Abstract
The rapid diagnosis and detection of respiratory bacteria at the early stage can effectively control the epidemic spread and bacterial infection. Here, we designed a rapid, ultrasensitive, and quantitative lateral flow immunoassay (LFA) strip for simultaneous detection of respiratory bacteria S. aureus and S. pneumoniae. In this assay, the surface enhanced Raman scattering (SERS) tags were designed through combining magnetite Raman enhancement nanoparticle Fe3O4@Au/DTNB and recognition element 4-mercaptophenylboronic acid (4-MPBA). Further, 4-MPBA could capture multiple bacteria in a complex environmental solution. Based on the strategies, Fe3O4@Au/DTNB-mediated magnetic enrichment and 4-MPBA-mediated universal capture capabilities improved the detection sensitivity, the limits of detection for S. aureus and S. pneumoniae were as low as 12 and 11 CFU mL−1, respectively, which were more sensitive than those of colloidal gold method. The Fe3O4@Au/DTNB/Au/4-MPBA-LFA also exhibited good reproducibility, excellent specificity, and high recovery rates in sputum samples, indicating its potential application in the detection of respiratory bacteria samples.
1. Introduction
Common respiratory bacteria, mainly including Staphylococcus aureus, Escherichia coli, Streptococcus pneumoniae, and Pseudomonas aeruginosa [1], infections caused by these bacteria can induce serious respiratory diseases [2]. Accurate and sensitive respiratory bacteria detection is crucial to controlling bacterial infections and human health. Numerous analytical methods for pathogen detection have been developed, conventional detection method standard plate colony counting generally takes several days and the sensitivity is low [3]. Common molecular biological detection methods, such as polymerase chain reaction (PCR) [4], enzyme-linked immunosorbent assay (ELISA) [5], surface plasmon resonance [6], and nucleic acid hybridization [7], however, all these methods typically have the disadvantages of time-consuming, laborious sample pretreatment, and expensive equipment requirements, which limit their application of bacterial detection [8,9,10]. Therefore, a rapid and ultrasensitive detection technology for respiratory pathogens is highly required.
SERS has been used in various biomarker or pathogens detection field owing to its high sensitivity, specificity, and high speed [11]. In particularly, SERS can provide spectroscopic fingerprint and nondestructive data acquisition even in complex sample environment with fewer background interference [12]. For instance, Zhang has reported a potential biomarker exosomal Programmed cell death 1 ligand 1 (PD−L1) for immunotherapy and provided SERS detection platform for the trace detection of clinical cancers, the detection limit for PD−L1 and exosomes based on PD−L1 were 0.1 ng mL−1 and 4.8 × 106 particles mL−1, respectively [13]. Cheng has fabricated SERS-based sensor based on Au-Ag nanocomplex-decorated ZnO nanopillars on paper for direct serological detection of liver diseases within 1 min [14]. Yang has reported a novel SERS strategy based on three-dimensional (3D) DNA walker for quantitative analysis of Salmonella typhimurium with a superior limit detection of 4 CFU mL−1 [15]. Lateral flow immunoassay (LFA) is a popular point-of-care testing (POCT) detection technology [16], which is simple operation, short analysis time, and low cost [17]. However, conventional LFA is mainly based on color visualization using colloidal gold as signal reporters, thus providing low sensitivity and only half-quantification detection ability [18]. SERS-based LFA strips were used for pathogen detection because of ultrasensitive and quantitative analysis capabilities, which mainly rely on the SERS tags consisting of a SERS-enhanced substrate, Raman reporters, and targeting biomolecules [19]. Recently, magnetic nanomaterials, such as Au-coated Fe3O4 magnetic nanoparticles (Fe3O4@Au MNPs), have been used in the detection of complex samples due to their excellent versatility and stability [20]. Therefore, after Fe3O4@Au MNPs are introduced into the biosensors, the magnetic SERS-based system can be endowed with magnetic enrichment capability in the complex environmental solutions, which avoid sample pretreatment process and shorten detection time.
Recognition elements are critical for the detection performance of target pathogen in typical SERS biosensor systems. The recognition elements such as antibody, aptamers, and antibiotic are used to graft on SERS tags to capture target pathogen [21]. Among them, antibodies are obtained from immunized animals, which are expensive and poor stability [22]. The screening process of antibodies is time-consuming, especially, the antibodies are lacking and difficult to prepare or screen in a short time for rare and sudden bacterial infection. The optimization process of aptamers is also difficult, and the binding between target bacteria and aptamers generally needs high-salt ions and a relatively long time [23,24]. A pair of antibody or aptamer is used to detect only one target bacterium. Antibiotics usually lack a high affinity for pathogens. Currently, 4-mercaptophenylboronic acid (4-MPBA) as a bacterial capture molecule is used in the field of biosensor detection. It can bind peptidoglycan on the cell walls of bacteria and thus capture various kinds of bacteria simultaneously, avoiding the process of screening antibodies or aptamers and possessing high affinity for the target bacteria [25,26,27], which appears to be a promising candidate and achieves broad-spectrum bacteria capture capability.
In this study, the magnetic SERS tags Fe3O4@Au/DTNB/Au/4-MPBA were fabricated using Fe3O4@Au MNPs as the substrate and 4-mercaptophenylboronic acid (4-MPBA) as Raman recognition element. The high-performance Fe3O4@Au/DTNB/Au/4-MPBA SERS tags possessed good stability, universal capture, magnetic enrichment, and excellent SERS activity. Based on these advantages, our results showed the bacterial capture efficiency of SERS tags was about 95.4% even at a low concentration and the limits of detection (LODs) were as low as 12 and 11 CFU mL−1 for respiratory bacteria S. aureus and S. pneumoniae, respectively.
2. Experimental
2.1. Materials and Chemicals
Phosphate buffered saline (PBS, 0.01 M, pH 7.4), polyethyleneimine (PEI), trisodium citrate dihydrate, polyvinylpyrrolidone (PVP, 40 kDa), hydroxylamine hydrochloride, 4-mercaptophenylboronic acid (4-MPBA), chloroauric acid tetrahydrate (HAuCl4·4H2O), fetal bovine serum (FBS), Tween-20, 5,5′-Dithiobis-(2-nitrobenzoic acid) (DTNB), and crystal violet (CV) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Mouse monoclonal antibodies (Catalog # MA1-10708, MA1-10709) were obtained from Thermo Fisher (Shanghai, China). Rabbit monoclonal antibodies (Catalog # Strpcpnu-001, Strpcpnu-002) were purchased from Xinxin Bio, Ltd. (Changzhou, China). Nitrocellulose (NC) membranes (Prima40 with 18 μm pore size, CN140 with 8 μm pore size, and CN95 with 15 μm pore size) were purchased from Sartorius (Gottingen, Germany), plastic backing card, sample loading pad, and absorbent pad were obtained from Jieyi Biotechnology Co., Ltd. (Shanghai, China). All analytical grade reagents were purchased from Sigma-Aldrich (USA).
2.2. Instruments
Transmission electron microscope (TEM) images were captured by a Hitachi H-7650 TEM (Tokyo, Japan) at 50 kV. High resolution transmission electron microscope (HRTEM) images were captured using the FEI Tecnai G2 F20 electron microscopy (Hillsboro, OR, USA) at 200 kV. Scanning electron microscope (SEM) images were obtained by a JEOL JSM-7001F instrument (Tokyo, Japan) at an operating voltage of 10 kV. Zeta potential data were tested by a Malvern Nano-ZS90 ZetaSizer (Malvern, UK) instrument. Raman spectra data were collected by a portable Raman system (B&W Tek, i-Raman Plus BWS465-785H spectrometer, Plainsboro, NJ, USA).
2.3. Fabrication of Fe3O4@Au/DTNB/Au/4-MPBA Tags
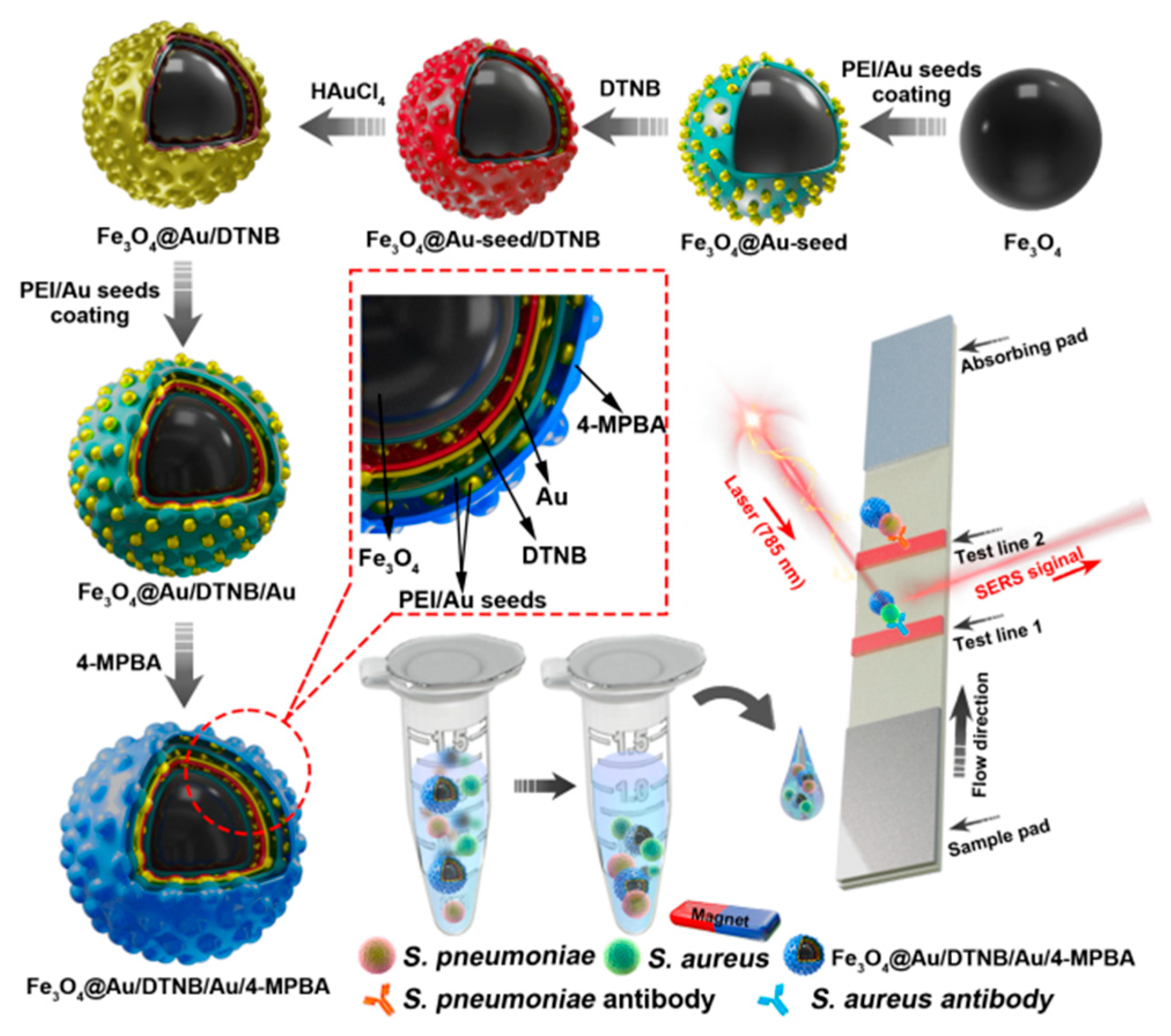
The design route of Fe3O4@Au/DTNB/Au/4-MPBA tags was demonstrated in Scheme 1. Fe3O4@Au/DTNB was fabricated by previously reported PEI-mediated seed growth method [28]. The specific synthesis details were provided in the Supporting Information S1. Fe3O4@Au/DTNB was dispersed in the PEI aqueous solution (2 mg mL−1) followed by sonication for 0.5 h and washed for three times, then mixed with colloidal 3-nm AuNPs and sonicated for another 0.5 h to obtain Fe3O4@Au/DTNB/Au. Finally, 200 μL of 4-MPBA (10 mM) was added to the prepared Fe3O4@Au/DTNB/Au and continuously sonicated for 2 h to fabricate Fe3O4@Au/DTNB/Au/4-MPBA tags. The Fe3O4@Au/DTNB/Au/4-MPBA tags were stored at room temperature for further use.
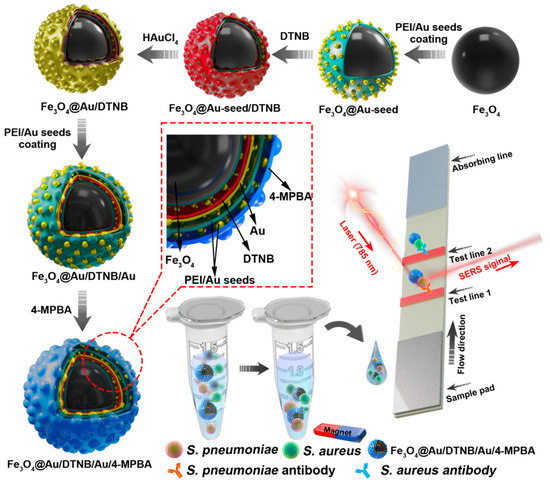
Scheme 1.
Fabrication of Fe3O4@Au/DTNB/Au/4-MPBA and schematic diagram of Fe3O4@Au/DTNB/Au/4-MPBA SERS strip for detecting two respiratory bacteria.
2.4. Fabrication of Lateral Flow Strip Bacteria Detection System
A LFA strip was prepared with absorbent pad, sample pad, and nitrocellulose membrane (NC membrane). S. aureus (0.6 mg mL−1) and S. pneumoniae (0.4 mg mL−1) antibodies were sprayed onto NC membrane as test line 1 (T1) and test line 2 (T2), respectively. NC membrane was placed at 37 °C for 2 h. Subsequently, absorption pad and sample pad were assembled onto the both ends of NC membrane to ensure liquid flow. The prepared card was then cut into the strips with a width of 3 mm and stored in a vacuum desiccator for further use.
2.5. Preparation of Bacterial Sample
Bacteria concentrations, including S. aureus and S. pneumoniae, were verified by plate counting method [29]. The bacteria were cultured using 5% sheep blood agar plates at 37 °C overnight. The initial bacterial solution was obtained by several bacterial colonies on the plates picked into 0.5 mL of sterile PBS solution. The initial solution was then diluted 104 to 106 times using PBS solution, 0.1 mL of diluted bacterial suspensions were withdrawn and dropped on blood agar plates followed by incubation at 37 °C overnight. The colony forming units (CFUs) on the plates were counted to calculate the concentration of initial bacterial solution, based on the calculated results, the testing concentration of 108 cells mL−1 was prepared for further use.
2.6. Detection of Respiratory Bacteria Using Fe3O4@Au/DTNB/Au/4-MPBA-Based LFA Strip
The detection of S. aureus and S. pneumoniae was conducted in a 2 mL tube. First, 4 μL of liquid Fe3O4@Au/DTNB/Au/4-MPBA tags was added into 1 mL of each bacteria suspension and magnetically collected after incubation for 20 min. The bacteria suspension in the tube was removed and 80 μL of PBS running buffer was added followed by ultrasonic for 1 min, the prepared strip was inserted into the tube for 20 min, then the strip was dried and SERS intensities of test lines were tested with a portable Raman spectrometer under 785 nm laser excitation.
2.7. Detection of Respiratory Pathogens S. aureus and S. pneumoniae in Biological Samples
The recovery rates (%) of bacteria were calculated using sputum specimens. The bacterial concentrations of 106 CFU mL−1 and 103 CFU mL−1 were respectively spiked to the diluted sputum specimens, the specific operation was conducted according to the protocol in Section 2.6. The recovery rates were determined according to the previously established calibration curves and the tests were conducted in triplicate for each sample.
3. Results and Discussion
3.1. Principle of Fe3O4@Au/DTNB/Au/4-MPBA-Based LFA Strip for the Simultaneous Detection of S. aureus and S. pneumoniae
In this detection system, Fe3O4@Au/DTNB/Au/4-MPBA was used as SERS tags to simultaneously capture two target bacteria and improve the sensitivity of LFA. Fe3O4@Au/DTNB/Au/4-MPBA tags presented good dispersity, magnetic responsiveness, universal capture, and strong SERS activity in a complex solution. The operating procedure of the simultaneous detection of respiratory bacteria was displayed in Scheme 1. The detection principle was based on antibody-antigen interaction. First, Fe3O4@Au/DTNB/Au/4-MPBA tags were added into the solution containing S. aureus and S. pneumoniae followed by shaking for about 20 min. The target bacteria were captured by Fe3O4@Au/DTNB/Au/4-MPBA tags during incubation, the Fe3O4@Au/DTNB/Au/4-MPBA-bacteria complexes were separated from the bacteria solution using a magnet, then resuspended in the prepared running buffer solution and ultrasonic for several seconds. Finally, LFA strips were inserted into the running buffer solution, the complexes Fe3O4@Au/DTNB/Au/4-MPBA-bacteria slowly flowed along the strip from the sample loading pad to the absorbent pad under capillary forces. Due to the interaction between antibody and antigen, the complexes were captured by the S. aureus and S. pneumoniae antibodies pre-coated on the T1 and T2 lines, respectively, and then the sandwich-like immunocomplexes Fe3O4@Au/DTNB/Au/4-MPBA-bacteria-detection antibodies were formed at the test lines. There was no control line (C line) on NC membrane owing to lack 4-MPBA antibody. Although C line was used to evaluate whether the tags and NC membrane were effective, our study revealed that the target bacteria were captured by fabricated Fe3O4@Au/DTNB/Au/4-MPBA tags, then the Fe3O4@Au/DTNB/Au/4-MPBA-bacteria complexes were captured by the antibodies on NC membranes, our results demonstrated the tags possessed excellent capture performance and the immune binding with antibodies on the test lines was normal, which further indicated that the prepared NC membranes were available and our detection system was successfully fabricated, relevant studies of lack of C line on NC membrane have also been reported [30,31]. Moreover, the Raman signal intensities became higher with the increase of tags and bacteria, which provided the quantification detection method, after reaction for 20 min, two dark bands appeared on the strip in the presence of S. aureus and S. pneumoniae, only one dark band (T1 line or T2 line) was observed in the presence of S. aureus or S. pneumoniae, while no line was observed in absent of bacteria, which indicated that the detection system was successfully fabricated. Finally, Raman intensities were measured by a portable Raman spectrometer.
3.2. Characterization of Fe3O4@Au/DTNB/Au/4-MPBA Tags
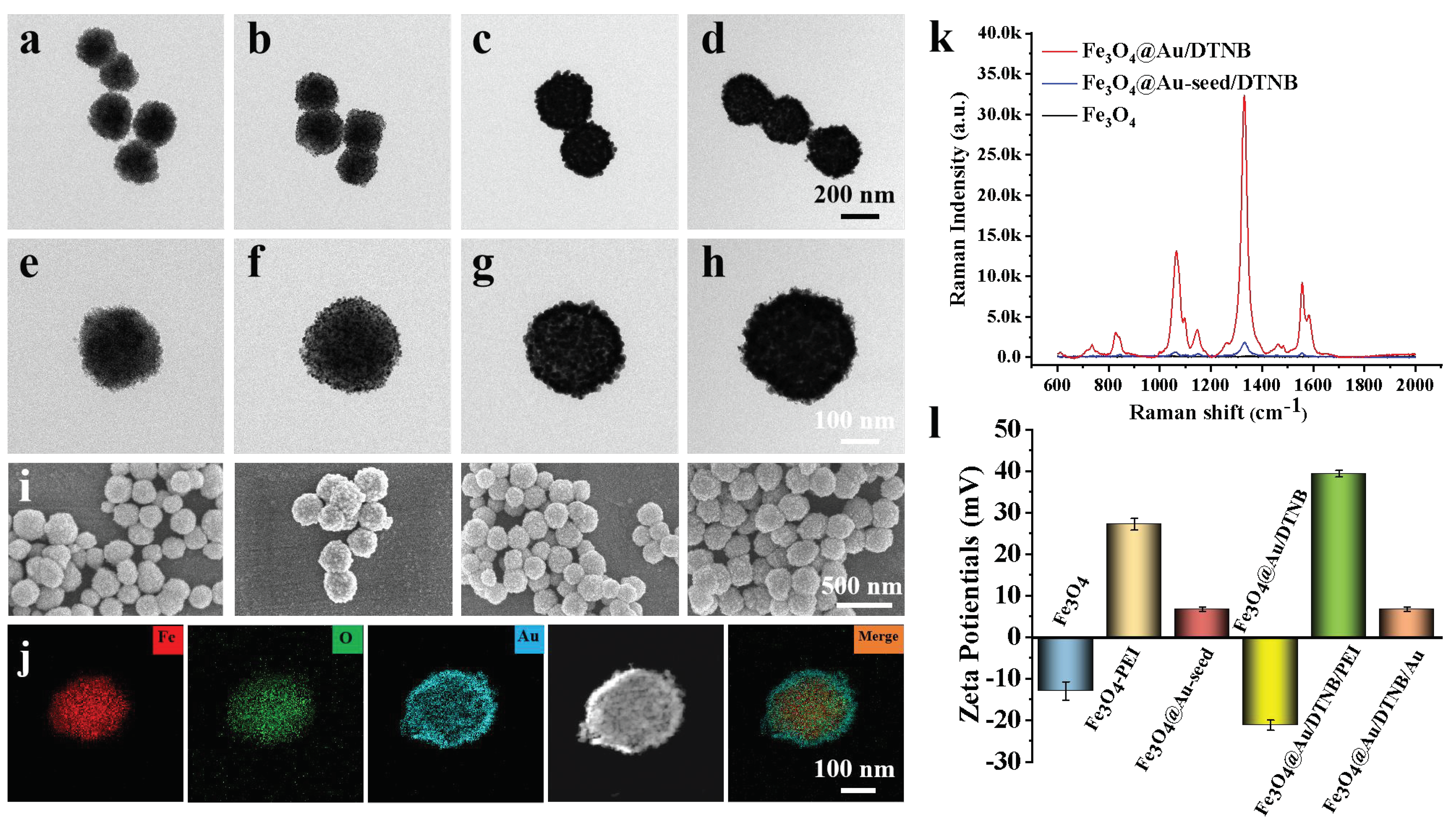
As shown in Figure 1a,e, Fe3O4 MNPs were successfully fabricated and the diameter was about 200 nm [32]. Au seeds adhered to the surface of Fe3O4 after 3-nm AuNPs were coated on synthetic Fe3O4−PEI (Figure 1b,f). A rough Au shell of Fe3O4@Au/DTNB with a size of 15 nm was decorated outside the Fe3O4 core under the reduction of HAuCl4 (Figure 1g) and the obtained Fe3O4@Au/DTNB was uniform with a diameter of approximately 280 nm (Figure 1c). Au seeds in Fe3O4@Au/DTNB/Au were observed again after the second layer of 3-nm AuNPs were modified with Fe3O4@Au/DTNB (Figure 1d,h). Moreover, SEM images further demonstrated that Fe3O4, Fe3O4@Au-seed/DTNB, Fe3O4@Au/DTNB, and Fe3O4@Au/DTNB/Au/4-MPBA MNPs were successfully synthesized (Figure 1i). X-ray mapping was conducted to confirm the basic element composition of Fe3O4@Au/DTNB/Au/4-MPBA, Fe and O elements were mainly distributed in the middle of MNPs, while Au element existed on the shell (Figure 1j).
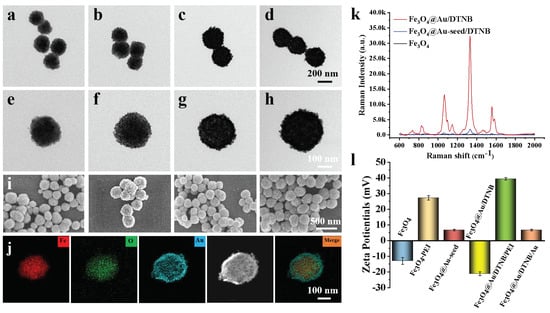
Figure 1.
Structure characterization of Fe3O4@Au/DTNB/Au/4-MPBA. TEM images of (a) Fe3O4, (b) Fe3O4@Au-seed/DTNB, (c) Fe3O4@Au/DTNB, (d) Fe3O4@Au/DTNB/Au/4-MPBA tags, and their corresponding magnified TEM images in (e), (f), (g), and (h), respectively. (i) SEM images of (a–d). (j) EDS elemental mapping images of Fe3O4@Au/DTNB/Au/4-MPBA. (k) SERS signal intensities of Fe3O4, Fe3O4@Au-seed/DTNB, and Fe3O4@Au/DTNB. (l) Zeta potentials of Fe3O4, Fe3O4−PEI, Fe3O4@Au-seed, Fe3O4@Au/DTNB, Fe3O4@Au/DTNB/PEI, and Fe3O4@Au/DTNB/Au.
The SERS activities of Fe3O4, Fe3O4@Au-seed/DTNB, and Fe3O4@Au/DTNB were tested, the feature peaks of DTNB at 736, 826, 1061, 1149, 1331, and 1557 cm−1 were detected, among them, 1331 cm−1 was the strongest peak (Figure 1k) and the corresponding enhancement factor (EF) was 1.3 × 106 (S4 and Figure S2), therefore, 1331 cm−1 was recognized as the characteristic peak for further analysis. The ability of Fe3O4@Au (without modification of DTNB) to enhance Raman signal and act as SERS were also validated using conventional dyes CV (S3 and Figure S1). As shown in Figure 1l, the zeta potential of Fe3O4 was −13.0 mV, the positively charged PEI and negatively charged 3-nm AuNPs were decorated onto the surface of Fe3O4 under electrostatic interaction, zeta potentials of Fe3O4−PEI and Fe3O4@Au-seed were +27.2 mV and +6.7 mV, respectively, while the potential of Fe3O4@Au/DTNB was decreased to −21.2 mV owing to the reduction of HAuCl4. Similarly, the potential of Fe3O4@Au/DTNB/PEI was +39.4 mV and Fe3O4@Au/DTNB/Au was +6.7 mV after the second layer of PEI and 3-nm AuNPs were coated on Fe3O4@Au/DTNB, which confirmed that Fe3O4@Au/DTNB/Au was successfully fabricated [33,34].
3.3. Optimization of the Fe3O4@Au/DTNB/Au/4-MPBA-Based LFA Strip
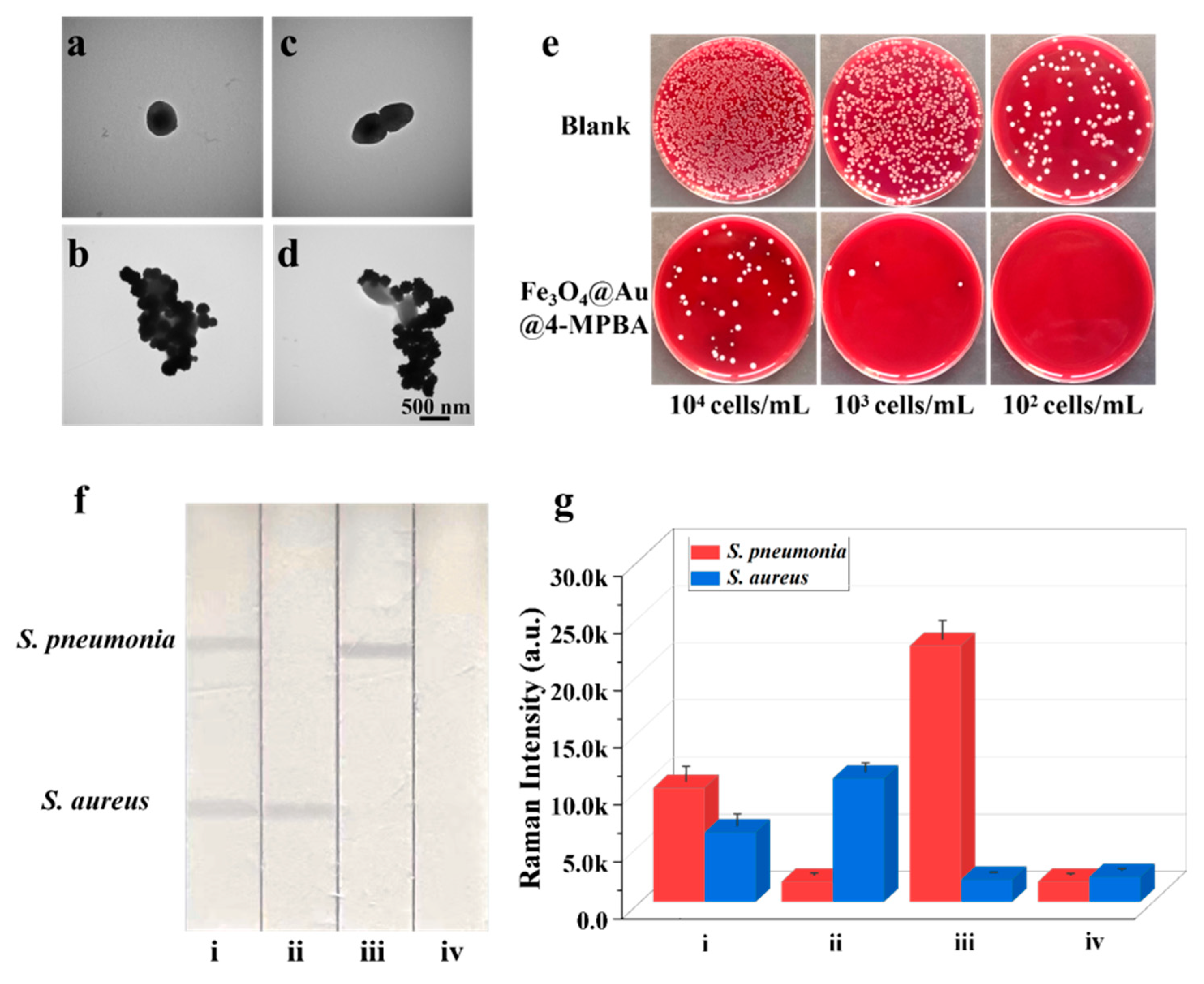
The Fe3O4@Au/DTNB/Au/4-MPBA tags can capture bacteria because 4-MPBA binds lipopolysaccharide that locates on the bacterial cell wall [35]. The TEM images clearly revealed that Fe3O4@Au/DTNB/Au/4-MPBA could effectively capture S. aureus and S. pneumoniae (Figure 2a–d). The bacteria capture efficiency was further determined by incubating Fe3O4@Au/DTNB/Au/4-MPBA in bacterial solutions of different concentrations. After magnetic enrichment of Fe3O4@Au/DTNB/Au/4-MPBA-bacteria complexes, the remaining bacteria in the supernatant were detected with traditional plate counting method and the capture efficiency of Fe3O4@Au/DTNB/Au/4-MPBA was about 95.4% for target bacteria from 104 cells mL−1 to 102 cells mL−1 (Figure 2e), demonstrating that Fe3O4@Au/DTNB/Au/4-MPBA SERS tags could capture almost all the bacteria in the sample solution.
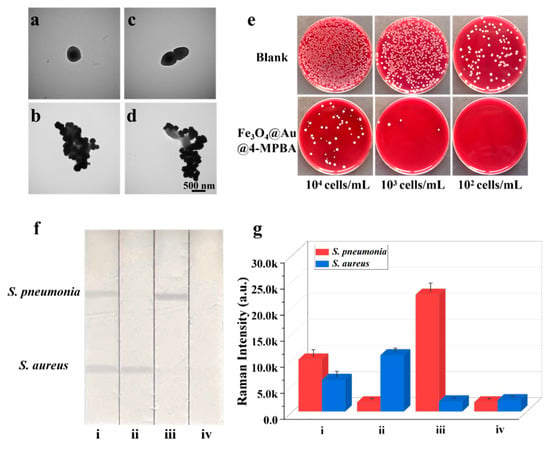
Figure 2.
TEM images of (a) S. aureus, (b) S. aureus captured by Fe3O4@Au/DTNB/Au/4-MPBA, (c) S. pneumonia, and (d) S. pneumonia captured by Fe3O4@Au/DTNB/Au/4-MPBA. (e) Photographs of bacteria capture, the colonies on blood agar plates represent the amounts of remaining S. aureus and S. pneumonia in the supernatant after being captured by Fe3O4@Au/DTNB/Au/4-MPBA MNPs, the concentrations of bacteria ranged from 104 cells mL−1 to 102 cells mL−1. (f) Images and (g) corresponding SERS intensities of Fe3O4@Au/DTNB/Au/4-MPBA-LFA with different concentrations of S. aureus and S. pneumonia: (i) 105, 105 cells mL−1; (ii) 105, 0 cells mL−1; (iii) 0, 105 cells mL−1; and (iv) 0, 0 cells mL−1.
Based on SERS strip, the specificity of antibodies was demonstrated by the solutions containing 105 CFU mL−1 of S. aureus, S. pneumoniae, and their mixture. As displayed in Figure 2f, two test lines of S. aureus and S. pneumoniae on NC membrane could be observed, indicating that there were two target bacteria in the detection sample, whereas only one test line was observed for S. aureus or S. pneumoniae that existed alone. Additionally, the SERS intensity values of S. aureus or S. pneumoniae alone were higher than that of S. aureus and S. pneumoniae simultaneous existence owing to broad-spectrum capture performance of 4-MPBA (Figure 2g), indicating an excellent specificity capability of S. aureus and S. pneumoniae antibodies. Here, bacteria captured by Fe3O4@Au/DTNB/Au/4-MPBA had little effect on Fe3O4@Au/DTNB/Au/4-MPBA-LFA, the generation of signals were mainly attributed to Fe3O4@Au/DTNB/Au/4-MPBA (S5 and Figure S3).
Considering the detection performance of the system, several critical parameters were optimized and provided in the supporting information. The NC membranes of Prima40 (18 μm pore size), CN140 (8 μm pore size), and CN95 (15 μm pore size) were evaluated using 106 CFU mL−1 of S. aureus and S. pneumoniae. The signal-to-noise ratio (SNR) was calculated by the ratio of the SERS signal intensities of positive and negative samples, CN95 membrane displayed the highest SNR for target bacteria (Figure S4). Owing to the immunobinding between Fe3O4@Au/DTNB/Au/4-MPBA-bacteria complexes and antibodies, the running buffer was further optimized, the results confirmed that PBS solution containing 2% (w/v) Tween and 15% (w/v) FBS could suppress the non-specific combination of Fe3O4@Au/DTNB/Au/4-MPBA on the test lines and obtained maximized SNR for target bacteria (Figure S5). A serial usage of Fe3O4@Au/DTNB/Au/4-MPBA tags 1 μL, 2 μL, 4 μL, 6 μL, and 8 μL were respectively incubated with S. aureus and S. pneumoniae at room temperature, the results showed that 4 μL of tags provided the highest SNR for target bacteria detection (Figure S6). Antibody concentrations of target bacteria were also optimized, 0.4 mg mL−1 of S. pneumoniae antibody and 0.6 mg mL−1 of S. aureus antibody could provide the highest SNR of SERS intensities (Figure S7). Finally, the reaction time was optimized, SNR increased from 0 to 20 min and then slightly lower after 20 min (Figure S8). Therefore, the reaction time of 20 min was enough for the whole process.
3.4. Analytical Performance of Fe3O4@Au/DTNB/Au/4-MPBA-Based LFA
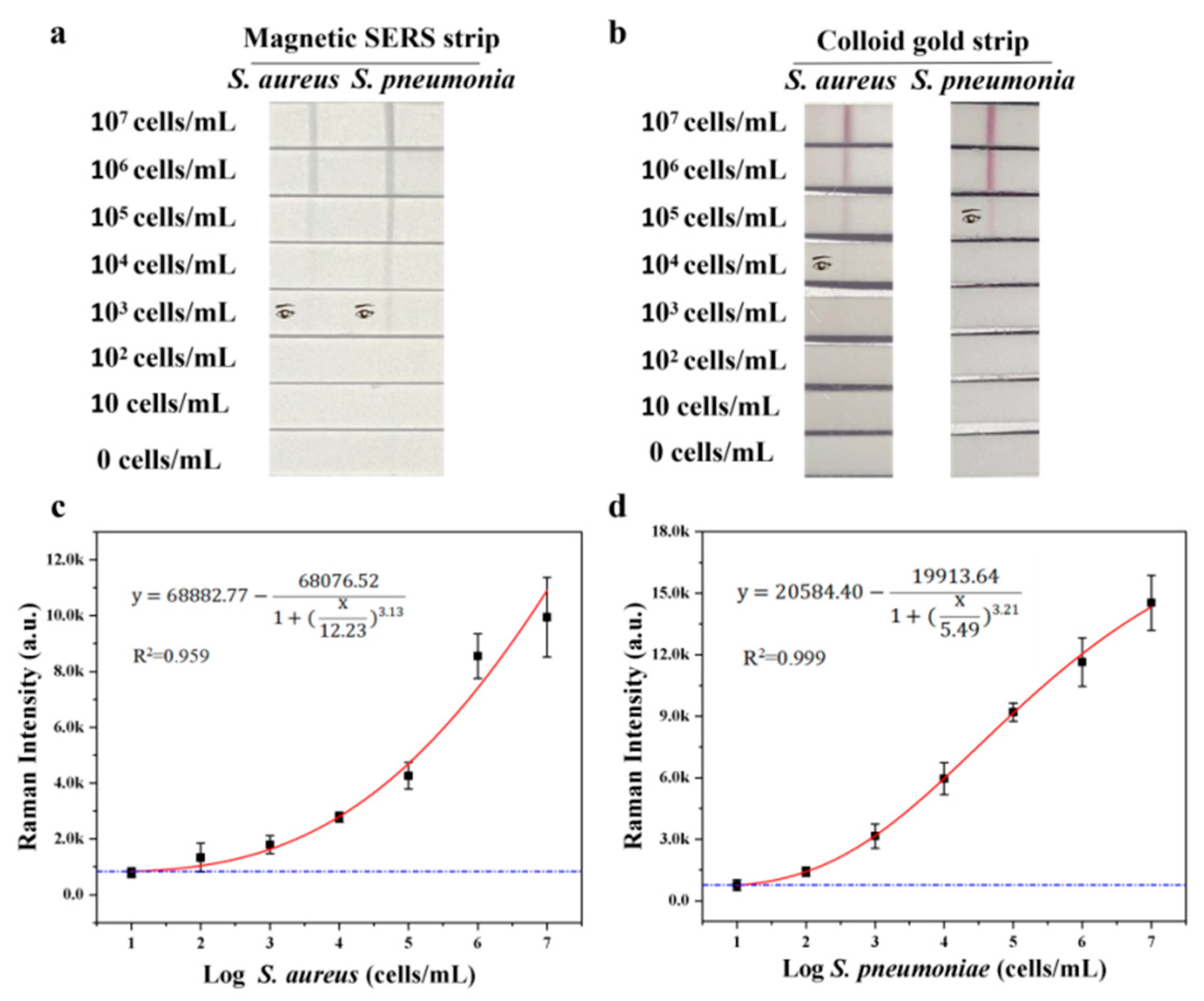
To access the sensitivity and selectivity of our system, different concentrations of S. aureus and S. pneumoniae from 0 to 107 CFU mL−1 were mixed with Fe3O4@Au/DTNB/Au/4-MPBA tags and then detected by Fe3O4@Au/DTNB/Au/4-MPBA-LFA strips. As shown in Figure 3a, with the decreasing concentration of target bacteria, the color of the test lines gradually shallowed. It could be observed with the naked eyes that the visualization concentrations of target bacteria were both 103 cells mL−1, respectively. Moreover, the SERS signal intensities of test lines at 1331 cm−1 increased with increasing bacterial concentration, the calibration curves of two test lines displayed dynamic relationships between SERS intensities and the concentrations of target bacteria and the correlation coefficients (R2) were 0.959 and 0.999 for S. aureus and S. pneumoniae, respectively (Figure 3c,d). Based on the calculation of blank SERS signal, LODs of Fe3O4@Au/DTNB/Au/4-MPBA were determined to be 12 and 11 cells mL−1 for S. aureus and S. pneumoniae, respectively, according to the International Union of Pure and Applied Chemistry (IUPAC) protocol (LOD = mean SERS signal intensities of blank groups + 3 × standard deviation of the control) [36]. We compared our method with standard colloidal gold (AuNP)-LFA method, the preparation of colloidal AuNPs was displayed in supporting information S2, the visualization concentrations of S. aureus and S. pneumoniae were 104 and 105 cells mL−1 for AuNP-LFA strip detection, respectively (Figure 3b). Therefore, our detection system Fe3O4@Au/DTNB/Au/4-MPBA-LFA presented excellent sensitivity.
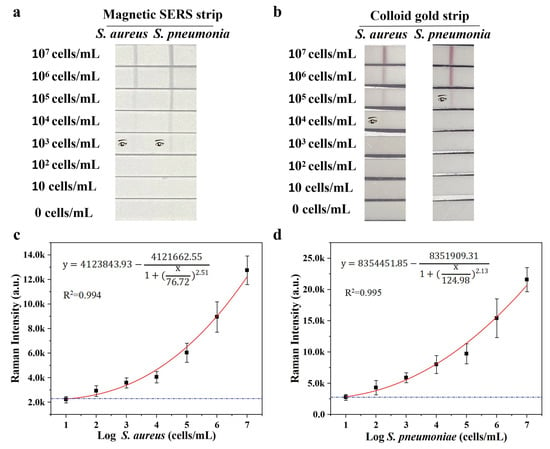
Figure 3.
(a) Photographs of Fe3O4@Au/DTNB/Au/4-MPBA-based LFA strips for the detection at different concentrations of target bacteria. The visualization concentrations of target bacteria are both 103 cells mL−1. (b) Photographs of colloidal gold-based LFA strips for target bacteria detection. The visualization concentrations of S. aureus and S. pneumoniae are 104 and 105 cells mL−1 for AuNP−LFA strip detection, respectively. (c,d) Corresponding calibration curves of target bacteria for Fe3O4@Au/DTNB/Au/4-MPBA-based LFA strips.
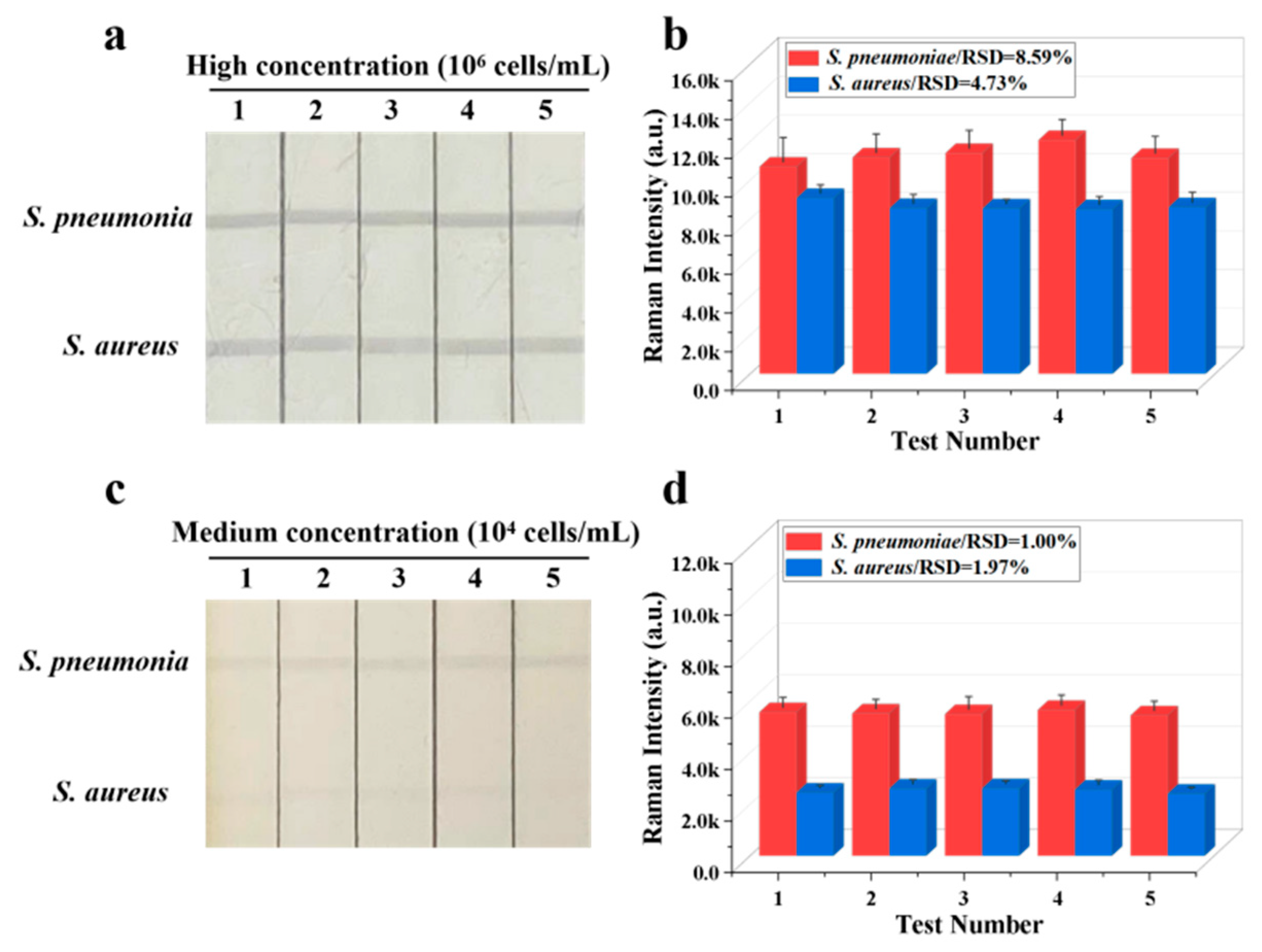
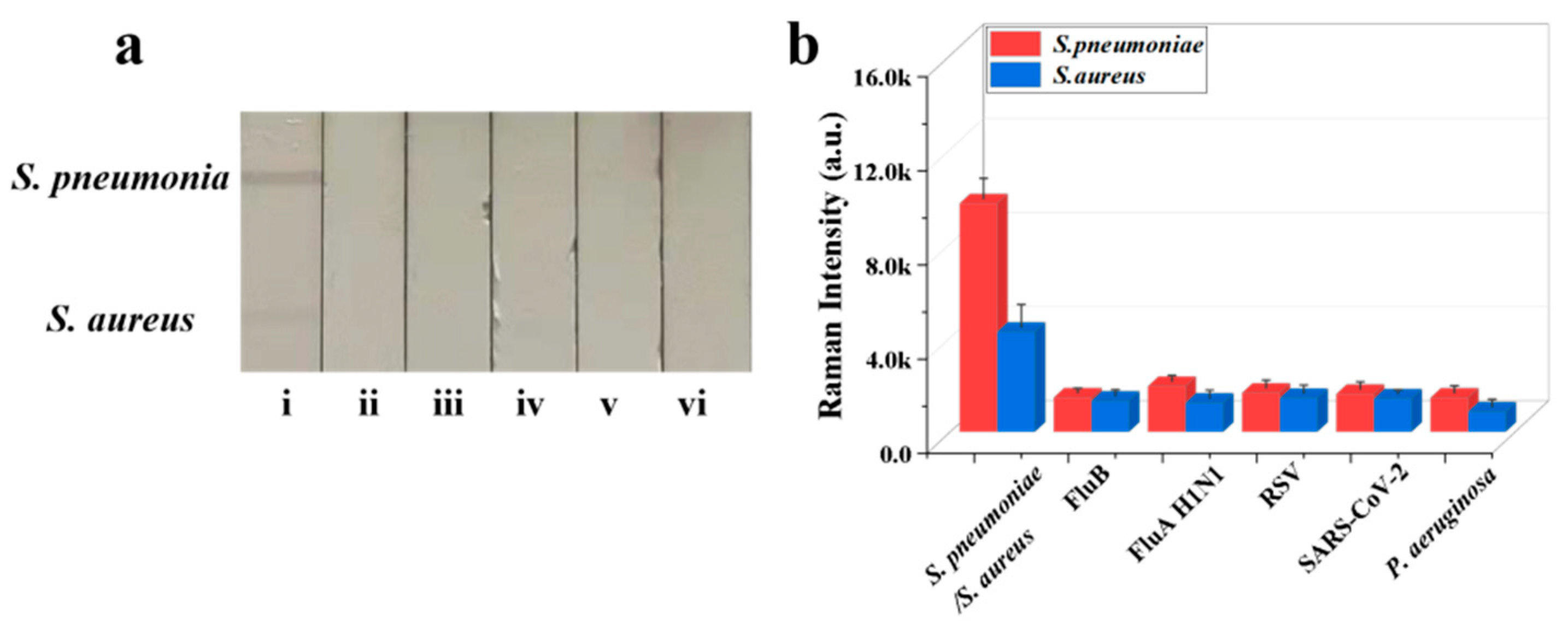
The reproducibility of Fe3O4@Au/DTNB/Au/4-MPBA-based LFA system is a crucial factor for its practical application. The bacteria S. pneumoniae and S. aureus were diluted into a high-dose group of 106 CFU mL−1, the results indicated good reproducibility with relative standard deviation (RSD) of 8.59% and 4.73% for S. pneumoniae and S. aureus, respectively (Figure 4a,b). The diluted medium-dose group of 104 CFU mL−1 was also conducted with RSD of 1.00% and 1.97% for S. pneumoniae and S. aureus, respectively (Figure 4c,d). The specificity of Fe3O4@Au/DTNB/Au/4-MPBA-based LFA was further evaluated, several respiratory viruses including FluB (105 copies mL−1), FluA H1N1 (105 copies mL−1), RSV (105 pfu mL−1), SARS-CoV-2 (100 ng mL−1), and respiratory bacteria P. aeruginosa (106 CFU mL−1) were used as negative samples [37,38], as displayed in Figure 5a,b, the positive groups containing target bacteria generated higher SERS intensities on their corresponding test lines, whereas all the negative samples displayed low SERS signal intensities, confirming Fe3O4@Au/DTNB/Au/4-MPBA-based LFA possessed good specificity. Finally, the recovery rates of Fe3O4@Au/DTNB/Au/4-MPBA-LFA strips were tested in the sputum specimens, as shown in Table 1, the average recoveries for S. aureus and S. pneumoniae ranged from 92.7% to 111.5%, and the RSD values of the SERS intensities on two test lines were less than 10%, therefore, Fe3O4@Au/DTNB/Au/4-MPBA-LFA strip had potential to detect S. aureus and S. pneumoniae in the real sputum specimens.
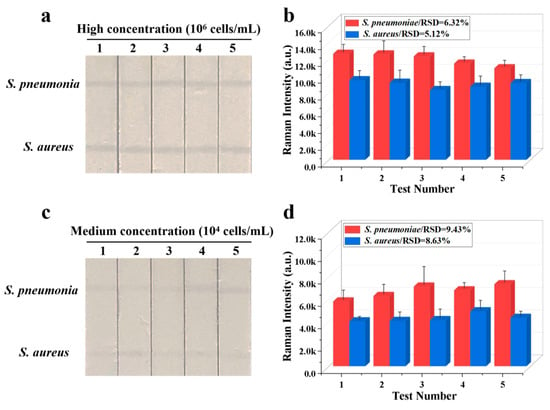
Figure 4.
Reproducibility of Fe3O4@Au/DTNB/Au/4-MPBA-LFA strips for target bacteria at concentrations of (a) 106 and (c) 104 cells mL−1, respectively, and corresponding SERS intensities (b,d) on the test lines.
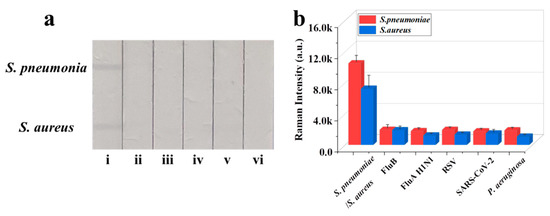
Figure 5.
(a) Specificity of Fe3O4@Au/DTNB/Au/4-MPBA-LFA strips for target bacteria at concentration of 105 cells mL−1: (i) S. pneumoniae/S. aureus; (ii) FluB; (iii) FluA H1N1; (iv) RSV; (v) SARS-CoV-2; and (vi) P. aeruginosa. (b) Corresponding SERS intensities for S. aureus, S. pneumoniae, and other negative samples.

Table 1.
Results of the recovery test in the sputum specimens (n = 3).
4. Conclusions
In this work, we fabricated a novel strip based on Fe3O4@Au/DTNB/Au/4-MPBA SERS nanotags for simultaneous detection of two respiratory bacteria. The SERS tags possessed magnetic enrichment and broad-spectrum bacteria capture capabilities. Our results showed that Fe3O4@Au/DTNB/Au/4-MPBA-LFA biosensor was ultrasensitive and the LODs of S. aureus and S. pneumoniae were as low as 12 and 11 CFU mL−1, respectively. Moreover, the biosensor displayed good reproducibility, excellent specificity, and high recovery rates in the sputum specimens. If the antibodies on NC membrane are altered, the strip can be used to detect other specific bacteria. Thus, the proposed multifunctional tool provided a universal platform for the detection of respiratory pathogens.
Supplementary Materials
The following supporting information [39,40] can be downloaded at: https://www.mdpi.com/article/10.3390/bios13020210/s1, S1. Preparation of Fe3O4@Au/DTNB; S2. Preparation of Colloidal AuNPs; S3. Evaluation of the Surface Enhancement Ability of Fe3O4@Au; S4. Calculation of Enhancement Factor (EF) for the SERS Detection; S5. Raman Spectra of S. aureus and S. pneumoniae with and without the Use of LFA.
Author Contributions
Conceptualization, J.L.; methodology, J.L., J.C. and Z.L.; software, Y.D.; validation, J.Z.; formal analysis, J.C.; investigation, Y.D.; resources, R.X.; data curation, J.L.; writing—original draft preparation, J.L.; writing—review and editing, R.X.; visualization, J.C.; supervision, S.L.; project administration, S.L.; funding acquisition, R.X. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by the National Key Research and Development Program of China (2021YFC2301102), the Major Project (JK20202A060431, BWS18J008), and Youth Cultivation Project (21QNPY151). This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors thank Yajie Wang of Beijing Ditan Hospital Affiliated to Capital Medical University for support in literature search and bacterial capture efficiency experiment.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ionescu, A.; Mattina, R.; Brambilla, E.; Ralli, M.; Passali, F.; Passali, D. Antibacterial and antibiofilm effects of radioactive thermal water. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 3576–3584. [Google Scholar] [PubMed]
- Beasley, V.; Joshi, P.; Singanayagam, A.; Molyneaux, P.; Johnston, S.; Mallia, P. Lung microbiology and exacerbations in COPD. Int. J. Chronic Obs. Pulmon Dis. 2012, 7, 555–569. [Google Scholar]
- Jasson, V.; Jacxsens, L.; Luning, P.; Rajkovic, A.; Uyttendaele, M. Alternative microbial methods: An overview and selection criteria. Food Microbiol. 2010, 27, 710–730. [Google Scholar] [CrossRef] [PubMed]
- Oeser, C.; Pond, M.; Butcher, P.; Russell, A.B.; Henneke, P.; Laing, K.; Planche, T.; Heath, P.T.; Harris, K. PCR for the detection of pathogens in neonatal early onset sepsis. PLoS ONE 2020, 15, e0226817. [Google Scholar] [CrossRef]
- Pang, B.; Zhao, C.; Li, L.; Song, X.; Xu, K.; Wang, J.; Liu, Y.; Fu, K.; Bao, H.; Song, D.; et al. Development of a low-cost paper-based ELISA method for rapid Escherichia coli O157:H7 detection. Anal. Biochem. 2018, 542, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, S.; Tiwari, U.; Pal, S.; Sinha, R. Rapid detection of Escherichia coli using fiber optic surface plasmon resonance immunosensor based on biofunctionalized Molybdenum disulfide (MoS2) nanosheets. Biosens. Bioelectron. 2019, 126, 501–509. [Google Scholar] [CrossRef]
- Azevedo, A.; Rocha, R.; Dias, N. Flow-FISH Using Nucleic Acid Mimic Probes for the Detection of Bacteria. Methods Mol. Biol. 2021, 2246, 263–277. [Google Scholar]
- Charalampous, T.; Kay, G.L.; Richardson, H.; Aydin, A.; Baldan, R.; Jeanes, C.; Rae, D.; Grundy, S.; Turner, D.J.; Wain, J.; et al. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat. Biotechnol. 2019, 37, 783–792. [Google Scholar] [CrossRef]
- Walter, A.; Marz, A.; Schumacher, W.; Rosch, P.; Popp, J. Towards a fast, high specific and reliable discrimination of bacteria on strain level by means of SERS in a microfluidic device. Lab Chip 2011, 11, 1013–1021. [Google Scholar] [CrossRef]
- Andrade, C.A.; Nascimento, J.M.; Oliveira, I.S.; de Oliveira, C.V.; de Melo, C.P.; Franco, O.L.; Oliveira, M.D. Nanostructured sensor based on carbon nanotubes and clavanin A for bacterial detection. Colloids Surf. B Biointerfaces 2015, 135, 833–839. [Google Scholar] [CrossRef]
- Pang, Y.; Wan, N.; Shi, L.; Wang, C.; Sun, Z.; Xiao, R.; Wang, S. Dual-recognition surface-enhanced Raman scattering(SERS)biosensor for pathogenic bacteria detection by using vancomycin-SERS tags and aptamer-Fe3O4@Au. Anal. Chim. Acta 2019, 1077, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Zhou, R.; Takei, K.; Hong, M. Toward Flexible Surface-Enhanced Raman Scattering (SERS) Sensors for Point-of-Care Diagnostics. Adv. Sci. 2019, 6, 1900925. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yi, K.; Zhang, W.; Huang, C.; Dong, S.; Wang, F.; Xiao, X. Trace Analysis of Multiple Tumor Exosomal PD-L1 Based on SERS Immunoassay Platform. Adv. Sens. Res. 2022, 20, 2200043. [Google Scholar] [CrossRef]
- Cheng, N.; Chen, D.; Lou, B.; Fu, J.; Wang, H. A biosensing method for the direct serological detection of liver diseases by integrating a SERS-based sensor and a CNN classifier. Biosens. Bioelectron. 2021, 186, 113246. [Google Scholar] [CrossRef]
- Yang, E.; Li, D.; Yin, P.; Xie, Q.; Li, Y.; Lin, Q.; Duan, Y. A novel surface-enhanced Raman scattering (SERS) strategy for ultrasensitive detection of bacteria based on three-dimensional (3D) DNA walker. Biosens. Bioelectron. 2021, 172, 112758. [Google Scholar] [CrossRef]
- Zhou, Y.; Wu, Y.; Ding, L.; Huang, X.; Xiong, Y. Point-of-care COVID-19 diagnostics powered by lateral flow assay. Trends Anal. Chem. 2021, 145, 116452. [Google Scholar] [CrossRef]
- Pang, R.; Zhu, Q.; Wei, J.; Wang, Y.; Xu, F.; Meng, X.; Wang, Z. Development of a gold-nanorod-based lateral flow immunoassay for a fast and dual-modal detection of C-reactive protein in clinical plasma samples. RSC Adv. 2021, 11, 28388–28394. [Google Scholar] [CrossRef]
- Li, D.; Huang, M.; Shi, Z.; Huang, L.; Jin, J.; Jiang, C.; Yu, W.; Guo, Z.; Wang, J. Ultrasensitive Competitive Lateral Flow Immunoassay with Visual Semiquantitative Inspection and Flexible Quantification Capabilities. Anal. Chem. 2022, 94, 2996–3004. [Google Scholar] [CrossRef]
- Liu, Y.-Q.; Zhu, W.; Hu, J.-M.; Shen, A.-G. Recent advances in plasmonic Prussian blue-based SERS nanotags for biological application. Nanoscale Adv. 2021, 3, 6568–6579. [Google Scholar] [CrossRef]
- Wang, Z.; Zong, S.; Wu, L.; Zhu, D.; Cui, Y. SERS-Activated Platforms for Immunoassay: Probes, Encoding Methods, and Applications. Chem. Rev. 2017, 117, 7910–7963. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, H.; Hu, Z.; Yu, G.; Yang, D.; Zhao, J. Label and label-free based surface-enhanced Raman scattering for pathogen bacteria detection: A review. Biosens. Bioelectron. 2017, 94, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Beier, R.; Shen, J. Immunoassays for the detection of macrocyclic lactones in food matrices—A review. TrAC Trends Anal. Chem. 2017, 92, 42–61. [Google Scholar] [CrossRef]
- Gupta, R.; Kumar, A.; Kumar, S.; Pinnaka, A.; Singhal, N. Naked eye colorimetric detection of Escherichia coli using aptamer conjugated graphene oxide enclosed Gold nanoparticles. Sens. Actuators B Chem. 2021, 329, 129100. [Google Scholar] [CrossRef]
- Renuka, R.; Achuth, J.; Chandan, H.; Venkataramana, M.; Kadirvelu, K. A fluorescent dual aptasensor for the rapid and sensitive onsite detection ofE. coliO157:H7 and its validation in various food matrices. New J. Chem. 2018, 42, 10807–10817. [Google Scholar] [CrossRef]
- Yuan, K.; Mei, Q.; Guo, X.; Xu, Y.; Yang, D.; Sánchez, B.J.; Sheng, B.; Liu, C.; Hu, Z.; Yu, G.; et al. Antimicrobial peptide based magnetic recognition elements and Au@Ag-GO SERS tags with stable internal standards: A three in one biosensor for isolation, discrimination and killing of multiple bacteria in whole blood. Chem. Sci. 2018, 9, 8781–8795. [Google Scholar] [CrossRef]
- Yue, X.; Su, Y.; Wang, X.; Li, L.; Ji, W.; Ozaki, Y. Reusable Silicon-Based SERS Chip for Ratiometric Analysis of Fluoride Ion in Aqueous Solutions. ACS Sens. 2019, 4, 2336–2342. [Google Scholar] [CrossRef]
- Wang, H.; Zhou, Y.; Jiang, X.; Sun, B.; Zhu, Y.; Wang, H.; Su, Y.; He, Y. Simultaneous capture, detection, and inactivation of bacteria as enabled by a surface-enhanced Raman scattering multifunctional chip. Angew. Chem. Int. Ed. Engl. 2015, 54, 5132–5136. [Google Scholar] [CrossRef]
- Zhou, Z.; Xiao, R.; Cheng, S.; Wang, S.; Shi, L.; Wang, C.; Qi, K.; Wang, S. A universal SERS-label immunoassay for pathogen bacteria detection based on Fe3O4@Au-aptamer separation and antibody-protein A orientation recognition. Anal. Chim. Acta 2021, 1160, 338421. [Google Scholar] [CrossRef]
- Li, J.; Wang, C.; Shi, L.; Shao, L.; Fu, P.; Wang, K.; Xiao, R.; Wang, S.; Gu, B. Rapid identification and antibiotic susceptibility test of pathogens in blood based on magnetic separation and surface-enhanced Raman scattering. Mikrochim. Acta 2019, 186, 475. [Google Scholar] [CrossRef]
- Yue, X.; Pan, Q.; Zhou, J.; Ren, H.; Peng, C.; Wang, Z.; Zhang, Y. A simplified fluorescent lateral flow assay for melamine based on aggregation induced emission of gold nanoclusters. Food Chem. 2022, 385, 132670. [Google Scholar] [CrossRef]
- Yang, H.; Wang, Y.; Liu, S.; Ouyang, H.; Lu, S.; Li, H.; Fu, Z. Lateral flow assay of methicillin-resistant Staphylococcus aureus using bacteriophage cellular wall-binding domain as recognition agent. Biosens. Bioelectron. 2021, 182, 113189. [Google Scholar] [CrossRef]
- Juang, R.-S.; Su, C.-J.; Wu, M.-C.; Lu, H.-C.; Wang, S.-F.; Sun, A.-C. Fabrication of Magnetic Fe₃O₄ Nanoparticles with Unidirectional Extension Pattern by a Facile and Eco-Friendly Microwave-Assisted Solvothermal Method. J. Nanosci. Nanotechnol. 2019, 19, 7645–7653. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Kang, H.; Gu, F.; Wang, C.; Cheng, S.; Gong, W.; Wang, L.; Gu, B.; Yang, Y. Rapid Detection Method for Pathogenic Candida Captured by Magnetic Nanoparticles and Identified Using SERS via AgNPs. Int. J. Nanomed. 2021, 16, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, J.; Li, M.; Qu, X.; Zhang, K.; Rong, Z.; Xiao, R.; Wang, S. A rapid SERS method for label-free bacteria detection using polyethylenimine-modified Au-coated magnetic microspheres and Au@Ag nanoparticles. Analyst 2016, 141, 6226–6238. [Google Scholar] [CrossRef]
- Zhou, S.; Guo, X.; Huang, H.; Huang, X.; Zhou, X.; Zhang, Z.; Sun, G.; Cai, H.; Zhou, H.; Sun, P. Triple-Function Au-Ag-Stuffed Nanopancakes for SERS Detection, Discrimination, and Inactivation of Multiple Bacteria. Anal. Chem. 2022, 94, 5785–5796. [Google Scholar] [CrossRef]
- Wang, X.; Choi, N.; Cheng, Z.; Ko, J.; Chen, L.; Choo, J. Simultaneous Detection of Dual Nucleic Acids Using a SERS-Based Lateral Flow Assay Biosensor. Anal. Chem. 2017, 89, 1163–1169. [Google Scholar] [CrossRef]
- Yun, J.; Park, J.H.; Kim, N.; Roh, E.Y.; Shin, S.; Yoon, J.H.; Kim, T.S.; Park, H. Evaluation of Three Multiplex Real-time Reverse Transcription PCR Assays for Simultaneous Detection of SARS-CoV-2, Influenza A/B, and Respiratory Syncytial Virus in Nasopharyngeal Swabs. J. Korean Med. Sci. 2021, 36, e328. [Google Scholar] [CrossRef]
- Jo, J.; Price-Whelan, A.; Cornell, W.; Dietrich, L. Interdependency of Respiratory Metabolism and Phenazine-Associated Physiology in Pseudomonas aeruginosa PA14. J. Bacteriol. 2020, 202, e00700-19. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, X.; Li, K.; Liu, H.; Xiao, R.; Wang, W.; Wang, C.; Wang, S. Fe3O4@Au SERS tags-based lateral flow assay for sim-ultaneous detection of serum amyloid A and C-reactive protein in unprocessed blood sample. Sens. Actuators B Chem. 2020, 320, 128350. [Google Scholar] [CrossRef]
- Li, J.F.; Tian, X.D.; Li, S.B.; Anema, J.R.; Yang, Z.L.; Ding, Y.; Wu, Y.F.; Zeng, Y.M.; Chen, Q.Z.; Ren, B.; et al. Surface analysis using shell-isolated nanoparticle-enhanced Raman spectroscopy. Nat. Protoc. 2013, 8, 52–65. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).