioelectron. X 2022, 12, 100232. [Google Scholar] [CrossRef]
- Falkowski, P.; Lukaszewski, Z.; Gorodkiewicz, E. Potential of Surface Plasmon Resonance Biosensors in Cancer Detection. J. Pharm. Biomed. Anal. 2021, 194, 113802. [Google Scholar] [CrossRef]
- Fu, Q.; Zhang, X.; Song, J.; Yang, H. Plasmonic Gold Nanoagents for Cancer Imaging and Therapy. View 2021, 2, 20200149. [Google Scholar] [CrossRef]
- Azzouz, A.; Hejji, L.; Kim, K.H.; Kukkar, D.; Souhail, B.; Bhardwaj, N.; Brown, R.J.C.; Zhang, W. Advances in Surface Plasmon Resonance–Based Biosensor Technologies for Cancer Biomarker Detection. Biosens. Bioelectron. 2022, 197, 113767. [Google Scholar] [CrossRef] [PubMed]
- Sugumaran, S.; Jamlos, M.F.; Ahmad, M.N.; Bellan, C.S.; Schreurs, D. Nanostructured Materials with Plasmonic Nanobiosensors for Early Cancer Detection: A Past and Future Prospect. Biosens. Bioelectron. 2018, 100, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Fattahi, Z.; Khosroushahi, A.Y.; Hasanzadeh, M. Recent Progress on Developing of Plasmon Biosensing of Tumor Biomarkers: Efficient Method towards Early Stage Recognition of Cancer. Biomed. Pharmacother. 2020, 132, 110850. [Google Scholar] [CrossRef] [PubMed]
- Bellassai, N.; D’Agata, R.; Jungbluth, V.; Spoto, G. Surface Plasmon Resonance for Biomarker Detection: Advances in Non-Invasive Cancer Diagnosis. Front. Chem. 2019, 7, 570. [Google Scholar] [CrossRef] [PubMed]
- Usman, F.; Dennis, J.O.; Aljameel, A.I.; Ali, M.K.M.; Aldaghri, O.; Ibnaouf, K.H.; Zango, Z.U.; Beygisangchin, M.; Alsadig, A.; Meriaudeau, F. Plasmonic Biosensors for the Detection of Lung Cancer Biomarkers: A Review. Chemosensors 2021, 9, 326. [Google Scholar] [CrossRef]
- Yin, B.; Ho, W.K.H.; Xia, X.; Chan, C.K.W.; Zhang, Q.; Ng, Y.M.; Lam, C.Y.K.; Cheung, J.C.W.; Wang, J.; Yang, M.; et al. A Multilayered Mesoporous Gold Nanoarchitecture for Ultraeffective Near-Infrared Light-Controlled Chemo/Photothermal Therapy for Cancer Guided by SERS Imaging. Small 2023, 2023, 2206762. [Google Scholar] [CrossRef] [PubMed]
- Constantinou, M.; Hadjigeorgiou, K.; Abalde-cela, S.; Andreou, C. Label-Free Sensing with Metal Nanostructure-Based Surface- Enhanced Raman Spectroscopy for Cancer Diagnosis. ACS Appl. Nano Mater. 2022, 5, 12276–12299. [Google Scholar] [CrossRef] [PubMed]
- Rajput, S.; Pink, D.; Findlay, S.; Woolner, E.; Lewis, J.D.; McDermott, M.T. Application of Surface-Enhanced Raman Spectroscopy to Guide Therapy for Advanced Prostate Cancer Patients. ACS Sens. 2022, 7, 827–838. [Google Scholar] [CrossRef]
- Avci, E.; Yilmaz, H.; Sahiner, N.; Tuna, B.G.; Cicekdal, M.B.; Eser, M.; Basak, K.; Altıntoprak, F.; Zengin, I.; Dogan, S.; et al. Label-Free Surface Enhanced Raman Spectroscopy for Cancer Detection. Cancers 2022, 14, 5021. [Google Scholar] [CrossRef]
- Guerrini, L.; Alvarez-puebla, R.A. Surface-Enhanced Raman Spectroscopy in Cancer Diagnosis, Prognosis and Monitoring. Cancers 2019, 11, 748. [Google Scholar] [CrossRef]
- Pollap, A.; Paweł, S. Recent Advances in Sandwich SERS Immunosensors for Cancer Detection. Int. J. Mol. Sci. 2022, 23, 4740. [Google Scholar] [CrossRef]
- Vendrell, M.; Maiti, K.K.; Dhaliwal, K.; Chang, Y.T. Surface-Enhanced Raman Scattering in Cancer Detection and Imaging. Trends Biotechnol. 2013, 31, 249–257. [Google Scholar] [CrossRef]
- Moisoiu, V.; Iancu, S.D.; Stefancu, A.; Moisoiu, T.; Pardini, B.; Dragomir, M.P.; Crisan, N.; Avram, L.; Crisan, D.; Andras, I.; et al. SERS Liquid Biopsy: An Emerging Tool for Medical Diagnosis. Colloids Surfaces B Biointerfaces 2021, 208, 112064. [Google Scholar] [CrossRef]
- Shanmugasundaram, K.B.; Li, J.; Sina, A.I.; Wuethrich, A.; Trau, M. Toward Precision Oncology: SERS Microfluidic Systems for Multiplex Biomarker Analysis in Liquid Biopsy. Mater. Adv. 2022, 3, 1459–1471. [Google Scholar] [CrossRef]
- Song, C.Y.; Yang, Y.J.; Yang, B.Y.; Sun, Y.Z.; Zhao, Y.P.; Wang, L.H. An Ultrasensitive SERS Sensor for Simultaneous Detection of Multiple Cancer-Related MiRNAs. Nanoscale 2016, 8, 17365–17373. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.N.; Crawford, B.M.; Norton, S.J.; Vo-Dinh, T. Direct and Label-Free Detection of MicroRNA Cancer Biomarkers Using SERS-Based Plasmonic Coupling Interference (PCI) Nanoprobes. J. Phys. Chem. B 2019, 123, 10245–10251. [Google Scholar] [CrossRef]
- Guerrini, L.; Garcia-Rico, E.; O’loghlen, A.; Giannini, V.; Alvarez-Puebla, R.A. Surface-Enhanced Raman Scattering (Sers) Spectroscopy for Sensing and Characterization of Exosomes in Cancer Diagnosis. Cancers. 2021, 13, 2179. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.U.; Kim, S.; Sim, S.J. SERS-Based Nanoplasmonic Exosome Analysis: Enabling Liquid Biopsy for Cancer Diagnosis and Monitoring Progression. BioChip J. 2020, 14, 231–241. [Google Scholar] [CrossRef]
- Vo-Dinh, T.; Allain, L.R.; Stokes, D.L. Cancer Gene Detection Using Surface-Enhanced Raman Scattering (SERS). J. Raman Spectrosc. 2002, 33, 511–516. [Google Scholar] [CrossRef]
- Zhu, D.; Li, A.; Di, Y.; Wang, Z.; Shi, J.; Ni, X.; Wang, Y. Interference-Free SERS Nanoprobes for Labeling and Imaging of MT1-MMP in Breast Cancer Cells. Nanotechnology 2022, 33, 115702. [Google Scholar] [CrossRef]
- Lin, J.; Chen, R.; Feng, S.; Pan, J.; Li, Y.; Chen, G.; Cheng, M.; Huang, Z.; Yu, Y.; Zeng, H. A Novel Blood Plasma Analysis Technique Combining Membrane Electrophoresis with Silver Nanoparticle-Based SERS Spectroscopy for Potential Applications in Noninvasive Cancer Detection. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 655–663. [Google Scholar] [CrossRef]
- Fabris, L. SERS Tags: The Next Promising Tool for Personalized Cancer Detection? ChemNanoMat 2016, 2, 249–258. [Google Scholar] [CrossRef]
- Davis, R.M.; Campbell, J.L.; Burkitt, S.; Qiu, Z.; Kang, S.; Mehraein, M.; Miyasato, D.; Salinas, H.; Liu, J.T.C.; Zavaleta, C. A Raman Imaging Approach Using CD47 Antibody-Labeled SERS Nanoparticles for Identifying Breast Cancer and Its Potential to Guide Surgical Resection. Nanomaterials 2018, 8, 953. [Google Scholar] [CrossRef]
- Yang, J.; Wang, Z.; Zong, S.; Song, C.; Zhang, R.; Cui, Y. Distinguishing Breast Cancer Cells Using Surface-Enhanced Raman Scattering. Anal. Bioanal. Chem. 2012, 402, 1093–1100. [Google Scholar] [CrossRef]
- Dinish, U.S.; Balasundaram, G.; Chang, Y.T.; Olivo, M. Actively Targeted in Vivo Multiplex Detection of Intrinsic Cancer Biomarkers Using Biocompatible SERS Nanotags. Sci. Rep. 2014, 4, 4075. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gao, X.; Xu, C.; Liu, D. SERS Tags for Biomedical Detection and Bioimaging. Theranostics 2022, 12, 1870–1903. [Google Scholar] [CrossRef] [PubMed]
- Report on Lung Cancer. Available online: https://www.chestnet.org/newsroom/chest-news/2020/07/world-lung-cancer-day-2020-fact-sheet (accessed on 30 January 2023).
- Mao, Y.; Sun, Y.; Xue, J.; Lu, W.; Cao, X. Ultra-Sensitive and High Efficiency Detection of Multiple Non-Small Cell Lung Cancer-Related MiRNAs on a Single Test Line in Catalytic Hairpin Assembly-Based SERS-LFA Strip. Anal. Chim. Acta 2021, 1178, 338800. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Liu, Y.; Ran, M.; Lu, D.; Cao, X.; Wang, Y. SERS Platform Based on Bimetallic Au-Ag Nanowires-Decorated Filter Paper for Rapid Detection of MiR-196ain Lung Cancer Patients Serum. J. Chem. 2020, 2020, 5073451. [Google Scholar] [CrossRef]
- Cao, X.; Sun, Y.; Mao, Y.; Ran, M.; Liu, Y.; Lu, D.; Bi, C. Rapid and Sensitive Detection of Dual Lung Cancer-Associated MiRNA Biomarkers by a Novel SERS-LFA Strip Coupling with Catalytic Hairpin Assembly Signal Amplification. J. Mater. Chem. C 2021, 9, 3661–3671. [Google Scholar] [CrossRef]
- Cao, X.; Mao, Y.; Gu, Y.; Ge, S.; Lu, W.; Gu, Y.; Li, Z. Highly Sensitive and Simultaneous Detection of CtDNAs Related to Non-Small Cell Lung Cancer in Serum Using a Catalytic Hairpin Assembly Strategy in a SERS Microfluidic Chip. J. Mater. Chem. B 2022, 10, 6194–6206. [Google Scholar] [CrossRef]
- Cao, X.; Ge, S.; Zhou, X.; Mao, Y.; Sun, Y.; Lu, W.; Ran, M. A Dual-Signal Amplification Strategy Based on Pump-Free SERS Microfluidic Chip for Rapid and Ultrasensitive Detection of Non-Small Cell Lung Cancer-Related Circulating Tumour DNA in Mice Serum. Biosens. Bioelectron. 2022, 205, 114110. [Google Scholar] [CrossRef]
- Ye, L.P.; Hu, J.; Liang, L.; Zhang, C.Y. Surface-Enhanced Raman Spectroscopy for Simultaneous Sensitive Detection of Multiple MicroRNAs in Lung Cancer Cells. Chem. Commun. 2014, 50, 11883–11886. [Google Scholar] [CrossRef]
- Guo, T.; Li, W.; Qian, L.; Yan, X.; Cui, D.; Zhao, J.; Ni, H.; Zhao, X.; Zhang, Z.; Li, X.; et al. Highly-Selective Detection of EGFR Mutation Gene in Lung Cancer Based on Surface Enhanced Raman Spectroscopy and Asymmetric PCR. J. Pharm. Biomed. Anal. 2020, 190, 113522. [Google Scholar] [CrossRef]
- Shin, H.; Oh, S.; Hong, S.; Kang, M.; Kang, D.; Ji, Y.G.; Choi, B.H.; Kang, K.W.; Jeong, H.; Park, Y.; et al. Early-Stage Lung Cancer Diagnosis by Deep Learning-Based Spectroscopic Analysis of Circulating Exosomes. ACS Nano 2020, 14, 5435–5444. [Google Scholar] [CrossRef]
- Lei, J.; Yang, D.; Li, R.; Dai, Z.X.; Zhang, C.; Yu, Z.; Wu, S.; Pang, L.; Liang, S.; Zhang, Y. Label-Free Surface-Enhanced Raman Spectroscopy for Diagnosis and Analysis of Serum Samples with Different Types Lung Cancer. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 261, 120021. [Google Scholar] [CrossRef]
- Wang, Z.; Hong, Y.; Yan, H.; Luo, H.; Zhang, Y.; Li, L.; Lu, S.; Chen, Y.; Wang, D.; Su, Y.; et al. Fabrication of Optoplasmonic Particles through Electroless Deposition and the Application in SERS-Based Screening of Nodule-Involved Lung Cancer. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 279, 121483. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dong, Y.; Zhu, W.; Xie, D.; Zhao, Y.; Yang, D.; Li, M. Ultrasensitive Detection of Circulating Tumor DNA of Lung Cancer via an Enzymatically Amplified SERS-Based Frequency Shift Assay. ACS Appl. Mater. Interfaces 2019, 11, 18145–18152. [Google Scholar] [CrossRef] [PubMed]
- Jonak, S.T.; Liu, Z.; Liu, J.; Li, T.; D’Souza, B.V.; Schiaffino, A.; Oh, S.; Xie, Y.-H. Analyzing Bronchoalveolar Fluid Derived Small Extracellular Vesicles Using Single-Vesicle SERS for Non-Small Cell Lung Cancer Detection. Sens. Diagn. 2023, 2, 90–99. [Google Scholar] [CrossRef]
- Park, J.; Hwang, M.; Choi, B.; Jeong, H.; Jung, J.H.; Kim, H.K.; Hong, S.; Park, J.H.; Choi, Y. Exosome Classification by Pattern Analysis of Surface-Enhanced Raman Spectroscopy Data for Lung Cancer Diagnosis. Anal. Chem. 2017, 89, 6695–6701. [Google Scholar] [CrossRef]
- Wen, H.; Wang, H.; Hai, J.; He, S.; Chen, F.; Wang, B. Photochemical Synthesis of Porous CuFeSe 2 /Au Heterostructured Nanospheres as SERS Sensor for Ultrasensitive Detection of Lung Cancer Cells and Their Biomarkers. ACS Sustain. Chem. Eng. 2019, 7, 5200–5208. [Google Scholar] [CrossRef]
- Qiao, X.; Su, B.; Liu, C.; Song, Q.; Luo, D.; Mo, G.; Wang, T. Selective Surface Enhanced Raman Scattering for Quantitative Detection of Lung Cancer Biomarkers in Superparticle@MOF Structure. Adv. Mater. 2018, 30, 1702275. [Google Scholar] [CrossRef]
- Perumal, J.; Lee, P.; Dev, K.; Lim, H.Q.; Dinish, U.S.; Olivo, M. Machine Learning Assisted Real-Time Label-Free SERS Diagnoses of Malignant Pleural Effusion Due to Lung Cancer. Biosensors 2022, 12, 940. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Liu, X.; Man, B.; Yang, C.; Zhang, C.; Liu, M.; Zhang, Y.; Liu, L.; Chen, C. Label-Free and Stable Serum Analysis Based on Ag-NPs/PSi Surface-Enhanced Raman Scattering for Noninvasive Lung Cancer Detection. Biomed. Opt. Express 2018, 9, 4345. [Google Scholar] [CrossRef]
- Zhang, K.; Hao, C.; Huo, Y.; Man, B.; Zhang, C.; Yang, C.; Liu, M.; Chen, C. Label-Free Diagnosis of Lung Cancer with Tissue-Slice Surface-Enhanced Raman Spectroscopy and Statistical Analysis. Lasers Med. Sci. 2019, 34, 1849–1855. [Google Scholar] [CrossRef]
- Chon, H.; Lee, S.; Yoon, S.Y.; Chang, S.I.; Lim, D.W.; Choo, J. Simultaneous Immunoassay for the Detection of Two Lung Cancer Markers Using Functionalized SERS Nanoprobes. Chem. Commun. 2011, 47, 12515–12517. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Gao, Y.; Lu, Y.; Zhang, H.; Cai, C. High Specific Detection and Near-Infrared Photothermal Therapy of Lung Cancer Cells with High SERS Active Aptamer-Silver-Gold Shell-Core Nanostructures. Analyst 2013, 138, 6501–6510. [Google Scholar] [CrossRef]
- Chen, Y.W.; Liu, T.Y.; Chen, P.J.; Chang, P.H.; Chen, S.Y. A High-Sensitivity and Low-Power Theranostic Nanosystem for Cell SERS Imaging and Selectively Photothermal Therapy Using Anti-EGFR-Conjugated Reduced Graphene Oxide/Mesoporous Silica/AuNPs Nanosheets. Small 2016, 12, 1458–1468. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ye, X.; Xu, G.; Jin, X.; Luan, M.; Lou, J.; Wang, L.; Huang, C.; Ye, J. Identification and Distinction of Non-Small-Cell Lung Cancer Cells by Intracellular SERS Nanoprobes. RSC Adv. 2016, 6, 5401–5407. [Google Scholar] [CrossRef]
- Huang, Y.; Xie, T.; Zou, K.; Gu, Y.; Yang, G.; Zhang, F.; Qu, L.L.; Yang, S. Ultrasensitive SERS Detection of Exhaled Biomarkers of Lung Cancer Using a Multifunctional Solid Phase Extraction Membrane. Nanoscale 2021, 13, 13344–13352. [Google Scholar] [CrossRef]
- Cai, C.; Liu, Y.; Li, J.; Wang, L.; Zhang, K. Serum Fingerprinting by Slippery Liquid-Infused Porous SERS for Non-Invasive Lung Cancer Detection. Analyst 2022, 147, 4426–4432. [Google Scholar] [CrossRef]
- Sivashanmugan, K.; Huang, W.L.; Lin, C.H.; Liao, J.D.; Lin, C.C.; Su, W.C.; Wen, T.C. Bimetallic Nanoplasmonic Gap-Mode SERS Substrate for Lung Normal and Cancer-Derived Exosomes Detection. J. Taiwan Inst. Chem. Eng. 2017, 80, 149–155. [Google Scholar] [CrossRef]
- Qian, K.; Wang, Y.; Hua, L.; Chen, A.; Zhang, Y. New Method of Lung Cancer Detection by Saliva Test Using Surface-Enhanced Raman Spectroscopy. Thorac. Cancer 2018, 9, 1556–1561. [Google Scholar] [CrossRef]
- Yang, T.; Guo, X.; Wu, Y.; Wang, H.; Fu, S.; Wen, Y.; Yang, H. Facile and Label-Free Detection of Lung Cancer Biomarker in Urine by Magnetically Assisted Surface-Enhanced Raman Scattering. ACS Appl. Mater. Interfaces 2014, 6, 20985–20993. [Google Scholar] [CrossRef]
- Breast Cancer Report. Available online: https://www.cancer.org/cancer/breast-cancer/about/how-common-is-breast-cancer.html (accessed on 30 January 2023).
- Moisoiu, T.; Iancu, S.D.; Burghelea, D.; Dragomir, M.P.; Iacob, G.; Stefancu, A.; Cozan, R.G.; Antal, O.; Bálint, Z.; Muntean, V.; et al. SERS Liquid Biopsy Profiling of Serum for the Diagnosis of Kidney Cancer. Biomedicines 2022, 10, 233. [Google Scholar] [CrossRef]
- Kim, S.; Kim, T.G.; Lee, S.H.; Kim, W.; Bang, A.; Moon, S.W.; Song, J.; Shin, J.H.; Yu, J.S.; Choi, S. Label-Free Surface-Enhanced Raman Spectroscopy Biosensor for On-Site Breast Cancer Detection Using Human Tears. ACS Appl. Mater. Interfaces 2020, 12, 7897–7904. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, R.A.R.; Lataliza, A.A.B.; Raposo, N.R.B.; Costa, L.A.S.; Sant’Ana, A.C. Insights on the Transport of Tamoxifen by Gold Nanoparticles for MCF-7 Breast Cancer Cells Based on SERS Spectroscopy. Colloids Surf. B Biointerfaces 2018, 170, 712–717. [Google Scholar] [CrossRef] [PubMed]
- Nargis, H.F.; Nawaz, H.; Bhatti, H.N.; Jilani, K.; Saleem, M. Comparison of Surface Enhanced Raman Spectroscopy and Raman Spectroscopy for the Detection of Breast Cancer Based on Serum Samples. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 246, 119034. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Su, X.; Wen, Y.; Zheng, C.; Li, M. Artificial Intelligent Label-Free SERS Profiling of Serum Exosomes for Breast Cancer Diagnosis and Postoperative Assessment. Nano Lett. 2022, 22, 7910–7918. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Truong, V.X.; Li, M. Real-Time Intraoperative Surface-Enhanced Raman Spectroscopy-Guided Thermosurgical Eradication of Residual Microtumors in Orthotopic Breast Cancer. Nano Lett. 2021, 21, 3066–3074. [Google Scholar] [CrossRef]
- Xiao, L.; Harihar, S.; Welch, D.R.; Zhou, A. Imaging of Epidermal Growth Factor Receptor on Single Breast Cancer Cells Using Surface-Enhanced Raman Spectroscopy. Anal. Chim. Acta 2014, 843, 73–82. [Google Scholar] [CrossRef]
- Liang, L.; Shen, Y.; Zhang, J.; Xu, S.; Xu, W.; Liang, C.; Han, B. Identification of Breast Cancer through Spectroscopic Analysis of Cell-Membrane Sialic Acid Expression. Anal. Chim. Acta 2018, 1033, 148–155. [Google Scholar] [CrossRef]
- Hernández-Arteaga, A.; de Jesús Zermeño Nava, J.; Kolosovas-Machuca, E.S.; Velázquez-Salazar, J.J.; Vinogradova, E.; José-Yacamán, M.; Navarro-Contreras, H.R. Diagnosis of Breast Cancer by Analysis of Sialic Acid Concentrations in Human Saliva by Surface-Enhanced Raman Spectroscopy of Silver Nanoparticles. Nano Res. 2017, 10, 3662–3670. [Google Scholar] [CrossRef]
- Han, Y.; Qiang, L.; Gao, Y.; Gao, J.; He, Q.; Liu, H.; Han, L.; Zhang, Y. Large-Area Surface-Enhanced Raman Spectroscopy Substrate by Hybrid Porous GaN with Au/Ag for Breast Cancer MiRNA Detection. Appl. Surf. Sci. 2021, 541, 148456. [Google Scholar] [CrossRef]
- Yarbakht, M.; Nikkhah, M.; Moshaii, A.; Weber, K.; Matthäus, C.; Cialla-May, D.; Popp, J. Simultaneous Isolation and Detection of Single Breast Cancer Cells Using Surface-Enhanced Raman Spectroscopy. Talanta 2018, 186, 44–52. [Google Scholar] [CrossRef]
- Zheng, Z.; Wu, L.; Li, L.; Zong, S.; Wang, Z.; Cui, Y. Simultaneous and Highly Sensitive Detection of Multiple Breast Cancer Biomarkers in Real Samples Using a SERS Microfluidic Chip. Talanta 2018, 188, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Hameed, M.K.; Parambath, J.B.M.; Gul, M.T.; Khan, A.A.; Park, Y.; Han, C.; Mohamed, A.A. Arylated Gold Nanostars Aided SERS Study of Breast Cancer Cells. Appl. Surf. Sci. 2022, 583, 152504. [Google Scholar] [CrossRef]
- Kapara, A.; Brunton, V.G.; Graham, D.; Faulds, K. Characterisation of Estrogen Receptor Alpha (ERα) Expression in Breast Cancer Cells and Effect of Drug Treatment Using Targeted Nanoparticles and SERS. Analyst 2020, 145, 7225–7233. [Google Scholar] [CrossRef] [PubMed]
- Kapara, A.; Brunton, V.; Graham, D.; Faulds, K. Investigation of Cellular Uptake Mechanism of Functionalised Gold Nanoparticles into Breast Cancer Using SERS. Chem. Sci. 2020, 11, 5819–5829. [Google Scholar] [CrossRef]
- Lee, S.; Chon, H.; Lee, J.; Ko, J.; Chung, B.H.; Lim, D.W.; Choo, J. Rapid and Sensitive Phenotypic Marker Detection on Breast Cancer Cells Using Surface-Enhanced Raman Scattering (SERS) Imaging. Biosens. Bioelectron. 2014, 51, 238–243. [Google Scholar] [CrossRef]
- Choi, N.; Dang, H.; Das, A.; Sim, M.S.; Chung, I.Y.; Choo, J. SERS Biosensors for Ultrasensitive Detection of Multiple Biomarkers Expressed in Cancer Cells. Biosens. Bioelectron. 2020, 164, 112326. [Google Scholar] [CrossRef]
- Meng, S.; Chen, R.; Xie, J.; Li, J.; Cheng, J.; Xu, Y.; Cao, H.; Wu, X.; Zhang, Q.; Wang, H. Surface-Enhanced Raman Scattering Holography Chip for Rapid, Sensitive and Multiplexed Detection of Human Breast Cancer-Associated MicroRNAs in Clinical Samples. Biosens. Bioelectron. 2021, 190, 113470. [Google Scholar] [CrossRef]
- Weng, S.; Lin, D.; Lai, S.; Tao, H.; Chen, T.; Peng, M.; Qiu, S.; Feng, S. Highly Sensitive and Reliable Detection of MicroRNA for Clinically Disease Surveillance Using SERS Biosensor Integrated with Catalytic Hairpin Assembly Amplification Technology. Biosens. Bioelectron. 2022, 208, 114236. [Google Scholar] [CrossRef]
- Li, Y.; Qi, X.; Lei, C.; Qifeng, Q.; Zhang, S. Simultaneous SERS Detection and Imaging of Two Biomarkers on the Cancer Cell Surface by Self-Assembly of Branched DNA-Gold Nanoaggregates. Chem. Commun. 2014, 50, 9907–9909. [Google Scholar] [CrossRef]
- Lee, J.U.; Kim, W.H.; Lee, H.S.; Park, K.H.; Sim, S.J. Quantitative and Specific Detection of Exosomal MiRNAs for Accurate Diagnosis of Breast Cancer Using a Surface-Enhanced Raman Scattering Sensor Based on Plasmonic Head-Flocked Gold Nanopillars. Small 2019, 15, 1804968. [Google Scholar] [CrossRef]
- Zhong, Q.; Zhang, K.; Huang, X.; Lu, Y.; Zhao, J.; He, Y.; Liu, B. In Situ Ratiometric SERS Imaging of Intracellular Protease Activity for Subtype Discrimination of Human Breast Cancer. Biosens. Bioelectron. 2022, 207, 114194. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liao, M.; Chen, Y.; Shan, B.; Li, M. Surface-Enhanced Raman Spectroscopy (SERS) Nanoprobes for Ratiometric Detection of Cancer Cells. J. Mater. Chem. B 2019, 7, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kang, S.; Khan, A.; Ruttner, G.; Leigh, S.Y.; Murray, M.; Abeytunge, S.; Peterson, G.; Rajadhyaksha, M.; Dintzis, S.; et al. Quantitative Molecular Phenotyping with Topically Applied SERS Nanoparticles for Intraoperative Guidance of Breast Cancer Lumpectomy. Sci. Rep. 2016, 6, 21242. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ma, R.; Zhang, Y.; Zhao, J.; Wang, Y.; Xu, Z. Dual-Aptamer-Assisted Ratiometric SERS Biosensor for Ultrasensitive and Precise Identification of Breast Cancer Exosomes. ACS Sens. 2023. [Google Scholar] [CrossRef]
- Shen, L.S.N.; Du, Y.; Wei, N.; Li, Q.; Li, S.M.; Sun, T.M.; Xu, S.; Wang, H.; Man, X.X.; Han, B. SERS Studies on Normal Epithelial and Cancer Cells Derived from Clinical Breast Cancer Specimens. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 237, 118364. [Google Scholar] [CrossRef]
- Lin, Y.; Gao, S.; Zheng, M.; Tang, S.; Lin, K.; Xie, S.; Yu, Y.; Lin, J. A Microsphere Nanoparticle Based-Serum Albumin Targeted Adsorption Coupled with Surface-Enhanced Raman Scattering for Breast Cancer Detection. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 261, 120039. [Google Scholar] [CrossRef]
- Lin, Y.; Gao, J.; Tang, S.; Zhao, X.; Zheng, M.; Gong, W.; Xie, S.; Gao, S.; Yu, Y.; Lin, J. Label-Free Diagnosis of Breast Cancer Based on Serum Protein Purification Assisted Surface-Enhanced Raman Spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 263, 120234. [Google Scholar] [CrossRef]
- Moisoiu, V.; Socaciu, A.; Stefancu, A.; Iancu, S.D.; Boros, I.; Alecsa, C.D.; Rachieriu, C.; Chiorean, A.R.; Eniu, D.; Leopold, N.; et al. Breast Cancer Diagnosis by Surface-Enhanced Raman Scattering (SERS) of Urine. Appl. Sci. 2019, 9, 806. [Google Scholar] [CrossRef]
- Akbar, S.; Majeed, M.I.; Nawaz, H.; Rashid, N.; Tariq, A.; Hameed, W.; Shakeel, S.; Dastgir, G.; Bari, R.Z.A.; Iqbal, M.; et al. Surface-Enhanced Raman Spectroscopic (SERS) Characterization of Low Molecular Weight Fraction of the Serum of Breast Cancer Patients with Principal Component Analysis (PCA) and Partial Least Square-Discriminant Analysis (PLS-DA). Anal. Lett. 2022, 55, 1588–1604. [Google Scholar] [CrossRef]
- Feng, S.; Huang, S.; Lin, D.; Chen, G.; Xu, Y.; Li, Y.; Huang, Z.; Pan, J.; Chen, R.; Zeng, H. Surface-Enhanced Raman Spectroscopy of Saliva Proteins for the Noninvasive Differentiation of Benign and Malignant Breast Tumors. Int. J. Nanomed. 2015, 10, 537–547. [Google Scholar] [CrossRef]
- Iancu, S.D.; Cozan, R.G.; Stefancu, A.; David, M.; Moisoiu, T.; Moroz-Dubenco, C.; Bajcsi, A.; Chira, C.; Andreica, A.; Leopold, L.F.; et al. SERS Liquid Biopsy in Breast Cancer. What Can We Learn from SERS on Serum and Urine? Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 273, 120992. [Google Scholar] [CrossRef]
- Lin, X.; Jia, X.; Lin, J.Y.; Wu, P.H.; Weng, Y.; Feng, S. A Comparative Study Based on Serum SERS Spectra in and on the Coffee Ring for High Precision Breast Cancer Detection. J. Raman Spectrosc. 2022, 53, 1371–1379. [Google Scholar] [CrossRef]
- Știufiuc, G.F.; Toma, V.; Buse, M.; Mărginean, R.; Morar-Bolba, G.; Culic, B.; Tetean, R.; Leopold, N.; Pavel, I.; Lucaciu, C.M.; et al. Solid Plasmonic Substrates for Breast Cancer Detection by Means of SERS Analysis of Blood Plasma. Nanomaterials 2020, 10, 1212. [Google Scholar] [CrossRef] [PubMed]
- Cervo, S.; Mansutti, E.; Del Mistro, G.; Spizzo, R.; Colombatti, A.; Steffan, A.; Sergo, V.; Bonifacio, A. SERS Analysis of Serum for Detection of Early and Locally Advanced Breast Cancer. Anal. Bioanal. Chem. 2015, 407, 7503–7509. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Wang, Y.; Wang, T.; Zhu, Y.; Lin, X.; Lin, Y.; Feng, S. Metabolite Profiling of Human Blood by Surface-Enhanced Raman Spectroscopy for Surgery Assessment and Tumor Screening in Breast Cancer. Anal. Bioanal. Chem. 2020, 412, 1611–1618. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Wang, Q.; Tang, J.; Yao, S.; Li, H.; Yue, X.; Fu, J.; Zhong, F.; Wang, T.; Wang, J. Non-Invasive SERS Serum Detection Technology Combined with Multivariate Statistical Algorithm for Simultaneous Screening of Cervical Cancer and Breast Cancer. Anal. Bioanal. Chem. 2021, 413, 4775–4784. [Google Scholar] [CrossRef]
- Vargas-Obieta, E.; Martínez-Espinosa, J.C.; Martínez-Zerega, B.E.; Jave-Suárez, L.F.; Aguilar-Lemarroy, A.; González-Solís, J.L. Breast Cancer Detection Based on Serum Sample Surface Enhanced Raman Spectroscopy. Lasers Med. Sci. 2016, 31, 1317–1324. [Google Scholar] [CrossRef]
- Ma, X.; Xiong, H.; Guo, J.; Liu, Z.; Han, Y.; Liu, M. Label-Free Breast Cancer Detection and Classification by Convolutional Neural Network-Based on Exosomes Surface-Enhanced Raman Scattering. J. Innov. Opt. Health Sci. 2022, 2022, 2244001. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, J.; Shanmugasundaram, K.B.; Yeo, B.; Möller, A.; Wuethrich, A.; Lin, L.L.; Trau, M. Tracking Drug-Induced Epithelial–Mesenchymal Transition in Breast Cancer by a Microfluidic Surface-Enhanced Raman Spectroscopy Immunoassay. Small 2020, 16, 1905614. [Google Scholar] [CrossRef]
- Zhu, J.; Zhou, J.; Guo, J.; Cai, W.; Liu, B.; Wang, Z.; Sun, Z. Surface-Enhanced Raman Spectroscopy Investigation on Human Breast Cancer Cells. Chem. Cent. J. 2013, 7, 37. [Google Scholar] [CrossRef]
- Brozek-Pluska, B.; Kopec, M.; Surmacki, J. Surface-Enhanced Raman Spectroscopy Analysis of Human Breast Cancer via Silver Nanoparticles: An Examination of Fabrication Methods. J. Spectrosc. 2018, 2018, 4893274. [Google Scholar] [CrossRef]
- Narayanan, N.; Kim, J.H.; Santhakumar, H.; Joseph, M.M.; Karunakaran, V.; Shamjith, S.; Saranya, G.; Sujai, P.T.; Jayasree, R.S.; Barman, I.; et al. Nanotheranostic Probe Built on Methylene Blue Loaded Cucurbituril [8] and Gold Nanorod: Targeted Phototherapy in Combination with SERS Imaging on Breast Cancer Cells. J. Phys. Chem. B 2021, 125, 13415–13424. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Chen, L.; Xia, Y.; Xing, J.; Li, Z.; Qian, Q.; Wang, Y.; Wu, A.; Zeng, L.; Zhou, Y. Bioconjugation of Gold Nanobipyramids for SERS Detection and Targeted Photothermal Therapy in Breast Cancer. ACS Biomater. Sci. Eng. 2017, 3, 608–618. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Pan, Y.; Wang, S.; Wang, X.; Zhao, X.; Ren, W.; Lu, G.; Wu, A. Raman Reporter-Coupled Agcore@Aushell Nanostars for in Vivo Improved Surface Enhanced Raman Scattering Imaging and Near-Infrared-Triggered Photothermal Therapy in Breast Cancers. ACS Appl. Mater. Interfaces 2015, 7, 16781–16791. [Google Scholar] [CrossRef] [PubMed]
- Xinyue, L.; Keshavarz, M.; Panagiotis, K.; Roddan, A.; Yeatman, E.; Thompson, A. SERS Detection of Breast Cancer-Derived Exosomes Using a Nanostructured Pt-Black Template. Adv. Sens. Res. 2023, 2023, 2200039. [Google Scholar] [CrossRef]
- Pramanik, A.; Mayer, J.; Patibandla, S.; Gates, K.; Gao, Y.; Davis, D.; Seshadri, R.; Ray, P.C. Mixed-Dimensional Heterostructure Material-Based SERS for Trace Level Identification of Breast Cancer-Derived Exosomes. ACS Omega 2020, 5, 16602–16611. [Google Scholar] [CrossRef]
- Li, G.; Zhu, N.; Zhou, J.; Kang, K.; Zhou, X.; Ying, B.; Yi, Q.; Wu, Y. A Magnetic Surface-Enhanced Raman Scattering Platform for Performing Successive Breast Cancer Exosome Isolation and Analysis. J. Mater. Chem. B 2021, 9, 2709–2716. [Google Scholar] [CrossRef]
- Yang, Z.; Su, H.S.; You, E.M.; Liu, S.; Li, Z.; Zhang, Y. High Uniformity and Enhancement Au@AgNS 3D Substrates for the Diagnosis of Breast Cancer. ACS Omega 2022, 7, 15223–15230. [Google Scholar] [CrossRef]
- Wang, X.P.; Walkenfort, B.; König, M.; König, L.; Kasimir-Bauer, S.; Schlücker, S. Fast and Reproducible ISERS Microscopy of Single HER2-Positive Breast Cancer Cells Using Gold Nanostars as SERS Nanotags. Faraday Discuss. 2017, 205, 377–386. [Google Scholar] [CrossRef]
- Chen, Z.; Shen, X.; Chen, S.; Dai, K. Gastric Cancer Prewarning and Early Diagnosis System; Springer: Berlin/Heidelberg, Germany, 2017; ISBN 9789402409499. [Google Scholar]
- Hunter, R.A.; Asare-Werehene, M.; Mandour, A.; Tsang, B.K.; Anis, H. Determination of Chemoresistance in Ovarian Cancer by Simultaneous Quantification of Exosomes and Exosomal Cisplatin with Surface Enhanced Raman Scattering. Sens. Actuators B Chem. 2022, 354, 131237. [Google Scholar] [CrossRef]
- Moothanchery, M.; Perumal, J.; Mahyuddin, A.P.; Singh, G.; Choolani, M.; Olivo, M. Rapid and Sensitive Detection of Ovarian Cancer Biomarker Using a Portable Single Peak Raman Detection Method. Sci. Rep. 2022, 12, 12459. [Google Scholar] [CrossRef]
- Sarkar, S.; Gogoi, M.; Mahato, M.; Joshi, A.B.; Baruah, A.J.; Kodgire, P.; Boruah, P. Biosensors for Detection of Prostate Cancer: A Review. Biomed. Microdevices 2022, 24, 32. [Google Scholar] [CrossRef]
- Turan, E.; Zengin, A.; Suludere, Z.; Kalkan, N.Ö.; Tamer, U. Construction of a Sensitive and Selective Plasmonic Biosensor for Prostate Specific Antigen by Combining Magnetic Molecularly-Imprinted Polymer and Surface-Enhanced Raman Spectroscopy. Talanta 2022, 237, 122926. [Google Scholar] [CrossRef]
- Haroon, M.; Tahir, M.; Nawaz, H.; Majeed, M.I.; Al-Saadi, A.A. Surface-Enhanced Raman Scattering (SERS) Spectroscopy for Prostate Cancer Diagnosis: A Review. Photodiagn. Photodyn. Ther. 2022, 37, 102690. [Google Scholar] [CrossRef] [PubMed]
- Ashrafizadeh, M.; Aghamiri, S.; Tan, S.C.; Zarrabi, A.; Sharifi, E.; Rabiee, N.; Kadumudi, F.B.; Pirouz, A.D.; Delfi, M.; Byrappa, K.; et al. Nanotechnological Approaches in Prostate Cancer Therapy: Integration of Engineering and Biology. Nano Today 2022, 45, 101532. [Google Scholar] [CrossRef]
- Gaba, F.; Tipping, W.J.; Salji, M.; Faulds, K.; Graham, D.; Leung, H.Y. Raman Spectroscopy in Prostate Cancer: Techniques, Applications and Advancements. Cancers. 2022, 14, 1535. [Google Scholar] [CrossRef]
- Pandey, A.; Sarkar, S.; Pandey, S.K.; Srivastava, A. Silica Nanospheres Coated Silver Islands as an Effective Opto-Plasmonic SERS Active Platform for Rapid and Sensitive Detection of Prostate Cancer Biomarkers. Molecules 2022, 27, 7821. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhu, Y.Y.; Wang, M.L. Surface-Enhanced Raman Spectroscopy of Gastric Cancer Serum with Gold Nanoparticles/Silicon Nanowire Arrays. Optik 2016, 127, 7902–7907. [Google Scholar] [CrossRef]
- Ito, H.; Inoue, H.; Hasegawa, K.; Hasegawa, Y.; Shimizu, T.; Kimura, S.; Onimaru, M.; Ikeda, H.; Kudo, S. ei Use of Surface-Enhanced Raman Scattering for Detection of Cancer-Related Serum-Constituents in Gastrointestinal Cancer Patients. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 599–608. [Google Scholar] [CrossRef]
- Ge, S.; Ran, M.; Mao, Y.; Sun, Y.; Zhou, X.; Li, L.; Cao, X. A Novel DNA Biosensor for the Ultrasensitive Detection of DNA Methyltransferase Activity Based on a High-Density ‘Hot Spot’ SERS Substrate and Rolling Circle Amplification Strategy. Analyst 2021, 146, 5326–5336. [Google Scholar] [CrossRef]
- Feng, S.; Chen, R.; Lin, J.; Pan, J.; Wu, Y.; Li, Y.; Chen, J.; Zeng, H. Gastric Cancer Detection Based on Blood Plasma Surface-Enhanced Raman Spectroscopy Excited by Polarized Laser Light. Biosens. Bioelectron. 2011, 26, 3167–3174. [Google Scholar] [CrossRef]
- Pan, H.; Dong, Y.; Gong, L.; Zhai, J.; Song, C.; Ge, Z.; Su, Y.; Zhu, D.; Chao, J.; Su, S.; et al. Sensing Gastric Cancer Exosomes with MoS2-Based SERS Aptasensor. Biosens. Bioelectron. 2022, 215, 114553. [Google Scholar] [CrossRef]
- Liu, Z.; Li, T.; Wang, Z.; Liu, J.; Huang, S.; Min, B.H.; An, J.Y.; Kim, K.M.; Kim, S.; Chen, Y.; et al. Gold Nanopyramid Arrays for Non-Invasive Surface-Enhanced Raman Spectroscopy-Based Gastric Cancer Detection via SEVs. ACS Appl. Nano Mater. 2022, 5, 12506–12517. [Google Scholar] [CrossRef]
- Gayoung, E.; Hongki, K.; Ahreum, H.; Hye-Young, S.; Yuna, C.; Jeong, M.; Donghyeong, K.; Miyeon, L.; Eun-Kyung, L.; Jinyoung, J.; et al. Nanogap-Rich Au Nanowire SERS Sensor for Ultrasensitive Telomerase Activity Detection. Adv. Funct. Mater. 2017, 27, 1701832. [Google Scholar] [CrossRef]
- Chen, Y.; Cheng, S.; Zhang, A.; Song, J.; Chang, J.; Wang, K.; Gao, G.; Zhang, Y.; Li, S.; Liu, H.; et al. Salivary Analysis Based on Surface Enhanced Raman Scattering Sensors Distinguishes Early and Advanced Gastric Cancer Patients from Healthy Persons. J. Biomed. Nanotechnol. 2018, 14, 1773–1784. [Google Scholar] [CrossRef]
- Cao, X.; Ge, S.; Hua, W.; Zhou, X.; Lu, W.; Gu, Y.; Li, Z.; Qian, Y. A Pump-Free and High-Throughput Microfluidic Chip for Highly Sensitive SERS Assay of Gastric Cancer-Related Circulating Tumor DNA via a Cascade Signal Amplification Strategy. J. Nanobiotechnol. 2022, 20, 271. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Y.; Pan, F.; Liu, J.; Wang, K.; Zhang, C.; Cheng, S.; Lu, L.; Zhang, W.; Zhang, Z.; et al. Breath Analysis Based on Surface-Enhanced Raman Scattering Sensors Distinguishes Early and Advanced Gastric Cancer Patients from Healthy Persons. ACS Nano 2016, 10, 8169–8179. [Google Scholar] [CrossRef]
- Huang, L.; Zhu, Y.; Xu, C.; Cai, Y.; Yi, Y.; Li, K.; Ren, X.; Jiang, D.; Ge, Y.; Liu, X.; et al. Noninvasive Diagnosis of Gastric Cancer Based on Breath Analysis with a Tubular Surface-Enhanced Raman Scattering Sensor. ACS Sens. 2022, 7, 1439–1450. [Google Scholar] [CrossRef]
- Cao, D.; Lin, H.; Liu, Z.; Qiu, J.; Ge, S.; Hua, W.; Cao, X.; Qian, Y.; Xu, H.; Zhu, X. PCA-TLNN-Based SERS Analysis Platform for Label-Free Detection and Identification of Cisplatin-Treated Gastric Cancer. Sens. Actuators B Chem. 2023, 375, 132903. [Google Scholar] [CrossRef]
- Guo, L.; Li, Y.; Huang, F.; Dong, J.; Li, F.; Yang, X.; Zhu, S.; Yang, M. Identification and Analysis of Serum Samples by Surface-Enhanced Raman Spectroscopy Combined with Characteristic Ratio Method and PCA for Gastric Cancer Detection. J. Innov. Opt. Health Sci. 2019, 12, 1950003. [Google Scholar] [CrossRef]
- Ma, J.; Zhou, H.; Gong, L.; Liu, S.; Zhou, Z.; Mao, W.; Zheng, R. Distinction of Gastric Cancer Tissue Based on Surface-Enhanced Raman Spectroscopy. Opt. Health Care Biomed. Opt. V 2012, 8553, 855328. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Chen, G.; Zheng, X.; He, C.; Feng, S.; Lin, X.; Chen, R.; Zeng, H. Discrimination of Gastric Cancer from Normal by Serum RNA Based on Surface-Enhanced Raman Spectroscopy (SERS) and Multivariate Analysis. Med. Phys. 2012, 39, 5664–5668. [Google Scholar] [CrossRef]
- Feng, S.Y.; Pan, J.J.; Wu, Y.A.; Lin, D.; Chen, Y.P.; Xi, G.Q.; Lin, J.Q.; Chen, R. Study on Gastric Cancer Blood Plasma Based on Surface-Enhanced Raman Spectroscopy Combined with Multivariate Analysis. Sci. China Life Sci. 2011, 54, 828–834. [Google Scholar] [CrossRef]
- Aslam, M.A.; Xue, C.; Wang, K.; Chen, Y.; Zhang, A.; Cai, W.; Ma, L.; Yang, Y.; Sun, X.; Liu, M.; et al. SVM Based Classification and Prediction System for Gastric Cancer Using Dominant Features of Saliva. Nano Biomed. Eng. 2020, 12, 1–13. [Google Scholar] [CrossRef]
- Aslam, M.A.; Xue, C.; Liu, M.; Wang, K.; Cui, D. Classification and Prediction of Gastric Cancer from Saliva Diagnosis Using Artificial Neural Network. Eng. Lett. 2020, 29, 2. [Google Scholar]
- Avram, L.; Iancu, S.D.; Stefancu, A.; Moisoiu, V.; Colnita, A.; Marconi, D.; Donca, V.; Buzdugan, E.; Craciun, R.; Leopold, N.; et al. SERS-Based Liquid Biopsy of Gastrointestinal Tumors Using a Portable Raman Device Operating in a Clinical Environment. J. Clin. Med. 2020, 9, 212. [Google Scholar] [CrossRef]
- Li, X.; Yang, T.; Li, S.; Wang, D.; Song, Y.; Yu, K. Different Classification Algorithms and Serum Surface Enhanced Raman Spectroscopy for Noninvasive Discrimination of Gastric Diseases. J. Raman Spectrosc. 2016, 47, 917–925. [Google Scholar] [CrossRef]
- Li, S.X.; Zhang, Y.J.; Zeng, Q.Y.; Li, L.F.; Guo, Z.Y.; Liu, Z.M.; Xiong, H.L.; Liu, S.H. Potential of Cancer Screening with Serum Surface-Enhanced Raman Spectroscopy and a Support Vector Machine. Laser Phys. Lett. 2014, 11, 065603. [Google Scholar] [CrossRef]
- Moisoiu, T.; Dragomir, M.P.; Iancu, S.D.; Schallenberg, S.; Birolo, G.; Ferrero, G.; Burghelea, D.; Stefancu, A.; Cozan, R.G.; Licarete, E.; et al. Combined MiRNA and SERS Urine Liquid Biopsy for the Point-of-Care Diagnosis and Molecular Stratification of Bladder Cancer. Mol. Med. 2022, 28, 39. [Google Scholar] [CrossRef]
- Gao, S.; Lin, Y.; Zhao, X.; Gao, J.; Xie, S.; Gong, W.; Yu, Y.; Lin, J. Label-Free Surface Enhanced Raman Spectroscopy Analysis of Blood Serum via Coffee Ring Effect for Accurate Diagnosis of Cancers. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 267, 120605. [Google Scholar] [CrossRef]
- Meng, C.; Li, H.; Chen, C.; Wu, W.; Gao, J.; Lai, Y.; Ka, M.; Zhu, M.; Lv, X.; Chen, F.; et al. Serum Raman Spectroscopy Combined with Gaussian—Convolutional Neural Network Models to Quickly Detect Liver Cancer Patients. Spectrosc. Lett. 2022, 55, 79–90. [Google Scholar] [CrossRef]
- Ni, J.-T.; Huang, M.-Y.; Ji, W.; Wang, L.; Sun, T.-D. Recent Advances in Surface-Enhanced Raman Scattering for Liver Cancer Detection. Chin. J. Anal. Chem. 2022, 50, 100180. [Google Scholar] [CrossRef]
- Zhang, Q.; Hou, D.; Wen, X.; Xin, M.; Li, Z.; Wu, L.; Pathak, J.L. Gold Nanomaterials for Oral Cancer Diagnosis and Therapy: Advances, Challenges, and Prospects. Mater. Today Bio 2022, 15, 100333. [Google Scholar] [CrossRef] [PubMed]
- Zadeh, F.A.; Shahhosseini, E.; Rasoolzadegan, S.; Özbolat, G.; Farahbod, F. Au Nanoparticles in the Diagnosis and Treatment of Ovarian Cancer: A New Horizon in the Personalized Medicine. Nanomed. Res. J. 2022, 7, 1–18. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Du, Q.; Cao, D.; Lu, X.; Meng, Z. Sensitive SERS Detection of Oral Squamous Cell Carcinoma-Related MiRNAs in Saliva via a Gold Nanohexagon Array Coupled with Hybridization Chain Reaction Amplification. Anal. Methods 2022, 14, 4563–4575. [Google Scholar] [CrossRef]
- Fălămaș, A.; Rotaru, H.; Hedeșiu, M. Surface-Enhanced Raman Spectroscopy (SERS) Investigations of Saliva for Oral Cancer Diagnosis. Lasers Med. Sci. 2020, 35, 1393–1401. [Google Scholar] [CrossRef]
- Wang, K.; Qiu, Y.; Wu, C.; Wen, Z.N.; Li, Y. Surface-Enhanced Raman Spectroscopy and Multivariate Analysis for the Diagnosis of Oral Squamous Cell Carcinoma. J. Raman Spectrosc. 2023. [Google Scholar] [CrossRef]
- Lin, Y.; Lin, J.; Zheng, M.; Gong, W.; Li, H.; Shu, Z.; Du, W.; Gao, S.; Yu, Y. Quantitative and Direct Serum Albumin Detection by Label-Free SERS Using Tunable Hydroxyapatite Nanostructure for Prostate Cancer Detection. Anal. Chim. Acta 2022, 1221, 340101. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, J.; Liu, Y.; Han, X.X.; Xu, B.; Ozaki, Y.; Zhao, B. Detection of Prostate Cancer Biomarkers via a SERS-Based Aptasensor. Biosens. Bioelectron. 2022, 216, 114660. [Google Scholar] [CrossRef]
- Lu, Y.; Zhan, C.; Yu, L.; Yu, Y.; Jia, H.; Chen, X.; Zhang, D.; Gao, R. Multifunctional Nanocone Array as Solid Immunoassay Plate and SERS Substrate for the Early Diagnosis of Prostate Cancer on Microfluidic Chip. Sens. Actuators B Chem. 2023, 376, 133046. [Google Scholar] [CrossRef]
- Munteanu, V.C.; Munteanu, R.A.; Gulei, D.; Mărginean, R.; Schițcu, V.H.; Onaciu, A.; Toma, V.; Știufiuc, G.F.; Coman, I.; Știufiuc, R.I. New Insights into the Multivariate Analysis of SER Spectra Collected on Blood Samples for Prostate Cancer Detection: Towards a Better Understanding of the Role Played by Different Biomolecules on Cancer Screening: A Preliminary Study. Cancers 2022, 14, 3227. [Google Scholar] [CrossRef] [PubMed]
- Stefancu, A.; Moisoiu, V.; Couti, R.; Andras, I.; Rahota, R.; Crisan, D.; Pavel, I.E.; Socaciu, C.; Leopold, N.; Crisan, N. Combining SERS Analysis of Serum with PSA Levels for Improving the Detection of Prostate Cancer. Nanomedicine 2018, 13, 2455–2467. [Google Scholar] [CrossRef]
- Liyanage, T.; Alharbi, B.; Quan, L.; Esquela-Kerscher, A.; Slaughter, G. Plasmonic-Based Biosensor for the Early Diagnosis of Prostate Cancer. ACS Omega 2022, 7, 2411–2418. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Xu, Q.; Lin, Y.; Du, W.; Bai, X.; Gao, J.; Li, T.; Huang, Y.; Yu, Y.; Wu, X.; et al. Label-free surface-enhanced Raman spectroscopy detectionof prostate cancer combined with multivariate statistical algorithm. J. Raman Spectrosc. 2022, 53, 1861–1870. [Google Scholar] [CrossRef]
- Sayyadi, N.; Justiniano, I.; Wang, Y.; Zheng, X.; Zhang, W.; Jiang, L.; Polikarpov, D.M.; Willows, R.D.; Gillatt, D.; Campbell, D.; et al. Detection of Rare Prostate Cancer Cells in Human Urine Offers Prospect of Non-Invasive Diagnosis. Sci. Rep. 2022, 12, 18452. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Lin, S.; Zhang, H.; Liang, L.; Shen, S. Methods of Respiratory Virus Detection: Advances towards Point-of-Care for Early Intervention. Micromachines 2021, 12, 697. [Google Scholar] [CrossRef] [PubMed]
- Omidifar, N.; Lankarani, K.B.; Moghadami, M.; Shokripour, M.; Chashmpoosh, M.; Mousavi, S.M.; Hashemi, S.A.; Gholami, A. Different Laboratory Diagnosis Methods of COVID-19: A Systematic Review. Arch. Clin. Infect. Dis. 2021, 16, e110667. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Hashemi, S.A.; Rahmanian, V.; Kalashgrani, M.Y. Highly Sensitive Flexible SERS-Based Sensing Platform for Detection of Biosensors Highly Sensitive Flexible SERS-Based Sensing Platform for Detection of COVID-19. Biosensors 2022, 12, 466. [Google Scholar] [CrossRef]
- Stöckel, S.; Kirchhoff, J.; Neugebauer, U.; Rösch, P.; Popp, J. The Application of Raman Spectroscopy for the Detection and Identification of Microorganisms. J. Raman Spectrosc. 2016, 47, 89–109. [Google Scholar] [CrossRef]
- Soler, M.; Estevez, M.C.; Cardenosa-Rubio, M.; Astua, A.; Lechuga, L.M. How Nanophotonic Label-Free Biosensors Can Contribute to Rapid and Massive Diagnostics of Respiratory Virus Infections: COVID-19 Case. ACS Sens. 2020, 5, 2663–2678. [Google Scholar] [CrossRef]
- Iravani, S. Nano- And Biosensors for the Detection of SARS-CoV-2: Challenges and Opportunities. Mater. Adv. 2020, 1, 3092–3103. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, X.; Ma, R.; Deng, S.; Wang, X.; Wang, X.; Zhang, X.; Huang, X.; Liu, Y.; Li, G.; et al. Ultra-Fast and Onsite Interrogation of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in Waters via Surface Enhanced Raman Scattering (SERS). Water Res. 2021, 200, 117243. [Google Scholar] [CrossRef] [PubMed]
- Joung, Y.; Kim, K.; Lee, S.; Chun, B.-S.; Lee, S.; Hwang, J.; Choi, S.; Kang, T.; Lee, M.-K.; Chen, L.; et al. Rapid and Accurate On-Site Immunodiagnostics of Highly Contagious Severe Acute Respiratory Syndrome Coronavirus 2 Using Portable Surface-Enhanced Raman Scattering-Lateral Flow Assay Reader. ACS Sens. 2022, 7, 3470–3480. [Google Scholar] [CrossRef] [PubMed]
- Saviñon-Flores, F.; Méndez, E.; López-Castaños, M.; Carabarin-Lima, A.; López-Castaños, K.A.; González-Fuentes, M.A.; Méndez-Albores, A. A Review on Sers-Based Detection of Human Virus Infections: Influenza and Coronavirus. Biosensors 2021, 11, 66. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.Y.; Nam, J.S.; Yang, S.E.; Shin, H.; Jang, Y.H.; Bae, G.U.; Kang, T.; Lim, K.I.; Choi, Y. Identification of Newly Emerging Influenza Viruses by Surface-Enhanced Raman Spectroscopy. Anal. Chem. 2015, 87, 11652–11659. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Wang, Z.; Liu, H.; Perea-López, N.; Ranasinghe, J.C.; Bepete, G.; Minns, A.M.; Rossi, R.M.; Lindner, S.E.; Huang, S.X.; et al. Understanding the Excitation Wavelength Dependence and Thermal Stability of the SARS-CoV-2 Receptor-Binding Domain Using Surface-Enhanced Raman Scattering and Machine Learning. ACS Photonics 2022, 9, 2963–2972. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, B.; Murray, J.; Haverstick, J.; Chen, X.; Tripp, R.A.; Zhao, Y. Rapid and Quantitative Detection of Respiratory Viruses Using Surface-Enhanced Raman Spectroscopy and Machine Learning. Biosens. Bioelectron. 2022, 217, 114721. [Google Scholar] [CrossRef]
- Ye, J.; Yeh, Y.; Xue, Y.; Wang, Z.; Zhang, N.; Liu, H.; Zhang, K.; Ricker, R.; Yu, Z.; Roder, A. Accurate Virus Identi Fi Cation with Interpretable Raman Signatures by Machine Learning. Proc. Natl. Acad. Sci. USA 2022, 199, e2118836119. [Google Scholar] [CrossRef] [PubMed]
- Carlomagno, C.; Bertazioli, D.; Gualerzi, A.; Picciolini, S.; Banfi, P.I.; Lax, A.; Messina, E.; Navarro, J.; Bianchi, L.; Caronni, A.; et al. COVID-19 Salivary Raman Fingerprint: Innovative Approach for the Detection of Current and Past SARS-CoV-2 Infections. Sci. Rep. 2021, 11, 4943. [Google Scholar] [CrossRef]
- Zavyalova, E.; Ambartsumyan, O.; Zhdanov, G.; Gribanyov, D.; Gushchin, V.; Tkachuk, A.; Rudakova, E.; Nikiforova, M.; Kuznetsova, N.; Popova, L.; et al. Sers-Based Aptasensor for Rapid Quantitative Detection of Sars-Cov-2. Nanomaterials 2021, 11, 1394. [Google Scholar] [CrossRef]
- Hwang, C.S.H.; Lee, S.; Lee, S.; Kim, H.; Kang, T.; Lee, D.; Jeong, K.H. Highly Adsorptive Au-TiO2 Nanocomposites for the SERS Face Mask Allow the Machine-Learning-Based Quantitative Assay of SARS-CoV-2 in Artificial Breath Aerosols. ACS Appl. Mater. Interfaces 2022, 14, 54550–54557. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Jiang, S.; Wang, X.; Dong, T.; Wang, Y.; Li, D.; Gao, X.; Qu, Z.; Li, Y. A Novel Enhanced Substrate for Label-Free Detection of SARS-CoV-2 Based on Surface-Enhanced Raman Scattering. Sens. Actuators B Chem. 2022, 359, 131568. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, C.; Wang, X.; Wang, K.; Zhu, Y.; Rong, Z.; Wang, W.; Xiao, R.; Wang, S. Magnetic SERS Strip for Sensitive and Simultaneous Detection of Respiratory Viruses. ACS Appl. Mater. Interfaces 2019, 11, 19495–19505. [Google Scholar] [CrossRef]
- Zhang, D.; Huang, L.; Liu, B.; Ge, Q.; Dong, J.; Zhao, X. Rapid and Ultrasensitive Quantification of Multiplex Respiratory Tract Infection Pathogen via Lateral Flow Microarray Based on SERS Nanotags. Theranostics 2019, 9, 4849–4859. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Peng, Y.; Lin, C.; Long, L.; Hu, J.; He, J.; Zeng, H.; Huang, Z.; Li, Z.Y.; Tanemura, M.; et al. Human ACE2-Functionalized Gold “Virus-Trap” Nanostructures for Accurate Capture of SARS-CoV-2 and Single-Virus SERS Detection. Nano-Micro Lett. 2021, 13, 109. [Google Scholar] [CrossRef]
- Peng, Y.; Lin, C.; Long, L.; Masaki, T.; Tang, M.; Yang, L.; Liu, J.; Huang, Z.; Li, Z.; Luo, X.; et al. Charge-Transfer Resonance and Electromagnetic Enhancement Synergistically Enabling MXenes with Excellent SERS Sensitivity for SARS-CoV-2 S Protein Detection. Nano-Micro Lett. 2021, 13, 52. [Google Scholar] [CrossRef]
- Gu, M.M.; Guan, P.C.; Xu, S.S.; Li, H.M.; Kou, Y.C.; Lin, X.D.; Kathiresan, M.; Song, Y.; Zhang, Y.J.; Jin, S.Z.; et al. Ultrasensitive Detection of SARS-CoV-2 S Protein with Aptamers Biosensor Based on Surface-Enhanced Raman Scattering. J. Chem. Phys. 2023, 158, 024203. [Google Scholar] [CrossRef]
- Lim, J.Y.; Nam, J.S.; Shin, H.; Park, J.; Song, H.I.; Kang, M.; Lim, K.I.; Choi, Y. Identification of Newly Emerging Influenza Viruses by Detecting the Virally Infected Cells Based on Surface Enhanced Raman Spectroscopy and Principal Component Analysis. Anal. Chem. 2019, 91, 5677–5684. [Google Scholar] [CrossRef]
- Eom, G.; Hwang, A.; Kim, H.; Yang, S.; Lee, D.K.; Song, S.; Ha, K.; Jeong, J.; Jung, J.; Lim, E.K.; et al. Diagnosis of Tamiflu-Resistant Influenza Virus in Human Nasal Fluid and Saliva Using Surface-Enhanced Raman Scattering. ACS Sens. 2019, 4, 2282–2287. [Google Scholar] [CrossRef]
- Zhang, M.; Li, X.; Pan, J.; Zhang, Y.; Zhang, L.; Wang, C.; Yan, X.; Liu, X.; Lu, G. Ultrasensitive Detection of SARS-CoV-2 Spike Protein in Untreated Saliva Using SERS-Based Biosensor. Biosens. Bioelectron. 2021, 190, 113421. [Google Scholar] [CrossRef]
- Yadav, S.; Sadique, M.A.; Ranjan, P.; Kumar, N.; Singhal, A.; Srivastava, A.K.; Khan, R. Sers Based Lateral Flow Immunoassay for Point-of-Care Detection of Sars-Cov-2 in Clinical Samples. ACS Appl. Bio Mater. 2021, 4, 2974–2995. [Google Scholar] [CrossRef]
- Chen, S.; Meng, L.; Wang, L.; Huang, X.; Ali, S.; Chen, X.; Yu, M.; Yi, M.; Li, L.; Chen, X.; et al. SERS-Based Lateral Flow Immunoassay for Sensitive and Simultaneous Detection of Anti-SARS-CoV-2 IgM and IgG Antibodies by Using Gap-Enhanced Raman Nanotags. Sens. Actuators B Chem. 2021, 348, 130706. [Google Scholar] [CrossRef]
- Liu, H.; Dai, E.; Xiao, R.; Zhou, Z.; Zhang, M.; Bai, Z.; Shao, Y.; Qi, K.; Tu, J.; Wang, C.; et al. Development of a SERS-Based Lateral Flow Immunoassay for Rapid and Ultra-Sensitive Detection of Anti-SARS-CoV-2 IgM/IgG in Clinical Samples. Sens. Actuators B Chem. 2021, 329, 129196. [Google Scholar] [CrossRef] [PubMed]
- Antoine, D.; Mohammadi, M.; Vitt, M.; Dickie, J.M.; Jyoti, S.S.; Tilbury, M.A.; Johnson, P.A.; Wawrousek, K.E.; Wall, J.G. Rapid, Point-of-Care ScFv-SERS Assay for Femtogram Level Detection of SARS-CoV-2. ACS Sens. 2022, 7, 866–873. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Luo, Y.; Song, Y.; Zhu, Q.; Xu, T.; Zhang, X. One-Click Investigation of Shape Influence of Silver Nanostructures on SERS Performance for Sensitive Detection of COVID-19. Anal. Chim. Acta 2022, 1234, 340523. [Google Scholar] [CrossRef]
- Kim, W.; Kim, S.; Han, J.; Kim, T.G.; Bang, A.; Choi, H.W.; Min, G.E.; Shin, J.H.; Moon, S.W.; Choi, S. An Excitation Wavelength-Optimized, Stable SERS Biosensing Nanoplatform for Analyzing Adenoviral and AstraZeneca COVID-19 Vaccination Efficacy Status Using Tear Samples of Vaccinated Individuals. Biosens. Bioelectron. 2022, 204, 114079. [Google Scholar] [CrossRef] [PubMed]
- Karunakaran, V.; Joseph, M.M.; Yadev, I.; Sharma, H.; Shamna, K.; Saurav, S.; Sreejith, R.P.; Anand, V.; Beegum, R.; Regi David, S.; et al. A Non-Invasive Ultrasensitive Diagnostic Approach for COVID-19 Infection Using Salivary Label-Free SERS Fingerprinting and Artificial Intelligence. J. Photochem. Photobiol. B Biol. 2022, 234, 112545. [Google Scholar] [CrossRef]
- Shanmukh, S.; Jones, L.; Driskell, J.; Zhao, Y.; Dluhy, R.; Tripp, R.A. Rapid and Sensitive Detection of Respiratory Virus Molecular Signatures Using a Silver Nanorod Array SERS Substrate. Nano Lett. 2006, 6, 2630–2636. [Google Scholar] [CrossRef] [PubMed]
- Dluhy, R.A.; Shanmukh, S.; Jones, L.; Zhao, Y.P.; Driskell, J.D.; Tripp, R.A. Identification and Classification of Respiratory Syncytial Virus (RSV) Strains by Surface-Enhanced Raman Spectroscopy and Multivariate Statistical Techniques. Anal. Bioanal. Chem. 2008, 390, 1551–1555. [Google Scholar] [CrossRef]
- Huang, J.; Wen, J.; Zhou, M.; Ni, S.; Le, W.; Chen, G.; Wei, L.; Zeng, Y.; Qi, D.; Pan, M.; et al. On-Site Detection of SARS-CoV-2 Antigen by Deep Learning-Based Surface-Enhanced Raman Spectroscopy and Its Biochemical Foundations. Anal. Chem. 2021, 93, 9174–9182. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.J.; Le, T.N.; Hsiao, W.W.W.; Tung, K.L.; Ostrikov, K.; Chiang, W.H. Plasmonic Nanostructure-Enhanced Raman Scattering for Detection of SARS-CoV-2 Nucleocapsid Protein and Spike Protein Variants. Anal. Chim. Acta 2023, 1239, 340651. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, C.; Zheng, S.; Yang, X.; Han, H.; Dai, Y.; Xiao, R. Simultaneously Ultrasensitive and Quantitative Detection of Influenza A Virus, SARS-CoV-2, and Respiratory Syncytial Virus via Multichannel Magnetic SERS-Based Lateral Flow Immunoassay. Nanomed. Nanotechnol. Biol. Med. 2023, 47, 102624. [Google Scholar] [CrossRef] [PubMed]
- Chisanga, M.; Williams, H.; Boudreau, D.; Pelletier, J.N.; Trottier, S. Label-Free SERS for Rapid Differentiation of SARS-CoV-2-Induced Serum Metabolic Profiles in Non-Hospitalized Adults. Anal. Chem. 2023, 95, 3638–3646. [Google Scholar] [CrossRef] [PubMed]
- Bacteria. Available online: https://microbiologysociety.org/why-microbiology-matters/what-is-microbiology/bacteria.html#:~:text=Bacteria%20are%20classified%20into%20five,)%20or%20corkscrew%20(spirochaetes) (accessed on 30 January 2023).
- Bacteria as Pathogens. Available online: https://sphweb.bumc.bu.edu/otlt/mph-modules/ph/ph709_infectiousagents/PH709_InfectiousAgents4.html#:~:text=While%20only%20about%205%25%20of,of%20human%20disease%20and%20death (accessed on 30 January 2023).
- Nanda, M.; Kumar, V.; Sharma, D.K. Multimetal Tolerance Mechanisms in Bacteria: The Resistance Strategies Acquired by Bacteria That Can Be Exploited to ‘Clean-up’ Heavy Metal Contaminants from Water. Aquat. Toxicol. 2019, 212, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Phytoremediation: Synergistic Use of Plants and Bacteria to Clean up the Environment. Biotechnol. Adv. 2003, 21, 383–393. [Google Scholar] [CrossRef]
- Kulshreshtha, A.; Agrawal, R.; Barar, M.; Saxena, S. A Review on Bioremediation of Heavy Metals in Contaminated Water. IOSR J. Environ. Sci. Toxicol. Food Technol. 2014, 8, 44–50. [Google Scholar] [CrossRef]
- Zhou, X.; Hu, Z.; Yang, D.; Xie, S.; Jiang, Z.; Niessner, R.; Haisch, C.; Zhou, H.; Sun, P. Bacteria Detection: From Powerful SERS to Its Advanced Compatible Techniques. Adv. Sci. 2020, 7, 2001739. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Kim, K.; Choi, N.; Wang, X.; Lee, J.; Jeon, J.H.; Rhie, G.E.; Choo, J. Highly Sensitive Detection of High-Risk Bacterial Pathogens Using SERS-Based Lateral Flow Assay Strips. Sens. Actuators B Chem. 2018, 270, 72–79. [Google Scholar] [CrossRef]
- Zhu, T.; Hu, Y.; Yang, K.; Dong, N.; Yu, M.; Jiang, N. A Novel SERS Nanoprobe Based on the Use of Core-Shell Nanoparticles with Embedded Reporter Molecule to Detect E. coli O157:H7 with High Sensitivity. Microchim. Acta 2018, 185, 30. [Google Scholar] [CrossRef]
- Chisanga, M.; Muhamadali, H.; Ellis, D.I.; Goodacre, R. Surface-Enhanced Raman Scattering (SERS) in Microbiology: Illumination and Enhancement of the Microbial World. Appl. Spectrosc. 2018, 72, 987–1000. [Google Scholar] [CrossRef]
- Kim, J.A.; Wales, D.J.; Thompson, A.J.; Yang, G.Z. Fiber-optic SERS probes fabricated using two-photon polymerization for rapid detection of bacteria. Adv. Opt. Mater. 2020, 8, 1901934. [Google Scholar] [CrossRef]
- Zhou, H.; Yang, D.; Ivleva, N.P.; Mircescu, N.E.; Niessner, R.; Haisch, C. SERS Detection of Bacteria in Water by in Situ Coating with Ag Nanoparticles. Anal. Chem. 2014, 86, 1525–1533. [Google Scholar] [CrossRef]
- Yang, Y.; Zeng, C.; Huang, J.; Wang, M.; Qi, W.; Wang, H.; He, Z. Specific and Quantitative Detection of Bacteria Based on Surface Cell Imprinted SERS Mapping Platform. Biosens. Bioelectron. 2022, 215, 114524. [Google Scholar] [CrossRef]
- Pearson, B.; Wang, P.; Mills, A.; Pang, S.; McLandsborough, L.; He, L. Innovative Sandwich Assay with Dual Optical and SERS Sensing Mechanisms for Bacterial Detection. Anal. Methods 2017, 9, 4732–4739. [Google Scholar] [CrossRef]
- Hudson, S.D.; Chumanov, G. Bioanalytical Applications of SERS (Surface-Enhanced Raman Spectroscopy). Anal. Bioanal. Chem. 2009, 394, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Mosier-Boss, P.A. Review on SERS of Bacteria. Biosensors 2017, 7, 51. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, R.M.; Goodacre, R. Characterisation and Identification of Bacteria Using SERS. Chem. Soc. Rev. 2008, 37, 931–936. [Google Scholar] [CrossRef]
- Xia, J.; Li, W.; Sun, M.; Wang, H. Application of SERS in the Detection of Fungi, Bacteria and Viruses. Nanomaterials 2022, 12, 3572. [Google Scholar] [CrossRef]
- Wang, C.; Meloni, M.M.; Wu, X.; Zhuo, M.; He, T.; Wang, J.; Wang, C.; Dong, P. Magnetic Plasmonic Particles for SERS-Based Bacteria Sensing: A Review. AIP Adv. 2019, 9, 010701. [Google Scholar] [CrossRef]
- Efrima, S.; Zeiri, L. Understanding SERS of Bacteria. J. Raman Spectrosc. 2009, 40, 277–288. [Google Scholar] [CrossRef]
- Liu, H.B.; Du, X.J.; Zang, Y.X.; Li, P.; Wang, S. SERS-Based Lateral Flow Strip Biosensor for Simultaneous Detection of Listeria Monocytogenes and Salmonella Enterica Serotype Enteritidis. J. Agric. Food Chem. 2017, 65, 10290–10299. [Google Scholar] [CrossRef]
- Mungroo, N.A.; Oliveira, G.; Neethirajan, S. SERS Based Point-of-Care Detection of Food-Borne Pathogens. Microchim. Acta 2016, 183, 697–707. [Google Scholar] [CrossRef]
- Lin, H.Y.; Huang, C.H.; Hsieh, W.H.; Liu, L.H.; Lin, Y.C.; Chu, C.C.; Wang, S.T.; Kuo, I.T.; Chau, L.K.; Yang, C.Y. On-Line SERS Detection of Single Bacterium Using Novel SERS Nanoprobes and a Microfl Uidic Dielectrophoresis Device. Small 2014, 10, 4700–4710. [Google Scholar] [CrossRef]
- Wang, Y.; Lee, K.; Irudayaraj, J. Silver Nanosphere SERS Probes for Sensitive Identification of Pathogens. J. Phys. Chem. C 2010, 114, 16122–16128. [Google Scholar] [CrossRef]
- Witkowska, E.; Korsak, D.; Kowalska, A.; Janeczek, A.; Kamińska, A. Strain-Level Typing and Identification of Bacteria—A Novel Approach for SERS Active Plasmonic Nanostructures. Anal. Bioanal. Chem. 2018, 410, 5019–5031. [Google Scholar] [CrossRef]
- Pang, Y.; Wan, N.; Shi, L.; Wang, C.; Sun, Z.; Xiao, R.; Wang, S. Dual-Recognition Surface-Enhanced Raman Scattering (SERS)Biosensor for Pathogenic Bacteria Detection by Using Vancomycin-SERS Tags and Aptamer-Fe3O4@Au. Anal. Chim. Acta 2019, 1077, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Yin, Y.; Wu, H.; Hao, Z.; Li, J.; Wang, S.; Liu, Y. Integrated SERS Platform for Reliable Detection and Photothermal Elimination of Bacteria in Whole Blood Samples. Anal. Chem. 2021, 93, 1569–1577. [Google Scholar] [CrossRef]
- Zhou, Z.; Xiao, R.; Cheng, S.; Wang, S.; Shi, L.; Wang, C.; Qi, K.; Wang, S. A Universal SERS-Label Immunoassay for Pathogen Bacteria Detection Based on Fe3O4@Au-Aptamer Separation and Antibody-Protein A Orientation Recognition. Anal. Chim. Acta 2021, 1160, 338421. [Google Scholar] [CrossRef]
- Wang, J.; Wu, X.; Wang, C.; Rong, Z.; Ding, H.; Li, H.; Li, S.; Shao, N.; Dong, P.; Xiao, R.; et al. Facile Synthesis of Au-Coated Magnetic Nanoparticles and Their Application in Bacteria Detection via a SERS Method. ACS Appl. Mater. Interfaces 2016, 8, 19958–19967. [Google Scholar] [CrossRef]
- Wang, C.; Wang, J.; Li, M.; Qu, X.; Zhang, K.; Rong, Z.; Xiao, R.; Wang, S. A Rapid SERS Method for Label-Free Bacteria Detection Using Polyethylenimine-Modified Au-Coated Magnetic Microspheres and Au@Ag Nanoparticles. Analyst 2016, 141, 6226–6238. [Google Scholar] [CrossRef]
- Huang, L.; Sun, D.W.; Wu, Z.; Pu, H.; Wei, Q. Reproducible, Shelf-Stable, and Bioaffinity SERS Nanotags Inspired by Multivariate Polyphenolic Chemistry for Bacterial Identification. Anal. Chim. Acta 2021, 1167, 338570. [Google Scholar] [CrossRef] [PubMed]
- Bi, L.; Wang, X.; Cao, X.; Liu, L.; Bai, C.; Zheng, Q.; Choo, J.; Chen, L. SERS-Active Au@Ag Core-Shell Nanorod (Au@AgNR) Tags for Ultrasensitive Bacteria Detection and Antibiotic-Susceptibility Testing. Talanta 2020, 220, 121397. [Google Scholar] [CrossRef] [PubMed]
- Hunter, R.; Sohi, A.N.; Khatoon, Z.; Berthiaume, V.R.; Alarcon, E.I.; Godin, M.; Anis, H. Optofluidic Label-Free SERS Platform for Rapid Bacteria Detection in Serum. Sens. Actuators B Chem. 2019, 300, 126907. [Google Scholar] [CrossRef]
- Sivanesan, A.; Witkowska, E.; Adamkiewicz, W.; Dziewit, Ł.; Kamińska, A.; Waluk, J. Nanostructured Silver-Gold Bimetallic SERS Substrates for Selective Identification of Bacteria in Human Blood. Analyst 2014, 139, 1037–1043. [Google Scholar] [CrossRef]
- Witkowska, E.; Szymborski, T.; Kamińska, A.; Waluk, J. Polymer Mat Prepared via ForcespinningTM as a SERS Platform for Immobilization and Detection of Bacteria from Blood Plasma. Mater. Sci. Eng. C 2017, 71, 345–350. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, J.; Mi, L.; Gong, H.; Jiang, S.; Yu, Q. Multifunctional Magnetic-Plasmonic Nanoparticles for Fast Concentration and Sensitive Detection of Bacteria Using SERS. Biosens. Bioelectron. 2012, 31, 130–136. [Google Scholar] [CrossRef]
- Krafft, B.; Tycova, A.; Urban, R.D.; Dusny, C.; Belder, D. Microfluidic device for concentration and SERS-based detection of bacteria in drinking water. Electrophoresis 2021, 42, 86–94. [Google Scholar] [CrossRef]
- Yang, E.; Li, D.; Yin, P.; Xie, Q.; Li, Y.; Lin, Q.; Duan, Y. A Novel Surface-Enhanced Raman Scattering (SERS) Strategy for Ultrasensitive Detection of Bacteria Based on Three-Dimensional (3D) DNA Walker. Biosens. Bioelectron. 2021, 172, 112758. [Google Scholar] [CrossRef]
- Wang, C.; Wang, C.; Li, J.; Tu, Z.; Gu, B.; Wang, S. Ultrasensitive and Multiplex Detection of Four Pathogenic Bacteria on a Bi-Channel Lateral Flow Immunoassay Strip with Three-Dimensional Membrane-like SERS Nanostickers. Biosens. Bioelectron. 2022, 214, 114525. [Google Scholar] [CrossRef]
- Kearns, H.; Goodacre, R.; Jamieson, L.E.; Graham, D.; Faulds, K. SERS Detection of Multiple Antimicrobial-Resistant Pathogens Using Nanosensors. Anal. Chem. 2017, 89, 12666–12673. [Google Scholar] [CrossRef]
- Gracie, K.; Correa, E.; Mabbott, S.; Dougan, J.A.; Graham, D.; Goodacre, R.; Faulds, K. Simultaneous Detection and Quantification of Three Bacterial Meningitis Pathogens by SERS. Chem. Sci. 2014, 5, 1030–1040. [Google Scholar] [CrossRef]
- Mosier-Boss, P.A.; Sorensen, K.C.; George, R.D.; Obraztsova, A. SERS Substrates Fabricated Using Ceramic Filters for the Detection of Bacteria. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2016, 153, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Walter, A.; März, A.; Schumacher, W.; Rösch, P.; Popp, J. Towards a Fast, High Specific and Reliable Discrimination of Bacteria on Strain Level by Means of SERS in a Microfluidic Device. Lab Chip 2011, 11, 1013–1021. [Google Scholar] [CrossRef]
- Wu, X.; Xu, C.; Tripp, R.A.; Huang, Y.W.; Zhao, Y. Detection and Differentiation of Foodborne Pathogenic Bacteria in Mung Bean Sprouts Using Field Deployable Label-Free SERS Devices. Analyst 2013, 138, 3005–3012. [Google Scholar] [CrossRef] [PubMed]
- Ankamwar, B.; Sur, U.K.; Das, P. SERS Study of Bacteria Using Biosynthesized Silver Nanoparticles as the SERS Substrate. Anal. Methods 2016, 8, 2335–2340. [Google Scholar] [CrossRef]
- Ciloglu, F.U.; Caliskan, A.; Saridag, A.M.; Kilic, I.H.; Tokmakci, M.; Kahraman, M.; Aydin, O. Drug-Resistant Staphylococcus Aureus Bacteria Detection by Combining Surface-Enhanced Raman Spectroscopy (SERS) and Deep Learning Techniques. Sci. Rep. 2021, 11, 18444. [Google Scholar] [CrossRef]
- Nerve Agents, BBC Report. Available online: https://www.bbc.com/news/uk-43431537 (accessed on 30 January 2023).
- Lister, A.P.; Sellors, W.J.; Howle, C.R.; Mahajan, S. Raman Scattering Techniques for Defense and Security Applications. Anal. Chem. 2021, 93, 417–429. [Google Scholar] [CrossRef]
- Sadayoshi, O.H.B.U.; Yamashina, A.; Takasu, N.; Yamaguchi, T.; Murai, T.; Nakano, K.; Hinohara, S. Sarin poisoning on Tokyo subway. South. Med. J. 1997, 90, 587–593. [Google Scholar]
- Inscore, F.E.; Gift, A.D.; Maksymiuk, P.; Farquharson, S. Characterization of Chemical Warfare G-Agent Hydrolysis Products by Surface-Enhanced Raman Spectroscopy. Chem. Biol. Point Sens. Homel. Def. II 2004, 5585, 46. [Google Scholar] [CrossRef]
- Mukherjee, S.; Gupta, R.D. Organophosphorus Nerve Agents: Types, Toxicity, and Treatments. J. Toxicol. 2020, 2020, 3007984. [Google Scholar] [CrossRef]
- Saylan, Y.; Akgönüllü, S.; Denizli, A. Plasmonic Sensors for Monitoring Biological and Chemical Threat Agents. Biosensors 2020, 10, 142. [Google Scholar] [CrossRef] [PubMed]
- Primera-Pedrozo, O.M.; Jerez-Rozo, J.I.; De La Cruz-Montoya, E.; Luna-Pineda, T.; Pacheco-Londoño, L.C.; Hernández-Rivera, S.P. Nanotechnology-Based Detection of Explosives and Biological Agents Simulants. IEEE Sens. J. 2008, 8, 963–973. [Google Scholar] [CrossRef]
- Yan, F.; Vo-Dinh, T. Surface-Enhanced Raman Scattering Detection of Chemical and Biological Agents Using a Portable Raman Integrated Tunable Sensor. Sens. Actuators B Chem. 2007, 121, 61–66. [Google Scholar] [CrossRef]
- Pearman, W.F.; Fountain, A.W. Classification of Chemical and Biological Warfare Agent Simulants by Surface-Enhanced Raman Spectroscopy and Multivariate Statistical Techniques. Appl. Spectrosc. 2006, 60, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Hakonen, A.; Rindzevicius, T.; Schmidt, M.S.; Andersson, P.O.; Juhlin, L.; Svedendahl, M.; Boisen, A.; Käll, M. Detection of Nerve Gases Using Surface-Enhanced Raman Scattering Substrates with High Droplet Adhesion. Nanoscale 2016, 8, 1305–1308. [Google Scholar] [CrossRef] [PubMed]
- Juhlin, L.; Mikaelsson, T.; Hakonen, A.; Stenbæk, M. Selective Surface-Enhanced Raman Scattering Detection of Tabun, VX and Cyclosarin Nerve Agents Using 4-Pyridine Amide Oxime Functionalized Gold Nanopillars Talanta Selective Surface-Enhanced Raman Scattering Detection of Tabun, VX and Cyclosarin Nerve A. Talanta 2020, 211, 120721. [Google Scholar] [CrossRef]
- Farquharson, S.; Gift, A.; Maksymiuk, P.; Inscore, F. Surface-Enhanced Raman Spectra of VX and Its Hydrolysis Products. Appl. Spectrosc. 2005, 59, 654–660. [Google Scholar] [CrossRef]
- Heleg-Shabtai, V.; Sharabi, H.; Zaltsman, A.; Ron, I.; Pevzner, A. Surface-Enhanced Raman Spectroscopy (SERS) for Detection of VX and HD in the Gas Phase Using a Hand-Held Raman Spectrometer. Analyst 2020, 145, 6334–6341. [Google Scholar] [CrossRef]
- Zhao, Q.; Liu, G.; Zhang, H.; Zhou, F.; Li, Y.; Cai, W. SERS-Based Ultrasensitive Detection of Organophosphorus Nerve Agents via Substrate’s Surface Modification. J. Hazard. Mater. 2017, 324, 194–202. [Google Scholar] [CrossRef]
- Spencer, K.M.; Sylvia, J.M.; Clauson, S.L.; Janni, J.A. Surface-Enhanced Raman as a Water Monitor for Warfare Agents. Vib. Spectrosc. Sens. Syst. 2002, 4577, 158. [Google Scholar] [CrossRef]
- Bertone, J.F.; Cordeiro, K.L.; Sylvia, J.M.; Spencer, K.M. A Nanoengineered Sensor to Detect Vibrational Modes of Warfare Agents/Explosives Using Surface-Enhanced Raman Scattering. Sens. Command. Control. Commun. Intell. Technol. Homel. Secur. Homel. Def. III 2004, 5403, 387. [Google Scholar] [CrossRef]
- Kim, Y.T.; Kim, D.; Park, S.; Zhexembekova, A.; Byeon, M.; Hong, T.E.; Lee, J.; Lee, C.Y. Aqueous Microlenses for Localized Collection and Enhanced Raman Spectroscopy of Gaseous Molecules. Adv. Opt. Mater. 2021, 9, 2101209. [Google Scholar] [CrossRef]
- Wu, J.; Zhu, Y.; Gao, J.; Chen, J.; Feng, J.; Guo, L.; Xie, J. A Simple and Sensitive Surface-Enhanced Raman Spectroscopic Discriminative Detection of Organophosphorous Nerve Agents. Anal. Bioanal. Chem. 2017, 409, 5091–5099. [Google Scholar] [CrossRef] [PubMed]
- Lafuente, M.; Berenschot, E.J.W.; Tiggelaar, R.M.; Mallada, R.; Tas, N.R.; Pina, M.P. 3D Fractals as SERS Active Platforms: Preparation and Evaluation for Gas Phase Detection of G-Nerve Agents. Micromachines 2018, 9, 60. [Google Scholar] [CrossRef] [PubMed]
- Goel, A.K. Anthrax: A Disease of Biowarfare and Public Health Importance. World J. Clin. Cases 2015, 3, 20. [Google Scholar] [CrossRef] [PubMed]
- Farrell, M.E.; Pellegrino, P.M. Army Relevant Biological Hazards Detection with Commercial SERS Substrates. Biosen. Nanomed. V 2012, 8460, 84600J. [Google Scholar] [CrossRef]
- Sajanlal, P.R.; Pradeep, T. Functional Hybrid Nickel Nanostructures as Recyclable SERS Substrates: Detection of Explosives and Biowarfare Agents. Nanoscale 2012, 4, 3427–3437. [Google Scholar] [CrossRef]
- Wang, T.; Dong, P.; Zhu, C.; Sha, P.; Gao, W.; Wu, Y.; Wu, X. Trace Detection of Anthrax Protective Antigens via a Competitive Method Based on Surface-Enhanced Raman Scattering. Sens. Actuators B Chem. 2021, 346, 130467. [Google Scholar] [CrossRef]
- Gao, R.; Ko, J.; Cha, K.; Ho Jeon, J.; Rhie, G.E.; Choi, J.; de Mello, A.J.; Choo, J. Fast and Sensitive Detection of an Anthrax Biomarker Using SERS-Based Solenoid Microfluidic Sensor. Biosens. Bioelectron. 2015, 72, 230–236. [Google Scholar] [CrossRef]
- Naqvi, T.K.; Bajpai, A.; Bharati, M.S.S.; Kulkarni, M.M.; Siddiqui, A.M.; Soma, V.R.; Dwivedi, P.K. Ultra-Sensitive Reusable SERS Sensor for Multiple Hazardous Materials Detection on Single Platform. J. Hazard. Mater. 2021, 407, 124353. [Google Scholar] [CrossRef]
- Sengupta, A.; Shende, C.; Farquharson, S.; Inscore, F. Detection of Bacillus Anthracis Spores Using Peptide Functionalized SERS-Active Substrates. Int. J. Spectrosc. 2012, 2012, 176851. [Google Scholar] [CrossRef]
- Yilmaz, M.; Senlik, E.; Biskin, E.; Yavuz, M.S.; Tamer, U.; Demirel, G. Combining 3-D Plasmonic Gold Nanorod Arrays with Colloidal Nanoparticles as a Versatile Concept for Reliable, Sensitive, and Selective Molecular Detection by SERS. Phys. Chem. Chem. Phys. 2014, 16, 5563–5570. [Google Scholar] [CrossRef]
- Li, B.; Wang, T.; Bai, W.; Su, Q.; Wu, X.; Dong, P. Label-Free and Rapid Detection of Anthrax Protective Antigen by Surface-Enhanced Raman Scattering on Au Nanorods. IEEE Sens. J. 2021, 21, 18425–18434. [Google Scholar] [CrossRef]
- Cheung, M.; Lee, W.W.Y.; Cowcher, D.P.; Goodacre, R.; Bell, S.E.J. SERS of Meso-Droplets Supported on Superhydrophobic Wires Allows Exquisitely Sensitive Detection of Dipicolinic Acid, an Anthrax Biomarker, Considerably below the Infective Dose. Chem. Commun. 2016, 52, 9925–9928. [Google Scholar] [CrossRef] [PubMed]
- Félix-Rivera, H.; González, R.; Rodríguez, G.D.M.; Primera-Pedrozo, O.M.; Ríos-Velázquez, C.; Hernández-Rivera, S.P. Improving SERS Detection of Bacillus Thuringiensis Using Silver Nanoparticles Reduced with Hydroxylamine and with Citrate Capped Borohydride. Int. J. Spectrosc. 2011, 2011, 989504. [Google Scholar] [CrossRef]
- FountainIII, A.W.; Pearman, W.F. Multivariate Statistical Classification of Surface Enhanced Raman Spectra of Chemical and Biological Warfare Agent Simulants. Chem. Biol. Sens. Ind. Environ. Secur. 2005, 5994, 180–193. [Google Scholar] [CrossRef]
- Arano-Martinez, J.A.; Martínez-González, C.L.; Salazar, M.I.; Torres-Torres, C. A Framework for Biosensors Assisted by Multiphoton Effects and Machine Learning. Biosensors 2022, 12, 710. [Google Scholar] [CrossRef]
- Luo, R.; Popp, J.; Bocklitz, T. Deep Learning for Raman Spectroscopy: A Review. Analytica 2022, 3, 287–301. [Google Scholar] [CrossRef]
- Ralbovsky, N.M.; Lednev, I.K. Towards Development of a Novel Universal Medical Diagnostic Method: Raman Spectroscopy and Machine Learning. Chem. Soc. Rev. 2020, 49, 7428–7453. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, Y.; Liu, C.; Jiang, Q.Y.; Chen, F.; Cao, Y. SERS-Based Biosensors Combined with Machine Learning for Medical Application. ChemistryOpen 2023, 12, e202200192. [Google Scholar] [CrossRef]
- Cui, F.; Yue, Y.; Zhang, Y.; Zhang, Z.; Zhou, H.S. Advancing Biosensors with Machine Learning. ACS Sens. 2020, 5, 3346–3364. [Google Scholar] [CrossRef]
- Lv, R.; Wang, Z.; Ma, Y.; Li, W.; Tian, J. Machine Learning Enhanced Optical Spectroscopy for Disease Detection. J. Phys. Chem. Lett. 2022, 13, 9238–9249. [Google Scholar] [CrossRef]
- Schackart, K.E.; Yoon, J.Y. Machine Learning Enhances the Performance of Bioreceptor-Free Biosensors. Sensors 2021, 21, 5519. [Google Scholar] [CrossRef]
- Lussier, F.; Missirlis, D.; Spatz, J.P.; Masson, J.F. Machine-Learning-Driven Surface-Enhanced Raman Scattering Optophysiology Reveals Multiplexed Metabolite Gradients Near Cells. ACS Nano 2019, 13, 1403–1411. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.H.L.; Hong, B.; Rubin, S.; Fainman, Y. Machine Learning for Composition Analysis of SsDNA Using Chemical Enhancement in SERS. Biomed. Opt. Express 2020, 11, 5092. [Google Scholar] [CrossRef] [PubMed]
- Narla, L.M.; Rao, S.V. Identification of Metals and Alloys Using Color CCD Images of Laser-Induced Breakdown Emissions Coupled with Machine Learning. Appl. Phys. B Lasers Opt. 2020, 126, 113. [Google Scholar] [CrossRef]
- Beeram, R.; Banerjee, D.; Narlagiri, L.M.; Soma, V.R. Machine Learning for Rapid Quantification of Trace Analyte Molecules Using SERS and Flexible Plasmonic Paper Substrates. Anal. Methods 2022, 14, 1788–1796. [Google Scholar] [CrossRef]
- Murthy, N.L.; Abdul Salam, S.; Rao, S.V. Stand-off Femtosecond Laser Induced Breakdown Spectroscopy of Metals, Soil, Plastics and Classification Studies. In Proceedings of the 2019 Workshop on Recent Advances in Photonics (WRAP), Guwahati, India, 13–14 December 2019; pp. 1–3. [Google Scholar] [CrossRef]
- Boehmke, B.; Greenwell, B. Hands-On Machine Learning with R; Chapman and Hall/CRC: Boca Raton, FL, USA, 2019; ISBN 9781492032649. [Google Scholar]
- Li, D.; Zhang, Q.; Deng, B.; Chen, Y.; Ye, L. Rapid, Sensitive Detection of Ganciclovir, Penciclovir and Valacyclovir-Hydrochloride by Artificial Neural Network and Partial Least Squares Combined with Surface Enhanced Raman Spectroscopy. Appl. Surf. Sci. 2021, 539, 148224. [Google Scholar] [CrossRef]
- Boulesteix, A.L.; Strimmer, K. Partial Least Squares: A Versatile Tool for the Analysis of High-Dimensional Genomic Data. Brief. Bioinform. 2007, 8, 32–44. [Google Scholar] [CrossRef]
- Deng, W.; Huang, Z.; Zhang, J.; Xu, J. A Data Mining Based System for Transaction Fraud Detection. In Proceedings of the 2021 IEEE International Conference on Consumer Electronics and Computer Engineering (ICCECE), Guangzhou, China, 15–17 January 2021; pp. 542–545. [Google Scholar] [CrossRef]
- Fan, X.; Ming, W.; Zeng, H.; Zhang, Z.; Lu, H. Deep Learning-Based Component Identification for the Raman Spectra of Mixtures. Analyst 2019, 144, 1789–1798. [Google Scholar] [CrossRef]
- Zhou, H.; Xu, L.; Ren, Z.; Zhu, J.; Lee, C. Machine Learning-Augmented Surface-Enhanced Spectroscopy toward next-Generation Molecular Diagnostics. Nanoscale Adv. 2023, 5, 538–570. [Google Scholar] [CrossRef]
- Malinick, A.S.; Stuart, D.D.; Lambert, A.S.; Cheng, Q. Surface Plasmon Resonance Imaging (SPRi) in Combination with Machine Learning for Microarray Analysis of Multiple Sclerosis Biomarkers in Whole Serum. Biosens. Bioelectron. X 2022, 10, 100127. [Google Scholar] [CrossRef]
- Pradhan, P.; Guo, S.; Ryabchykov, O.; Popp, J.; Bocklitz, T.W. Deep Learning a Boon for Biophotonics? J. Biophotonics 2020, 13, e201960186. [Google Scholar] [CrossRef] [PubMed]
- Moon, G.; Lee, J.; Lee, H.; Yoo, H.; Ko, K.; Im, S.; Kim, D. Machine Learning and Its Applications for Plasmonics in Biology. Cell Rep. Phys. Sci. 2022, 3, 101042. [Google Scholar] [CrossRef]
- Sun, Y.; Shi, L.; Mi, L.; Guo, R.; Li, T. Recent Progress of SERS Optical Nanosensors for MiRNA Analysis. J. Mater. Chem. B 2020, 8, 5178–5183. [Google Scholar] [CrossRef] [PubMed]
- Raji, H.; Tayyab, M.; Sui, J.; Mahmoodi, S.R.; Javanmard, M. Biosensors and Machine Learning for Enhanced Detection, Stratification, and Classification of Cells: A Review. Biomed. Microdevices 2022, 24, 26. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Maity, S.; Mastrangelo, C.H. Nanostructures for biosensing, with a brief overview on Cancer Detection, IoT, and the Role of Machine Learning In Smart Biosensors. Sensors 2021, 21, 1253. [Google Scholar] [CrossRef]
- Beeram, R.; Vendamani, V.S.; Soma, V.R. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy Deep Learning Approach to Overcome Signal Fluctuations in SERS for Efficient On-Site Trace Explosives Detection. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 289, 122218. [Google Scholar] [CrossRef] [PubMed]
- Moon, G.; Son, T.; Lee, H.; Kim, D. Deep Learning Approach for Enhanced Detection of Surface Plasmon Scattering. Anal. Chem. 2019, 91, 9538–9545. [Google Scholar] [CrossRef]
- Gupta, A.K.; Hsu, C.H.; Lai, C.S. Enhancement of the Au/ZnO-NA Plasmonic SERS Signal Using Principal Component Analysis as a Machine Learning Approach. IEEE Photonics J. 2020, 12, 1–11. [Google Scholar] [CrossRef]
- Vendamani, V.S.; Beeram, R.; Neethish, M.M.; Rao, S.V.S.N.; Rao, S.V. Wafer-Scale Silver Nanodendrites with Homogeneous Distribution of Gold Nanoparticles for Biomolecules Detection. iScience 2022, 25, 104849. [Google Scholar] [CrossRef]
- Erzina, M.; Trelin, A.; Guselnikova, O.; Dvorankova, B.; Strnadova, K.; Perminova, A.; Ulbrich, P.; Mares, D.; Jerabek, V.; Elashnikov, R.; et al. Precise Cancer Detection via the Combination of Functionalized SERS Surfaces and Convolutional Neural Network with Independent Inputs. Sens. Actuators B Chem. 2020, 308, 127660. [Google Scholar] [CrossRef]
- Wang, S.; Dong, H.; Shen, W.; Yang, Y.; Li, Z.; Liu, Y.; Wang, C.; Gu, B.; Zhang, L. Rapid SERS Identification of Methicillin-Susceptible and Methicillin-Resistant: Staphylococcus Aureus via Aptamer Recognition and Deep Learning. RSC Adv. 2021, 11, 34425–34431. [Google Scholar] [CrossRef]
- Kazemzadeh, M.; Hisey, C.L.; Dauros-Singorenko, P.; Swift, S.; Zargar-Shoshtari, K.; Xu, W.; Broderick, N.G.R. Label-Free Classification of Bacterial Extracellular Vesicles by Combining Nanoplasmonic Sensors with Machine Learning. IEEE Sens. J. 2022, 22, 1128–1137. [Google Scholar] [CrossRef]
- Dong, R.; Weng, S.; Yang, L.; Liu, J. Detection and Direct Readout of Drugs in Human Urine Using Dynamic Surface-Enhanced Raman Spectroscopy and Support Vector Machines. Anal. Chem. 2015, 87, 2937–2944. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Liang, S.; Li, Y.; Peng, Y.; Huang, Z.; Li, Z.; Yang, Y.; Luo, X. Localized Plasmonic Sensor for Direct Identifying Lung and Colon Cancer from the Blood. Biosens. Bioelectron. 2022, 211, 114372. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Lu, D.; Zhang, B.; You, R.; Chen, J.; Xu, H.; Lu, Y. Machine Learning—Assisted Internal Standard Calibration Label—Free SERS Strategy for Colon Cancer Detection. Anal. Bioanal. Chem. 2023. [Google Scholar] [CrossRef] [PubMed]
- Seifert, S.; Merk, V.; Kneipp, J. Identification of Aqueous Pollen Extracts Using Surface Enhanced Raman Scattering (SERS) and Pattern Recognition Methods. J. Biophotonics 2016, 9, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Hassoun, M.; Rüger, J.; Kirchberger-Tolstik, T.; Schie, I.W.; Henkel, T.; Weber, K.; Cialla-May, D.; Krafft, C.; Popp, J. A Droplet-Based Microfluidic Chip as a Platform for Leukemia Cell Lysate Identification Using Surface-Enhanced Raman Scattering. Anal. Bioanal. Chem. 2018, 410, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Mühlig, A.; Bocklitz, T.; Labugger, I.; Dees, S.; Henk, S.; Richter, E.; Andres, S.; Merker, M.; Stöckel, S.; Weber, K.; et al. LOC-SERS: A Promising Closed System for the Identification of Mycobacteria. Anal. Chem. 2016, 88, 7998–8004. [Google Scholar] [CrossRef] [PubMed]
- Bratchenko, L.A.; Al-Sammarraie, S.Z.; Tupikova, E.N.; Konovalova, D.Y.; Lebedev, P.A.; Zakharov, V.P.; Bratchenko, I.A. Analyzing the Serum of Hemodialysis Patients with End-Stage Chronic Kidney Disease by Means of the Combination of SERS and Machine Learning. Biomed. Opt. Express 2022, 13, 4926. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Zhu, H.; Charron, B.; Mochizuki, T.; Dong, C.; Ding, H.; Cui, Y.; Lu, M.; Peng, W.; Zhu, S.; et al. Combining Dense Au Nanoparticle Layers and 2D Surface-Enhanced Raman Scattering Arrays for the Identification of Mutant Cyanobacteria Using Machine Learning. J. Phys. Chem. C 2022, 126, 9446–9455. [Google Scholar] [CrossRef]
- Ikponmwoba, E.; Ukorigho, O.; Moitra, P.; Pan, D.; Gartia, M.R.; Owoyele, O. A Machine Learning Framework for Detecting COVID-19 Infection Using Surface-Enhanced Raman Scattering. Biosensors 2022, 12, 589. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Jiménez, A.I.; Lyu, D.; Lu, Z.; Liu, G.; Ren, B. Surface-Enhanced Raman Spectroscopy: Benefits, Trade-Offs and Future Developments. Chem. Sci. 2020, 11, 4563–4577. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.; Huang, Y.; Ma, L.; Zhang, Z. Quantitative Analysis of Single and Mix Food Antiseptics Basing on SERS Spectra with PLSR Method. Nanoscale Res. Lett. 2016, 11, 296. [Google Scholar] [CrossRef]
- Yan, S.; Liu, C.; Fang, S.; Ma, J.; Qiu, J.; Xu, D.; Li, L.; Yu, J.; Li, D.; Liu, Q. SERS-Based Lateral Flow Assay Combined with Machine Learning for Highly Sensitive Quantitative Analysis of Escherichia coli O157:H7. Anal. Bioanal. Chem. 2020, 412, 7881–7890. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, C.Q.; Thrift, W.J.; Bhattacharjee, A.; Ranjbar, S.; Gallagher, T.; Darvishzadeh-Varcheie, M.; Sanderson, R.N.; Capolino, F.; Whiteson, K.; Baldi, P.; et al. Longitudinal Monitoring of Biofilm Formation via Robust Surface-Enhanced Raman Scattering Quantification of Pseudomonas Aeruginosa -Produced Metabolites. ACS Appl. Mater. Interfaces 2018, 10, 12364–12373. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.M.; Herrman, T.J. Determination and Prediction of Fumonisin Contamination in Maize by Surface–Enhanced Raman Spectroscopy (SERS). Food Bioprocess Technol. 2016, 9, 588–603. [Google Scholar] [CrossRef]
- Kuligowski, J.; El-Zahry, M.R.; Sánchez-Illana, Á.; Quintás, G.; Vento, M.; Lendl, B. Surface Enhanced Raman Spectroscopic Direct Determination of Low Molecular Weight Biothiols in Umbilical Cord Whole Blood. Analyst 2016, 141, 2165–2174. [Google Scholar] [CrossRef]
- Tan, A.; Zhao, Y.; Sivashanmugan, K.; Squire, K.; Wang, A.X. Quantitative TLC-SERS Detection of Histamine in Seafood with Support Vector Machine Analysis. Food Control 2019, 103, 111–118. [Google Scholar] [CrossRef]
- Rahman, A.; Kang, S.; Wang, W.; Huang, Q.; Kim, I.; Vikesland, P.J. Lectin-Modified Bacterial Cellulose Nanocrystals Decorated with Au Nanoparticles for Selective Detection of Bacteria Using Surface-Enhanced Raman Scattering Coupled with Machine Learning. ACS Appl. Nano Mater. 2022, 5, 259–268. [Google Scholar] [CrossRef]
- Banaei, N.; Moshfegh, J.; Mohseni-Kabir, A.; Houghton, J.M.; Sun, Y.; Kim, B. Machine Learning Algorithms Enhance the Specificity of Cancer Biomarker Detection Using SERS-Based Immunoassays in Microfluidic Chips. RSC Adv. 2019, 9, 1859–1868. [Google Scholar] [CrossRef]
- Cheng, N.; Chen, D.; Lou, B.; Fu, J.; Wang, H. A Biosensing Method for the Direct Serological Detection of Liver Diseases by Integrating a SERS-Based Sensor and a CNN Classifier. Biosens. Bioelectron. 2021, 186, 113246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liang, B.; Zhang, J.; Hao, X.; Xu, X.; Chang, H.M.; Leung, P.C.K.; Tan, J. Raman Spectroscopy of Follicular Fluid and Plasma with Machine-Learning Algorithms for Polycystic Ovary Syndrome Screening. Mol. Cell. Endocrinol. 2021, 523, 111139. [Google Scholar] [CrossRef] [PubMed]
- Barucci, A.; D’Andrea, C.; Farnesi, E.; Banchelli, M.; Amicucci, C.; De Angelis, M.; Hwang, B.; Matteini, P. Label-Free SERS Detection of Proteins Based on Machine Learning Classification of Chemo-Structural Determinants. Analyst 2021, 146, 674–682. [Google Scholar] [CrossRef]
- Kazemzadeh, M.; Hisey, C.L.; Zargar-Shoshtari, K.; Xu, W.; Broderick, N.G.R. Deep Convolutional Neural Networks as a Unified Solution for Raman Spectroscopy-Based Classification in Biomedical Applications. Opt. Commun. 2022, 510, 127977. [Google Scholar] [CrossRef]
- Othman, N.H.; Yoot Lee, K.; Mohd Radzol, A.R.; Mansor, W.; Amanina Yusoff, N. PCA-Polynomial-ELM Model Optimal for Detection of NS1 Adulterated Salivary SERS Spectra. J. Phys. Conf. Ser. 2019, 1372, 012064. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, B.; Haverstick, J.; Ibtehaz, N.; Muszyński, A.; Chen, X.; Chowdhury, M.E.H.; Zughaier, S.; Zhao, Y. Differentiation and Classification of Bacterial Endotoxins Based on Surface Enhanced Raman Scattering and Advanced Machine Learning. Nanoscale 2022, 14, 8806–8817. [Google Scholar] [CrossRef]
- Lin, D.; Hsieh, C.L.; Hsu, K.C.; Liao, P.H.; Qiu, S.; Gong, T.; Yong, K.T.; Feng, S.; Kong, K.V. Geometrically Encoded SERS Nanobarcodes for the Logical Detection of Nasopharyngeal Carcinoma-Related Progression Biomarkers. Nat. Commun. 2021, 12, 3430. [Google Scholar] [CrossRef]
- Wang, G.; Lipert, R.J.; Jain, M.; Kaur, S.; Chakraboty, S.; Torres, M.P.; Batra, S.K.; Brand, R.E.; Porter, M.D. Detection of the Potential Pancreatic Cancer Marker MUC4 in Serum Using Surface-Enhanced Raman Scattering. Anal. Chem. 2011, 83, 2554–2561. [Google Scholar] [CrossRef]
- Lu, L.; Guan, S.; Guan, Y.; Hong, M. Dual-Modal Fluorescence-SERS Detection of Blood Glucose Engineered by Hierarchical Laser-Induced Micro/Nano Structures for Diabetes Screening. Adv. Mater. Interfaces 2022, 9, 2102532. [Google Scholar] [CrossRef]
- Sun, J.; Gong, L.; Wang, W.; Gong, Z.; Wang, D.; Fan, M. Surface-Enhanced Raman Spectroscopy for on-Site Analysis: A Review of Recent Developments. Luminescence 2020, 35, 808–820. [Google Scholar] [CrossRef] [PubMed]
- Kho, K.W.; Fu, C.Y.; Dinish, U.S.; Olivo, M. Clinical SERS: Are We There Yet? J. Biophotonics 2011, 4, 667–684. [Google Scholar] [CrossRef] [PubMed]

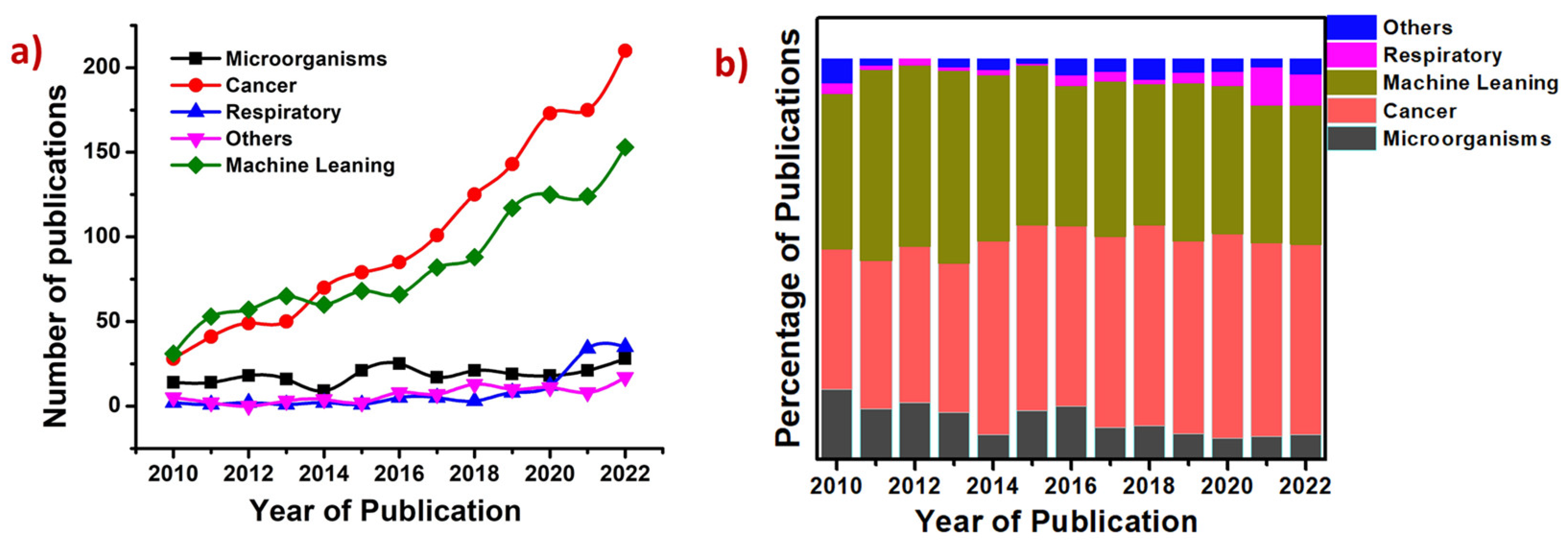
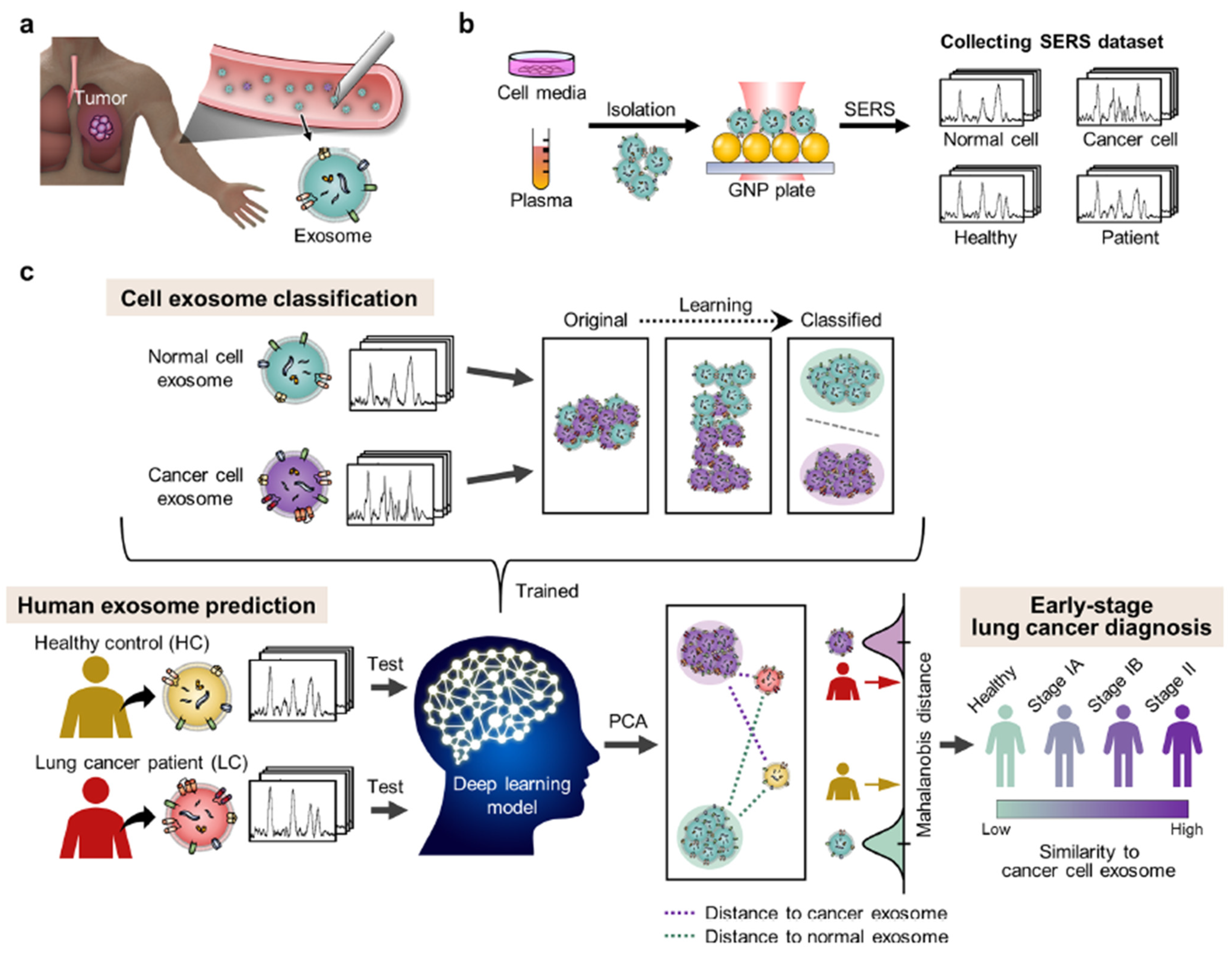

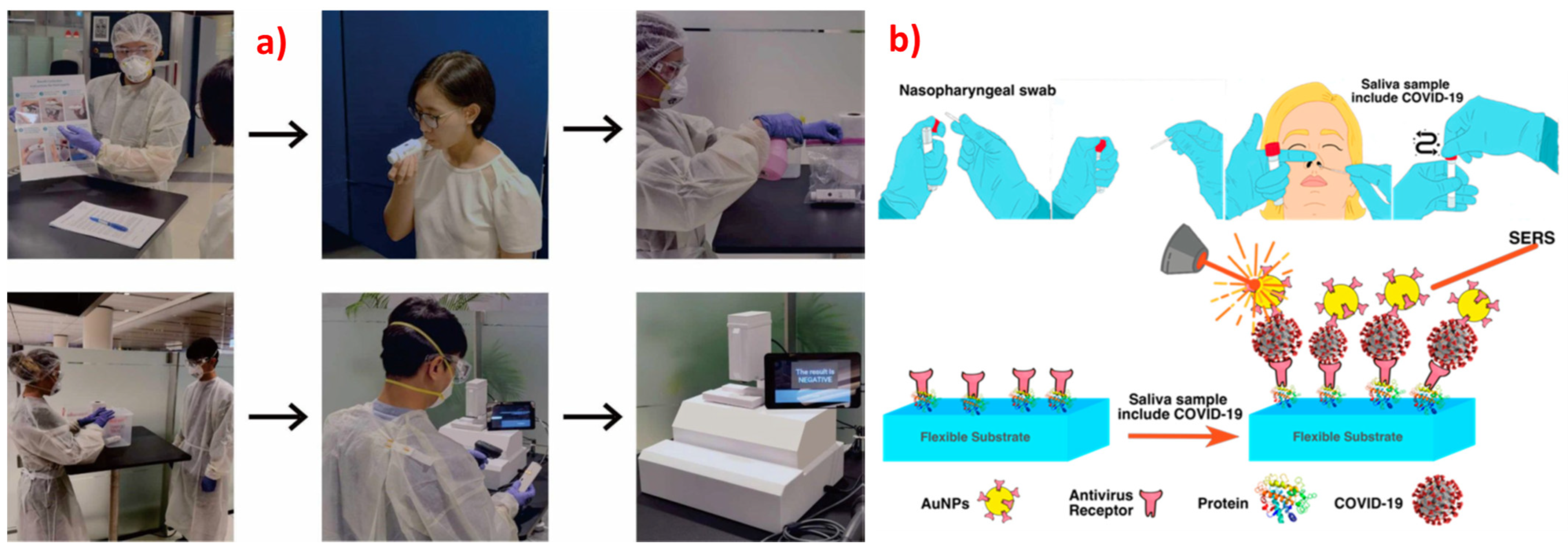
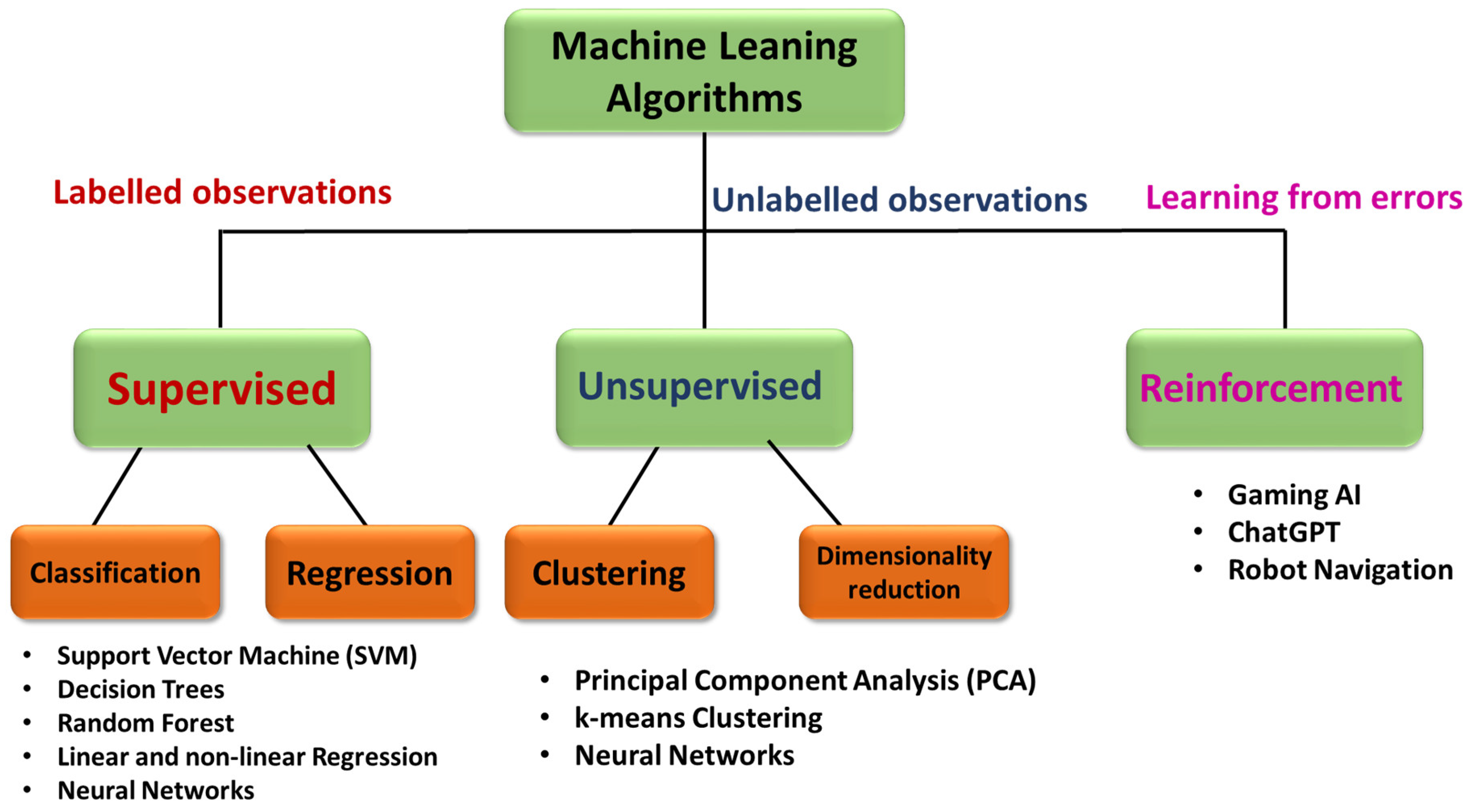
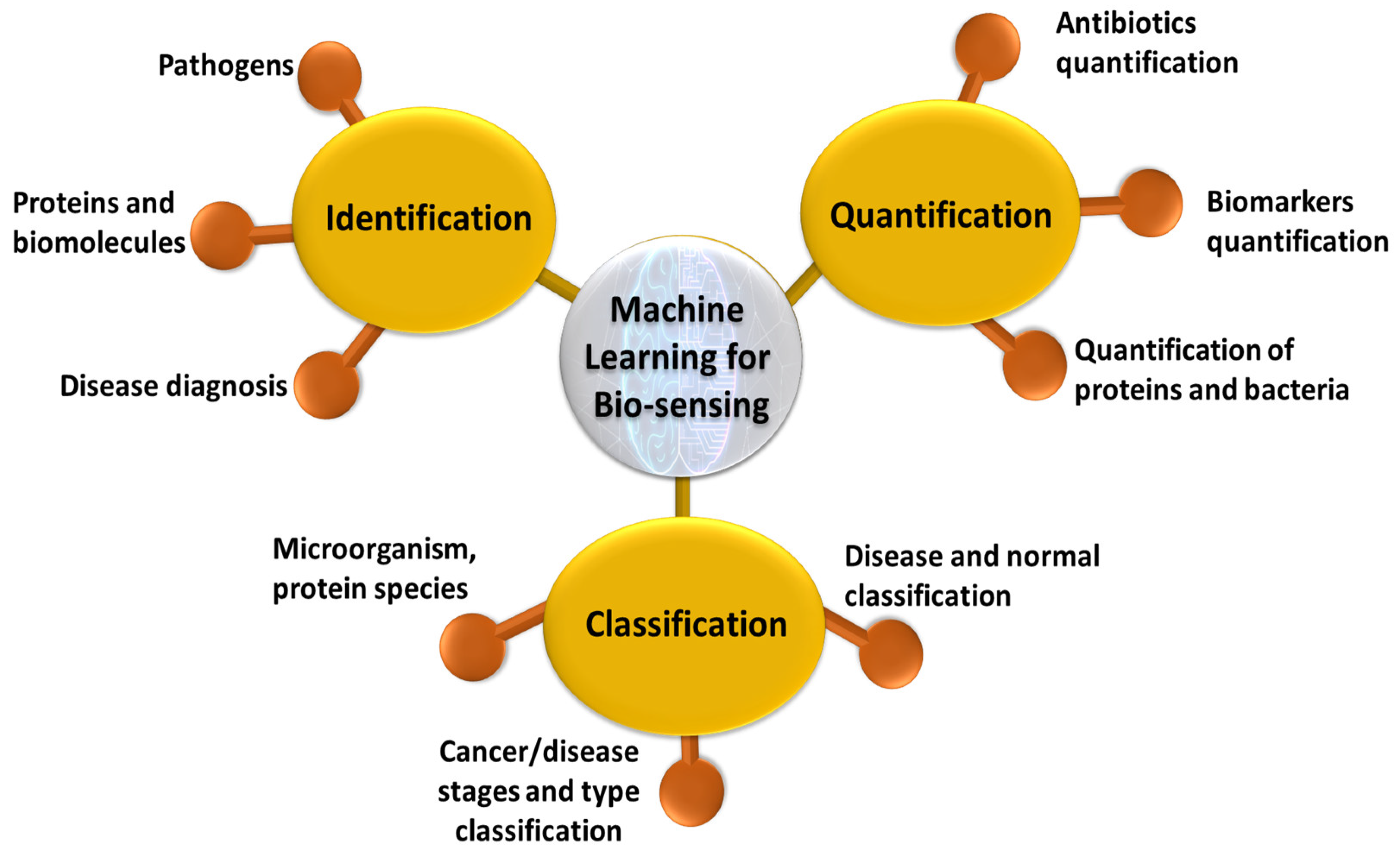
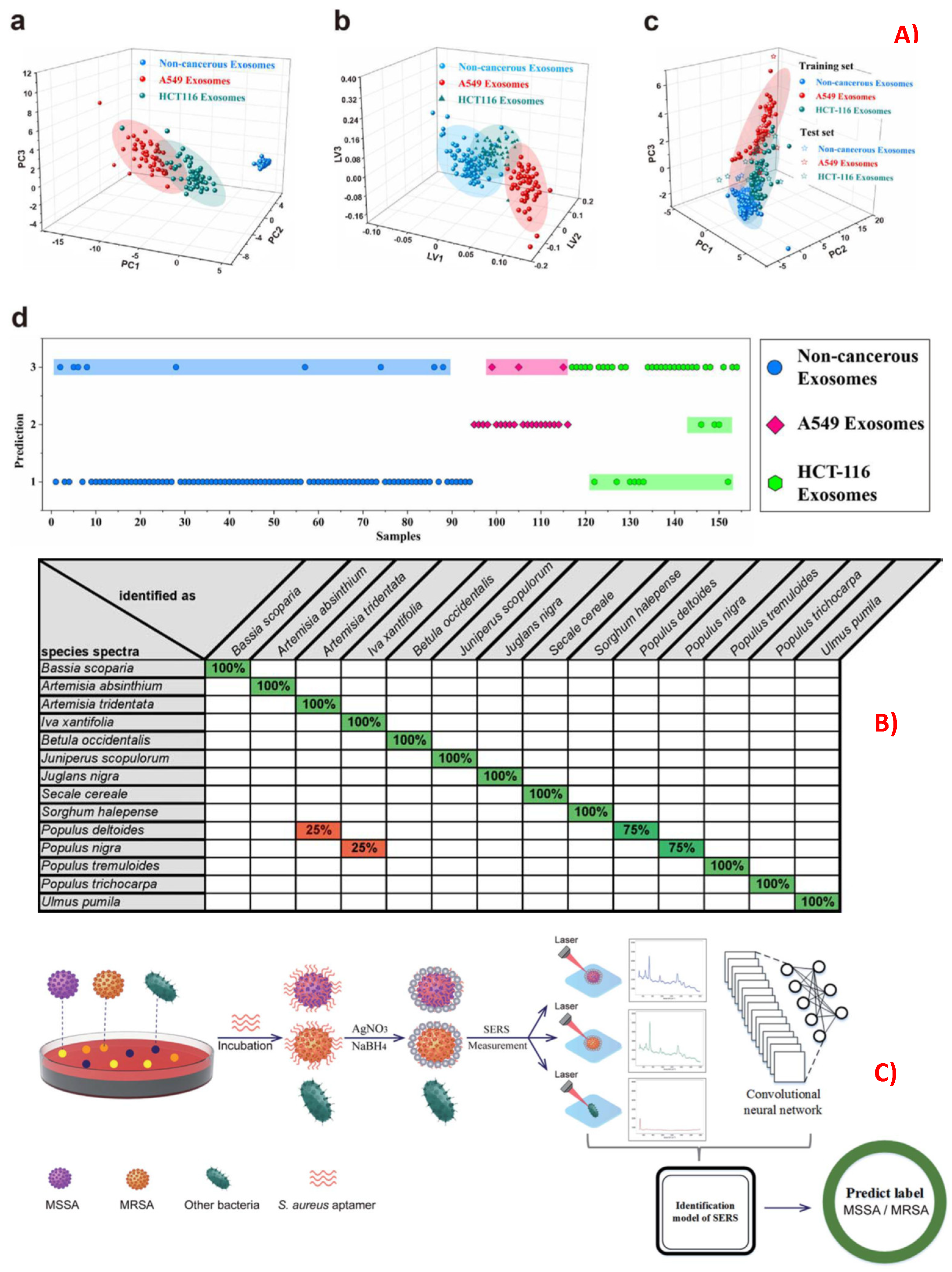
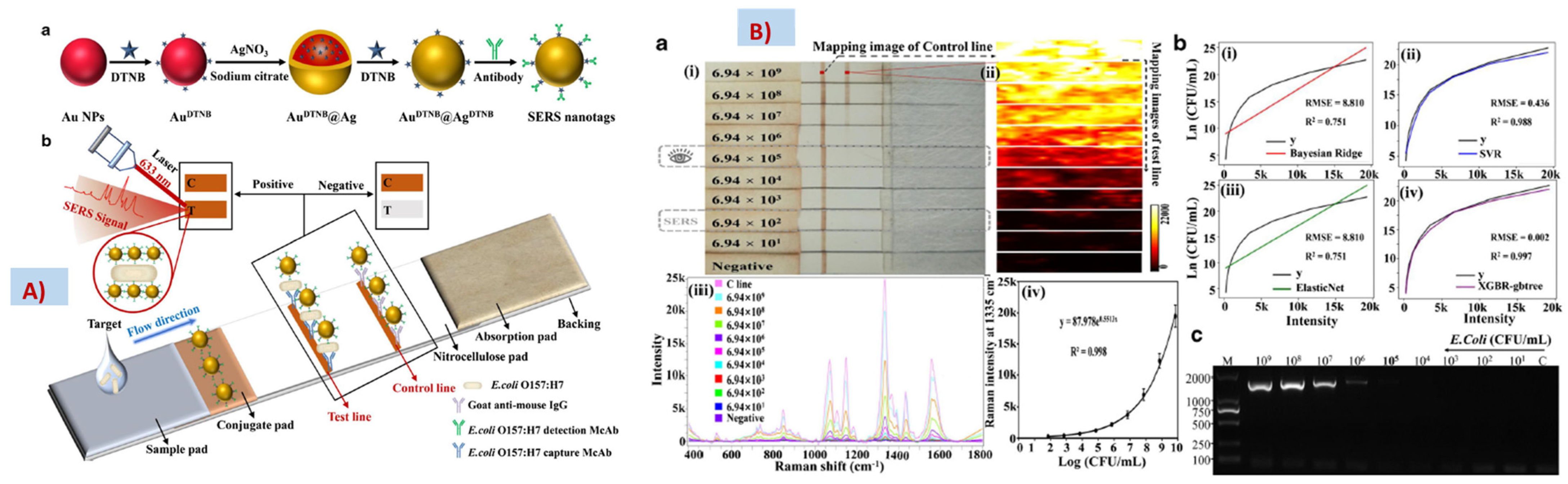
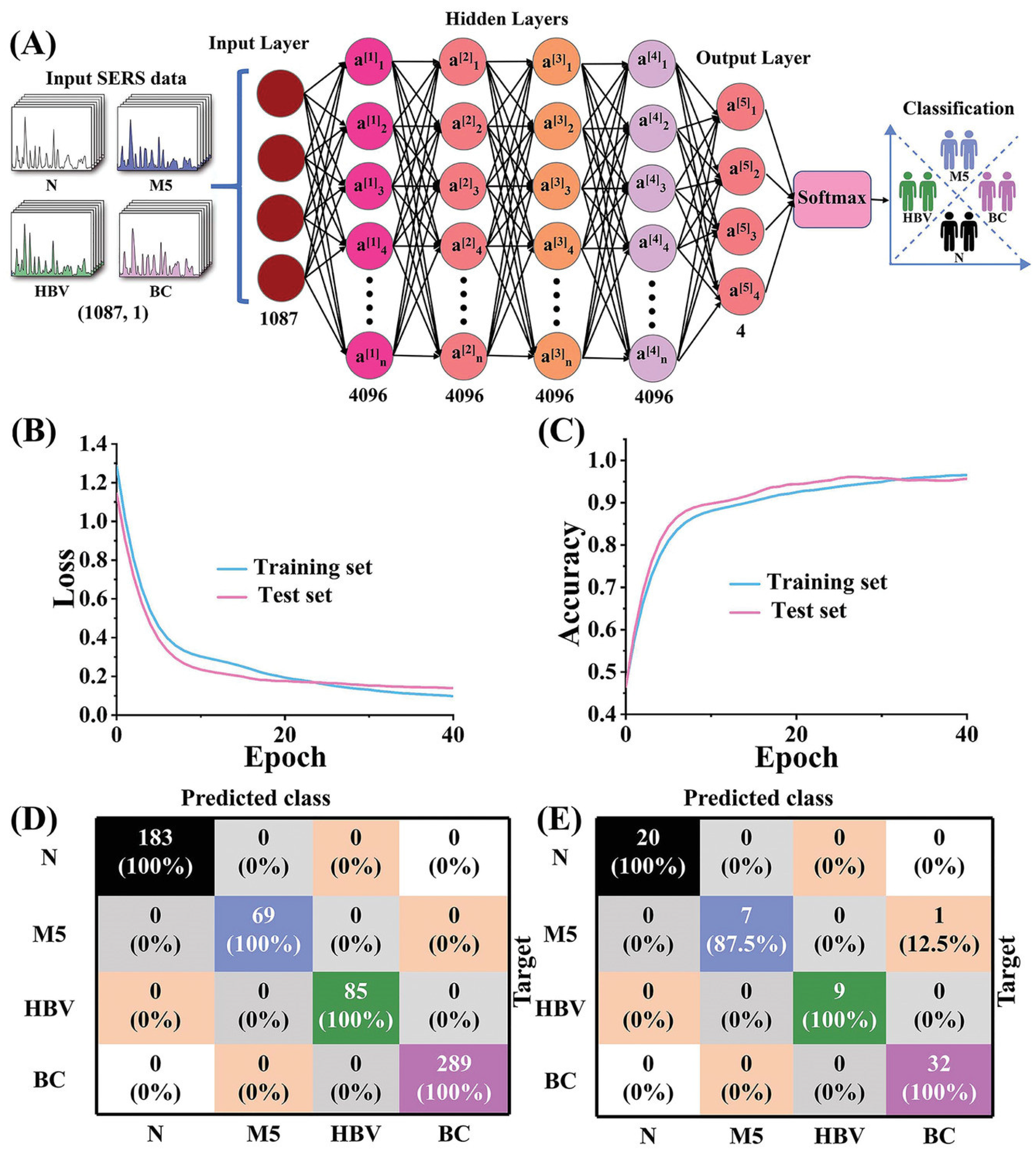
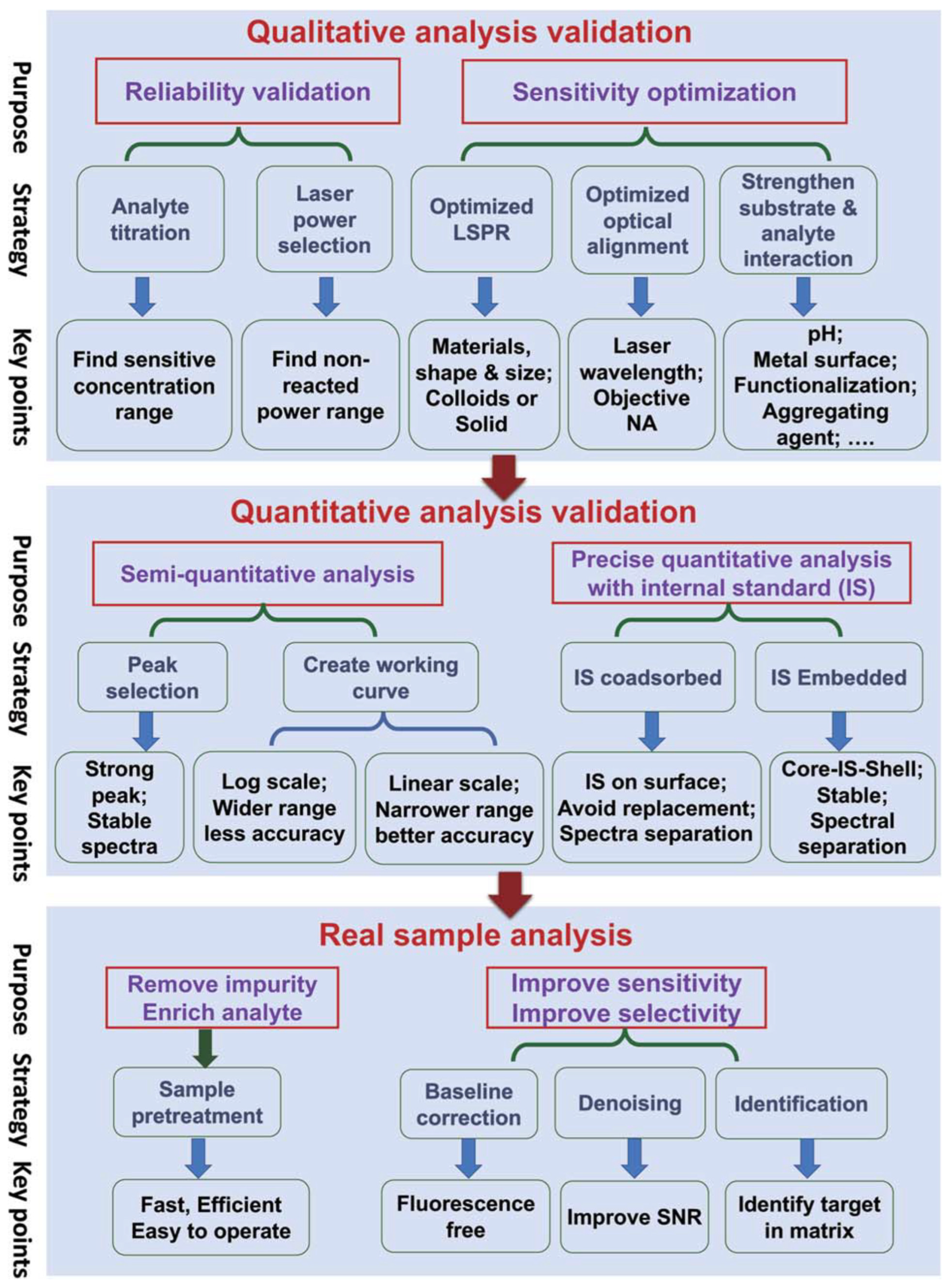
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beeram, R.; Vepa, K.R.; Soma, V.R. Recent Trends in SERS-Based Plasmonic Sensors for Disease Diagnostics, Biomolecules Detection, and Machine Learning Techniques. Biosensors 2023, 13, 328. https://doi.org/10.3390/bios13030328
Beeram R, Vepa KR, Soma VR. Recent Trends in SERS-Based Plasmonic Sensors for Disease Diagnostics, Biomolecules Detection, and Machine Learning Techniques. Biosensors. 2023; 13(3):328. https://doi.org/10.3390/bios13030328
Chicago/Turabian StyleBeeram, Reshma, Kameswara Rao Vepa, and Venugopal Rao Soma. 2023. "Recent Trends in SERS-Based Plasmonic Sensors for Disease Diagnostics, Biomolecules Detection, and Machine Learning Techniques" Biosensors 13, no. 3: 328. https://doi.org/10.3390/bios13030328
APA StyleBeeram, R., Vepa, K. R., & Soma, V. R. (2023). Recent Trends in SERS-Based Plasmonic Sensors for Disease Diagnostics, Biomolecules Detection, and Machine Learning Techniques. Biosensors, 13(3), 328. https://doi.org/10.3390/bios13030328