An Ultrasensitive miRNA-Based Genosensor for Detection of MicroRNA 21 in Gastric Cancer Cells Based on Functional Signal Amplifier and Synthesized Perovskite-Graphene Oxide and AuNPs
Abstract
1. Introduction
2. Experimental
2.1. Apparatus and Instrumentation
2.2. Reagents and Materials
2.3. Nanocomposite Preparation Steps
2.3.1. Preparation of Perovskite and Chemically-Synthesis of Graphene Oxide (GO)
2.3.2. Preparing Perovskite/Graphene Oxide Nanocomposite
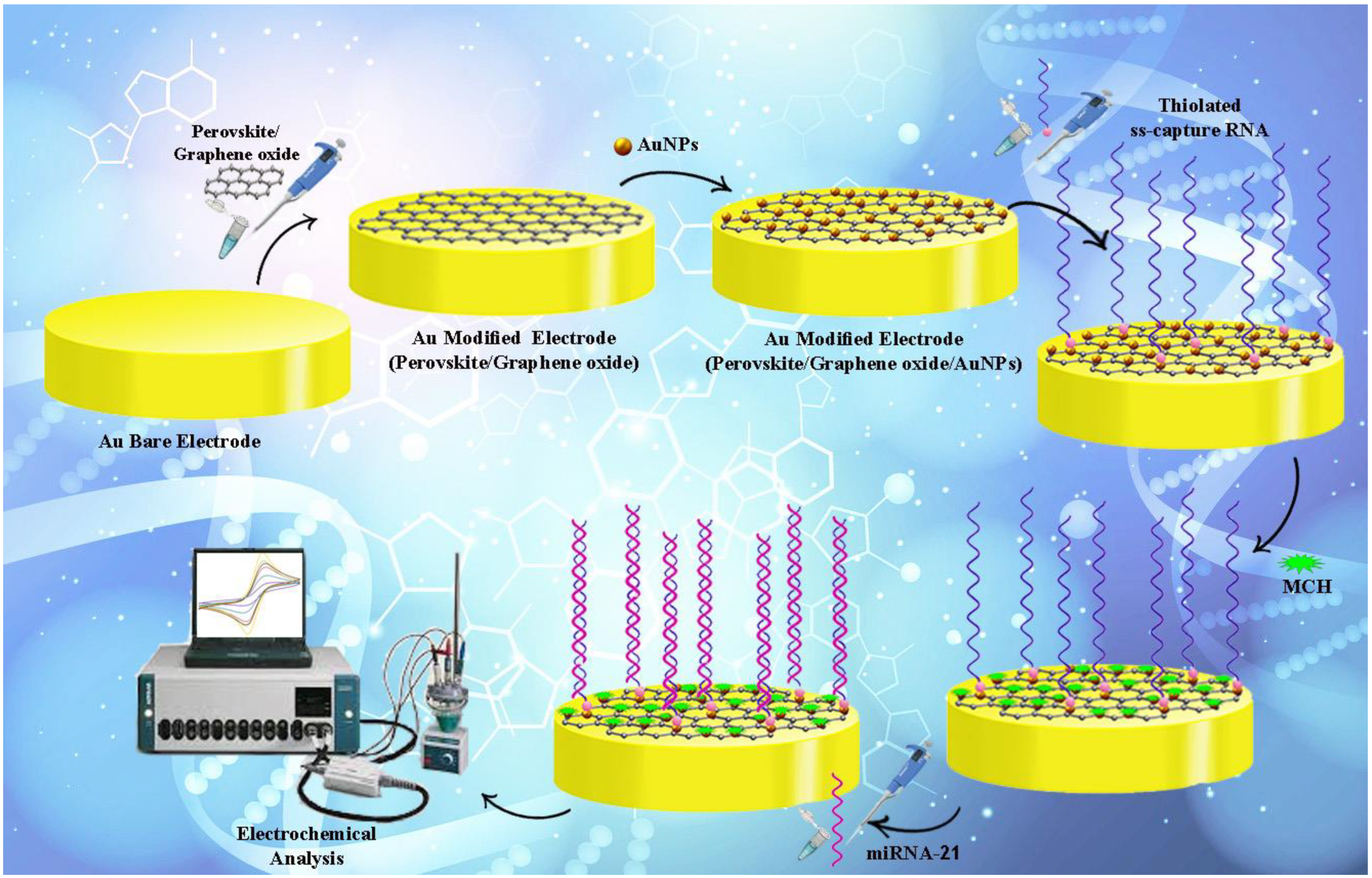
2.4. Fabricating Electrochemical Genosensing Assessment
2.5. Electrochemically Detecting miRNA-21
2.6. Real Samples
2.6.1. Cell Culture
2.6.2. MicroRNA Transfection
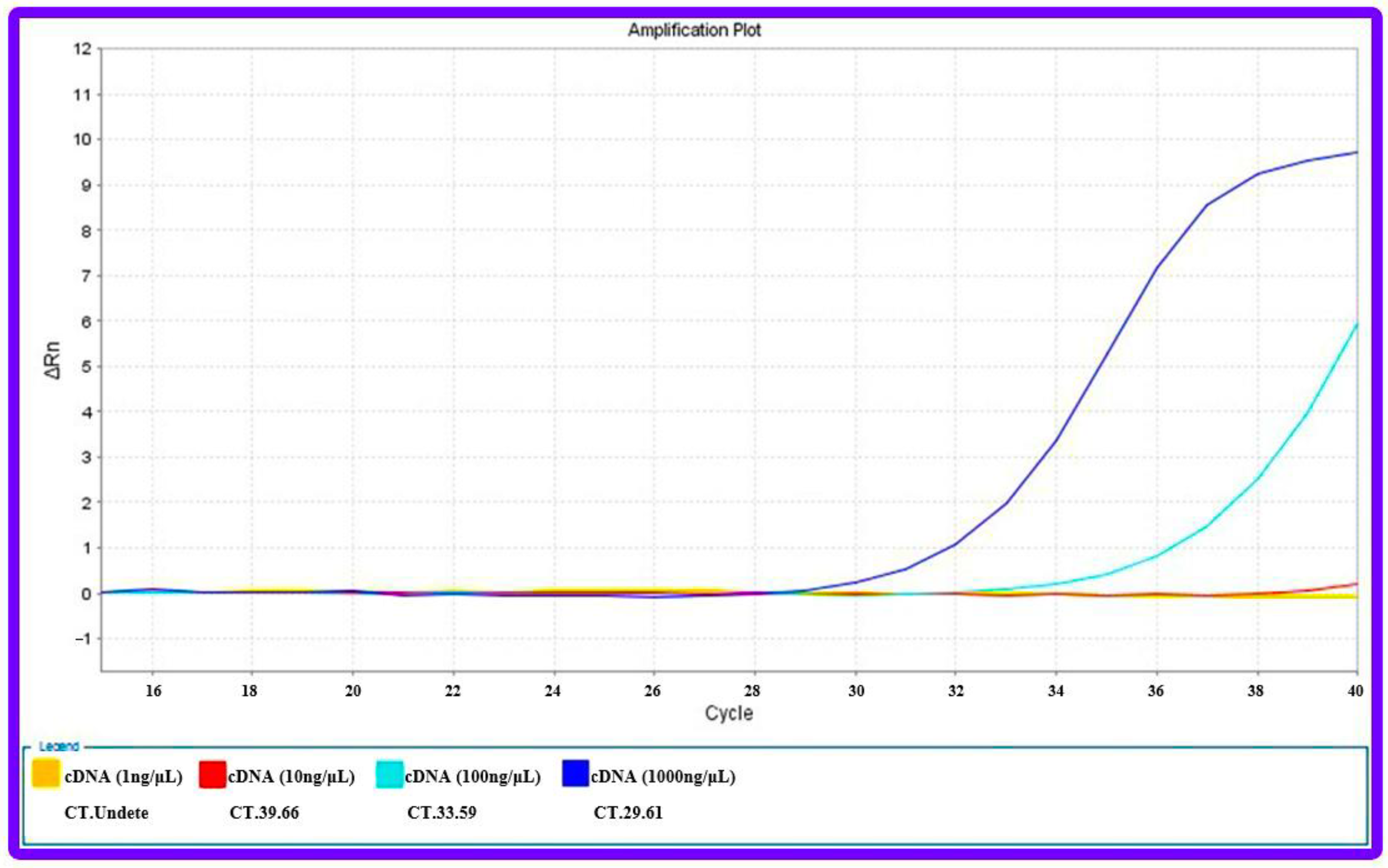
2.6.3. Quantitative Real Time-Polymerase Chain Reaction (qRT-PCR)
3. Results and Discussion
3.1. Surface Morphology of the Gold Electrode
3.1.1. Field Emission Scanning Electron Microscope
3.1.2. Energy-Dispersive X-ray Spectroscopy (EDS)
3.1.3. Raman Spectroscopy
3.1.4. X-ray Powder Diffraction (XRD)
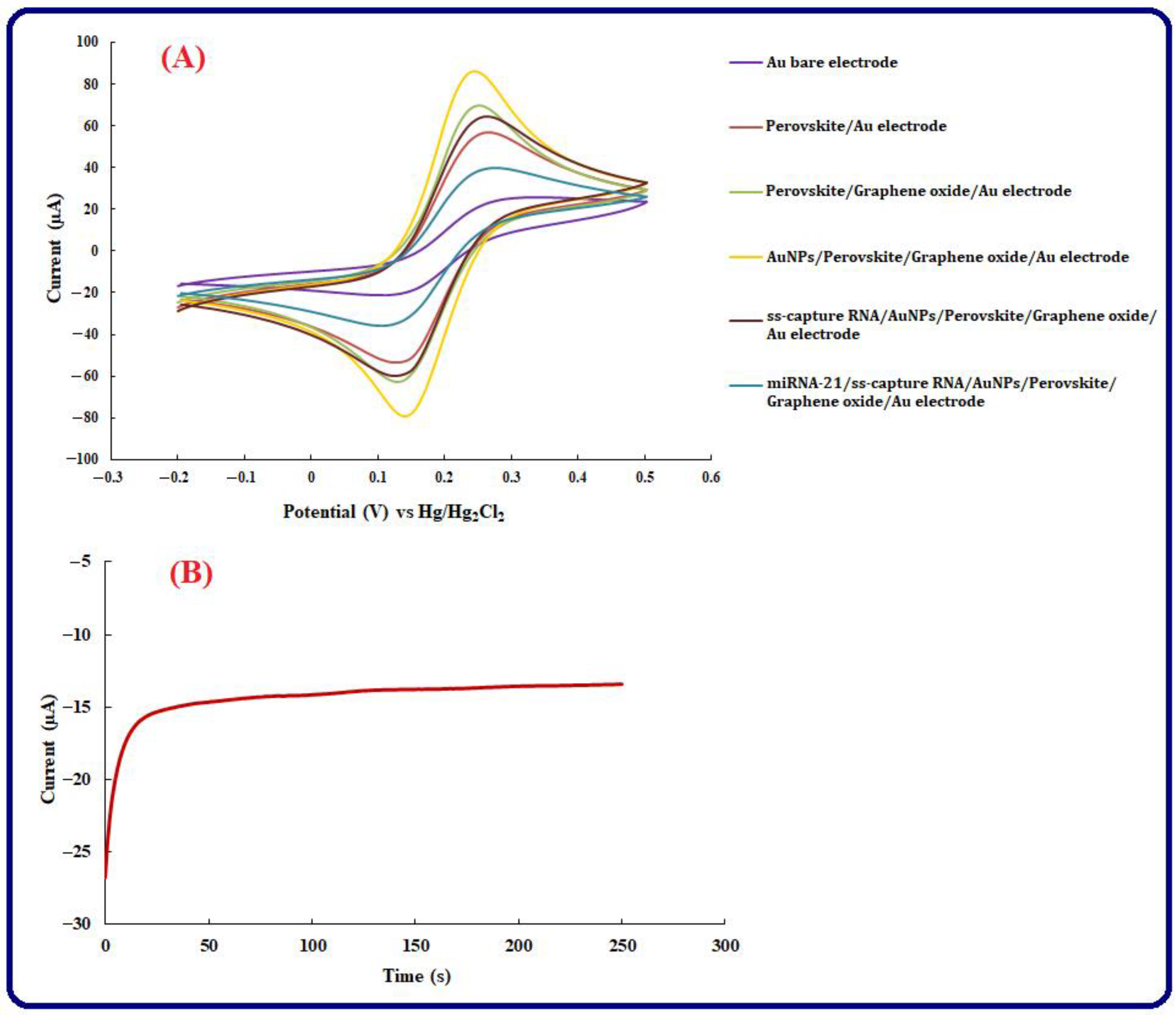
3.2. Electrochemical Investigation of the Engineered Genosensing Assay
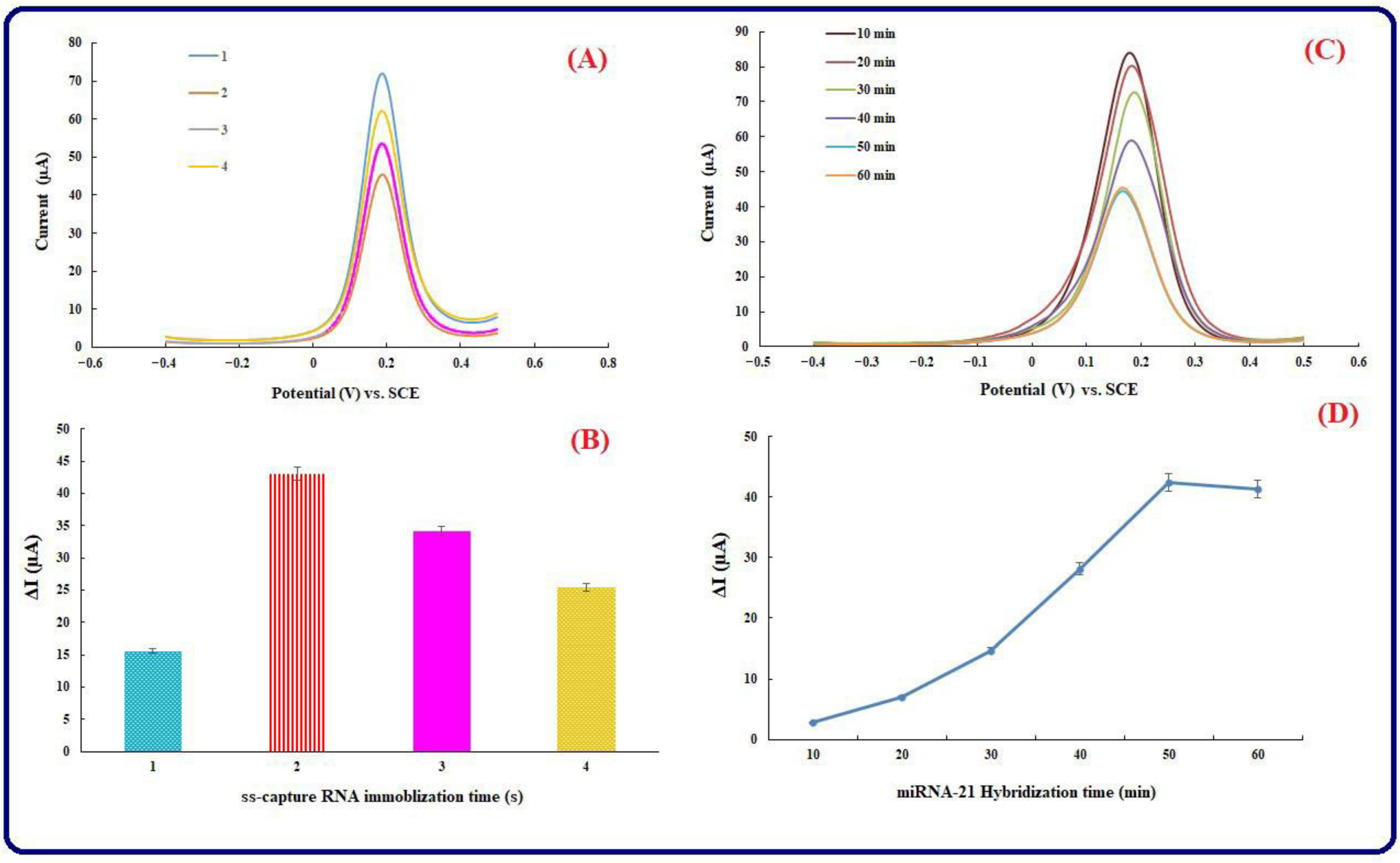
3.3. ss-Capture RNA Immobilization Time and Hybridization Time
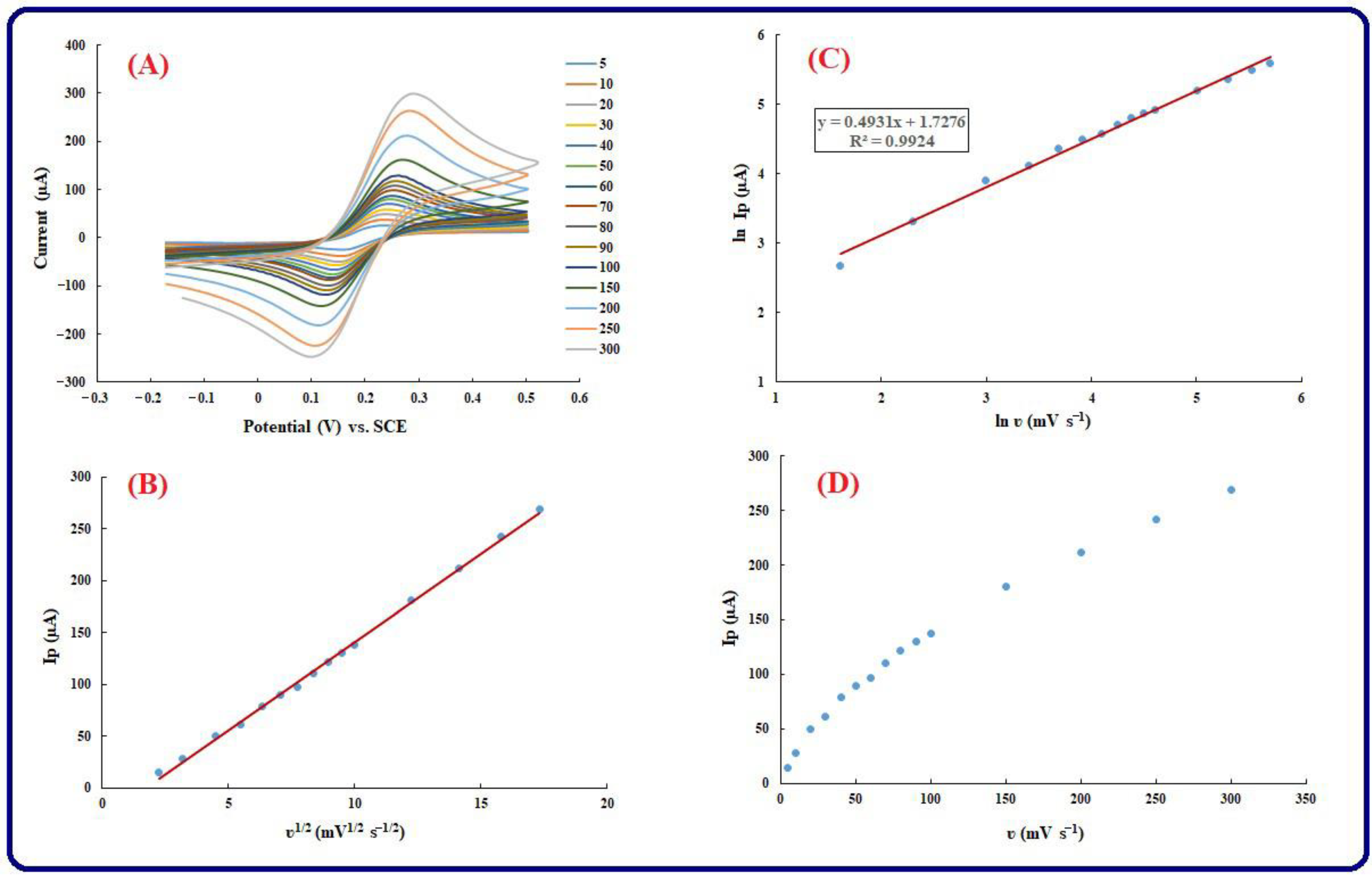
3.4. Kinetic Investigation
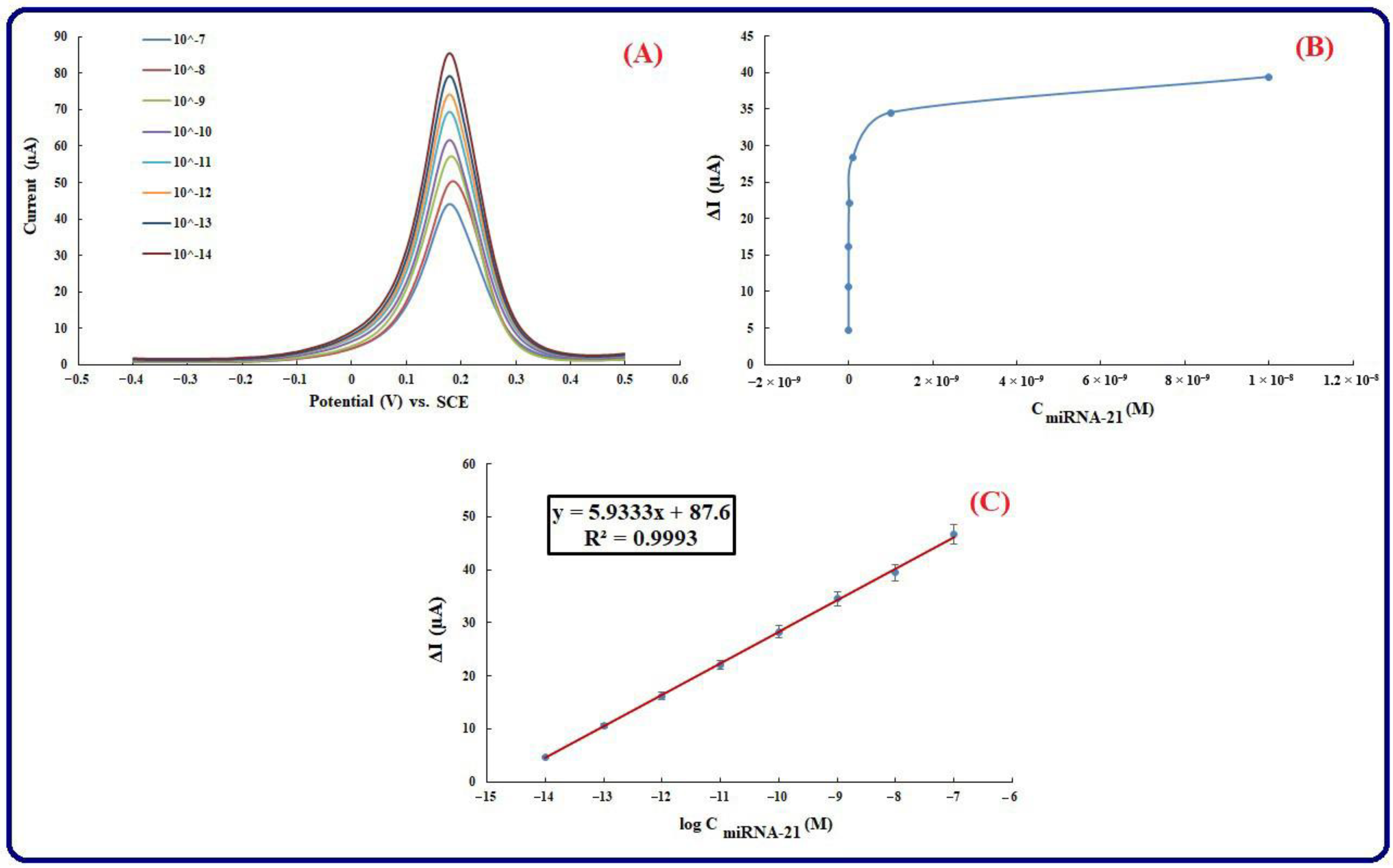
3.5. Calibration Plot and Analytical Features
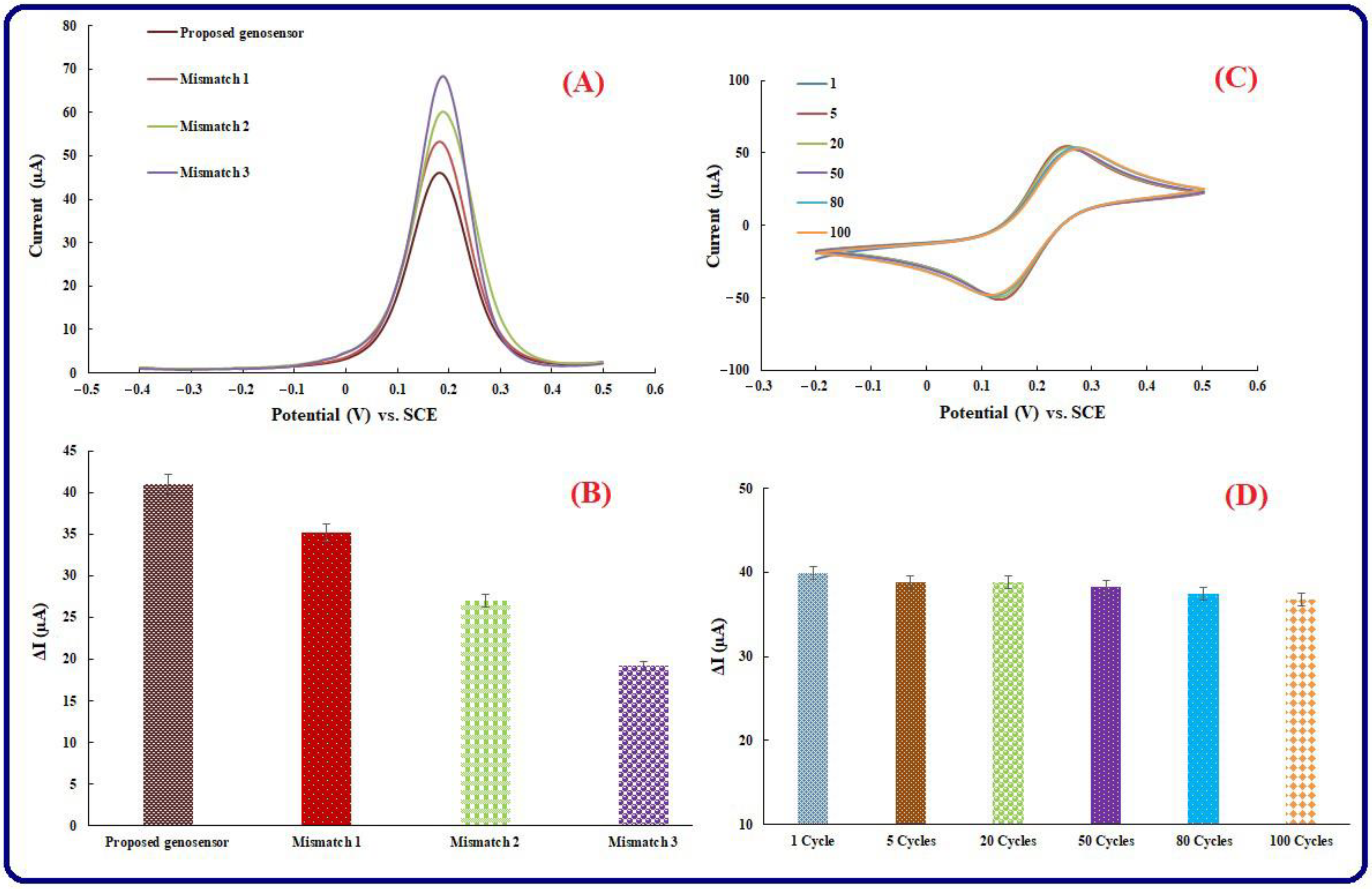
3.6. Selectivity Investigation
3.7. Repeatability, Reproducibility, and Stability of Genosensing Bio-Assay
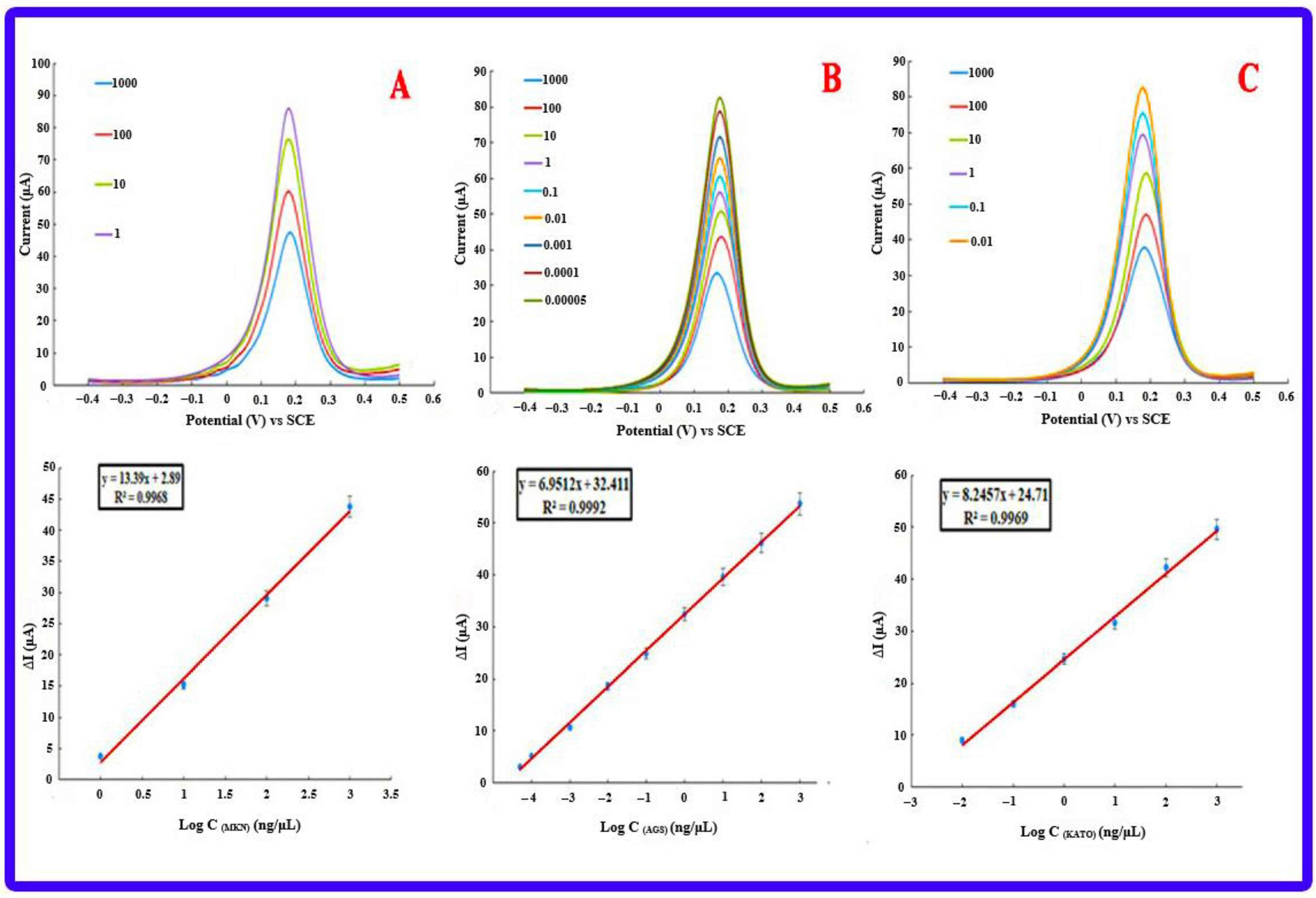
3.8. Investigating the Performance of Bioassays in Real Samples
3.8.1. Gastric Cancer Cell Lines
3.8.2. Anti-miR-21 Treatment
4. Conclusions, Challenges, and Future Outlooks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jafari, S.H.; Saadatpour, Z.; Salmaninejad, A.; Momeni, F.; Mokhtari, M.; Nahand, J.S.; Rahmati, M.; Mirzaei, H.; Kianmehr, M. Breast cancer diagnosis: Imaging techniques and biochemical markers. J. Cell. Physiol. 2018, 233, 5200–5213. [Google Scholar] [CrossRef] [PubMed]
- Keshavarzi, M.; Darijani, M.; Momeni, F.; Moradi, P.; Ebrahimnejad, H.; Masoudifar, A.; Mirzaei, H. Molecular imaging and oral cancer diagnosis and therapy. J. Cell. Biochem. 2017, 118, 3055–3060. [Google Scholar] [CrossRef] [PubMed]
- Keshavarzi, M.; Sorayayi, S.; Rezaei, M.J.; Mohammadi, M.; Ghaderi, A.; Rostamzadeh, A.; Masoudifar, A.; Mirzaei, H. MicroRNAs-based imaging techniques in cancer diagnosis and therapy. J. Cell. Biochem. 2017, 118, 4121–4128. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Yekta, S.; Shih, I.-h.; Bartel, D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science 2004, 304, 594–596. [Google Scholar] [CrossRef]
- Rezaei, T.; Amini, M.; Hashemi, Z.S.; Mansoori, B.; Rezaei, S.; Karami, H.; Mosafer, J.; Mokhtarzadeh, A.; Baradaran, B. Microrna-181 serves as a dual-role regulator in the development of human cancers. Free. Radic. Biol. Med. 2020, 152, 432–454. [Google Scholar] [CrossRef] [PubMed]
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef]
- Farazi, T.A.; Hoell, J.I.; Morozov, P.; Tuschl, T. MicroRNAs in human cancer. MicroRNA Cancer Regul. 2013, 774, 1–20. [Google Scholar]
- Montagner, S.; Dehó, L.; Monticelli, S. MicroRNAs in hematopoietic development. BMC Immunol. 2014, 15, 1–12. [Google Scholar] [CrossRef]
- Moreno-Moya, J.M.; Vilella, F.; Simón, C. MicroRNA: Key gene expression regulators. Fertil. Steril. 2014, 101, 1516–1523. [Google Scholar] [CrossRef]
- Piubelli, C.; Meraviglia, V.; Pompilio, G.; D’Alessandra, Y.; Colombo, G.I.; Rossini, A. MicroRNAs and cardiac cell fate. Cells 2014, 3, 802–823. [Google Scholar] [CrossRef] [PubMed]
- Ardekani, A.M.; Naeini, M.M. The role of microRNAs in human diseases. Avicenna J. Med. Biotechnol. 2010, 2, 161. [Google Scholar] [PubMed]
- Hejazi, M.; Baghbani, E.; Amini, M.; Rezaei, T.; Aghanejad, A.; Mosafer, J.; Mokhtarzadeh, A.; Baradaran, B. MicroRNA-193a and taxol combination: A new strategy for treatment of colorectal cancer. J. Cell. Biochem. 2020, 121, 1388–1399. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Kilic, T.; Erdem, A.; Ozsoz, M.; Carrara, S. MicroRNA biosensors: Opportunities and challenges among conventional and commercially available techniques. Biosens. Bioelectron. 2018, 99, 525–546. [Google Scholar] [CrossRef] [PubMed]
- Jebelli, A.; Oroojalian, F.; Fathi, F.; Mokhtarzadeh, A.; de la Guardia, M. Recent advances in surface plasmon resonance biosensors for microRNAs detection. Biosens. Bioelectron. 2020, 169, 112599. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Sohrabi, H.; Bolandi, N.; Hemmati, A.; Eyvazi, S.; Ghasemzadeh, S.; Baradaran, B.; Oroojalian, F.; Majidi, M.R.; de la Guardia, M.; Mokhtarzadeh, A. State-of-the-art cancer biomarker detection by portable (Bio) sensing technology: A critical review. Microchem. J. 2022, 177, 107248. [Google Scholar] [CrossRef]
- Yousefi, M.; Dehghani, S.; Nosrati, R.; Zare, H.; Evazalipour, M.; Mosafer, J.; Tehrani, B.S.; Pasdar, A.; Mokhtarzadeh, A.; Ramezani, M. Aptasensors as a new sensing technology developed for the detection of MUC1 mucin: A review. Biosens. Bioelectron. 2019, 130, 1–19. [Google Scholar] [CrossRef]
- Mokhtarzadeh, A.; Nazhad Dolatabadi, J.E.; Abnous, K.; de la Guardia, M.; Ramezani, M. Nanomaterial-based cocaine aptasensors. Biosens. Bioelectron. 2015, 68, 95–106. [Google Scholar] [CrossRef]
- Guo, Y.K.; Yang, Z.G.; Li, Y.; Ma, E.S.; Deng, Y.P.; Min, P.Q.; Yin, L.L.; Hu, J.; Zhang, X.C.; Chen, T.W. Addison’s disease due to adrenal tuberculosis: Contrast-enhanced CT features and clinical duration correlation. Eur. J. Radiol. 2007, 62, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zou, X.; Chen, W. A new X-ray activated nanoparticle photosensitizer for cancer treatment. J. Biomed. Nanotechnol. 2014, 10, 1501–1508. [Google Scholar] [CrossRef] [PubMed]
- Næser, E.; Møller, H.; Fredberg, U.; Frystyk, J.; Vedsted, P. Routine blood tests and probability of cancer in patients referred with non-specific serious symptoms: A cohort study. BMC Cancer 2017, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Yuan, C.; Yan, Q.; Duan, Q.; Li, X.; Yi, G. An electrochemical biosensor for microRNA-196a detection based on cyclic enzymatic signal amplification and template-free DNA extension reaction with the adsorption of methylene blue. Biosens. Bioelectron. 2018, 105, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Lautner, G.; Gyurcsányi, R.E. Electrochemical detection of miRNAs. Electroanalysis 2014, 26, 1224–1235. [Google Scholar] [CrossRef]
- Liu, C.; Chen, C.; Li, S.; Dong, H.; Dai, W.; Xu, T.; Liu, Y.; Yang, F.; Zhang, X. Target-triggered catalytic hairpin assembly-induced core–satellite nanostructures for high-sensitive “Off-to-On” sers detection of intracellular MicroRNA. Anal. Chem. 2018, 90, 10591–10599. [Google Scholar] [CrossRef] [PubMed]
- Sohrabi, H.; Sani, P.S.; Zolfaghari, R.; Majidi, M.R.; Yoon, Y.; Khataee, A. MOF-Based Mycotoxin Nanosensors for Food Quality and Safety Assessment through Electrochemical and Optical Methods. Molecules 2022, 27, 7511. [Google Scholar] [CrossRef]
- Sohrabi, H.; Majidi, M.R.; Khaki, P.; Jahanban-Esfahlan, A.; de la Guardia, M.; Mokhtarzadeh, A. State of the art: Lateral flow assays toward the point-of-care foodborne pathogenic bacteria detection in food samples. Compr. Rev. Food Sci. Food Saf. 2022, 21, 1868–1912. [Google Scholar] [CrossRef]
- Arora, N. Recent advances in biosensors technology: A review. Octa J. Biosci. 2013, 1, 147–150. [Google Scholar]
- Majidi, M.R.; Sohrabi, H. Chiral Conductive Polymers. In Conductive Polymers in Analytical Chemistry; ACS Publications: Washington, DC, USA, 2022; pp. 287–312. [Google Scholar]
- Damborský, P.; Švitel, J.; Katrlík, J. Optical biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar]
- Eivazzadeh-Keihan, R.; Pashazadeh-Panahi, P.; Baradaran, B.; de la Guardia, M.; Hejazi, M.; Sohrabi, H.; Mokhtarzadeh, A.; Maleki, A. Recent progress in optical and electrochemical biosensors for sensing of Clostridium botulinum neurotoxin. TrAC Trends Anal. Chem. 2018, 103, 184–197. [Google Scholar] [CrossRef]
- Pohanka, M. Overview of piezoelectric biosensors, immunosensors and DNA sensors and their applications. Materials 2018, 11, 448. [Google Scholar] [CrossRef] [PubMed]
- Sohrabi, H.; Majidi, M.R.; Asadpour-Zeynali, K.; Khataee, A.; Mokhtarzadeh, A. Bimetallic Fe/Mn MOFs/MβCD/AuNPs stabilized on MWCNTs for developing a label-free DNA-based genosensing bio-assay applied in the determination of Salmonella typhimurium in milk samples. Chemosphere 2022, 287, 132373. [Google Scholar] [CrossRef]
- Sohrabi, H.; Majidi, M.R.; Asadpour-Zeynali, K.; Khataee, A.; Dastborhan, M.; Mokhtarzadeh, A. A PCR-free genosensing platform for detection of Shigella dysenteriae in human plasma samples by porous and honeycomb-like biochar decorated with ultrathin flower-like MoS2 nanosheets incorporated with Au nanoparticles. Chemosphere 2022, 288, 132531. [Google Scholar] [CrossRef] [PubMed]
- Erdem, A. Electrochemical sensor technology based on nanomaterials for biomolecular recognitions. In Functionalized Nanoscale Materials, Devices and Systems; Springer: Berlin/Heidelberg, Germany, 2008; pp. 273–278. [Google Scholar]
- Johnson, B.N.; Mutharasan, R. Sample preparation-free, real-time detection of microRNA in human serum using piezoelectric cantilever biosensors at attomole level. Anal. Chem. 2012, 84, 10426–10436. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, S.; Verma, A.; Tyagi, N.; Kumar, V. Biosensors: Principle, types and applications. Int. J. Adv. Res. Innov. Ideas Educ. 2017, 3, 3639–3644. [Google Scholar]
- Sohrabi, H.; Majidi, M.R.; Arbabzadeh, O.; Khaaki, P.; Pourmohammad, S.; Khataee, A.; Orooji, Y. Recent advances in the highly sensitive determination of zearalenone residues in water and environmental resources with electrochemical biosensors. Environ. Res. 2022, 204, 112082. [Google Scholar] [CrossRef] [PubMed]
- Govindasamy, M.; Jian, C.-R.; Kuo, C.-F.; Hsieh, A.-H.; Sie, J.-L.; Huang, C.-H. A chemiresistive biosensor for detection of cancer biomarker in biological fluids using CVD-grown bilayer graphene. Microchim. Acta 2022, 189, 1–12. [Google Scholar] [CrossRef]
- Sohrabi, H.; Arbabzadeh, O.; Khaaki, P.; Majidi, M.R.; Khataee, A.; Joo, S.W. Emerging electrochemical sensing and biosensing approaches for detection of Fumonisins in food samples. Crit. Rev. Food Sci. Nutr. 2021, 62, 8761–8776. [Google Scholar] [CrossRef]
- Sohrabi, H.; Arbabzadeh, O.; Khaaki, P.; Khataee, A.; Majidi, M.R.; Orooji, Y. Patulin and Trichothecene: Characteristics, occurrence, toxic effects and detection capabilities via clinical, analytical and nanostructured electrochemical sensing/biosensing assays in foodstuffs. Crit. Rev. Food Sci. Nutr. 2021, 62, 5540–5568. [Google Scholar] [CrossRef]
- Sohrabi, H.; Hemmati, A.; Majidi, M.R.; Eyvazi, S.; Jahanban-Esfahlan, A.; Baradaran, B.; Adlpour-Azar, R.; Mokhtarzadeh, A.; de la Guardia, M. Recent advances on portable sensing and biosensing assays applied for detection of main chemical and biological pollutant agents in water samples: A critical review. TrAC Trends Anal. Chem. 2021, 143, 116344. [Google Scholar] [CrossRef]
- Hasanzadeh, M.; Baghban, H.N.; Mokhtarzadeh, A.; Shadjou, N.; Mahboob, S. An innovative immunosensor for detection of tumor suppressor protein p53 in unprocessed human plasma and cancer cell lysates. Int. J. Biol. Macromol. 2017, 105, 1337–1348. [Google Scholar] [CrossRef]
- Zeng, Y.; Cullen, B.R. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004, 32, 4776–4785. [Google Scholar] [CrossRef] [PubMed]
- Mokhtarzadeh, A.; Eivazzadeh-Keihan, R.; Pashazadeh, P.; Hejazi, M.; Gharaatifar, N.; Hasanzadeh, M.; Baradaran, B.; de la Guardia, M. Nanomaterial-based biosensors for detection of pathogenic virus. TrAC Trends Anal. Chem. 2017, 97, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-W.; Rajaji, U.; Chen, S.-M.; Govindasamy, M.; Selvin, S.S.P.; Manavalan, S.; Arumugam, R. Sonochemical synthesis of graphene oxide sheets supported Cu2S nanodots for high sensitive electrochemical determination of caffeic acid in red wine and soft drinks. Compos. Part B Eng. 2019, 158, 419–427. [Google Scholar] [CrossRef]
- Tseng, T.-W.; Rajaji, U.; Chen, T.-W.; Chen, S.-M.; Huang, Y.-C.; Mani, V.; Jothi, A.I. Sonochemical synthesis and fabrication of perovskite type calcium titanate interfacial nanostructure supported on graphene oxide sheets as a highly efficient electrocatalyst for electrochemical detection of chemotherapeutic drug. Ultrason. Sonochemistry 2020, 69, 105242. [Google Scholar] [CrossRef]
- Arul, P.; Huang, S.-T.; Mani, V.; Huang, C.-H. Graphene quantum dots-based nanocomposite for electrocatalytic application of L-cysteine in whole blood and live cells. Electrochim. Acta 2022, 428, 140937. [Google Scholar] [CrossRef]
- Gargari, M.R.; Mahmoudi, E.; Majidi, M.M.; Sohrabi, H.; Majidi, M.R.; Niaei, A. Mesoporous perovskite-type La0. 8Sr0. 2Cu0. 7Mn0. 3O3/SiO2 nanocomposite-decorated-graphene-oxide nanosheets: Green synthesis and application in the sensitive determination of Morin in kiwi fruit samples. Synth. Met. 2023, 293, 117257. [Google Scholar] [CrossRef]
- Hu, T.; Zhang, L.; Wen, W.; Zhang, X.; Wang, S. Enzyme catalytic amplification of miRNA-155 detection with graphene quantum dot-based electrochemical biosensor. Biosens. Bioelectron. 2016, 77, 451–456. [Google Scholar] [CrossRef]
- Peng, Y.; Jiang, J.; Yu, R. A sensitive electrochemical biosensor for microRNA detection based on streptavidin–gold nanoparticles and enzymatic amplification. Anal. Methods 2014, 6, 2889–2893. [Google Scholar] [CrossRef]
- Hosseini, S.A.; Mehri, B.; Niaei, A.; Izadkhah, B.; Alvarez-Galvan, C.; Fierro, J.G.L. Selective catalytic reduction of NO x by CO over LaMnO3 nano perovskites prepared by microwave and ultrasound assisted sol–gel method. J. Sol-Gel Sci. Technol. 2018, 85, 647–656. [Google Scholar] [CrossRef]
- Mohammadi, A.; Farzi, A.; Thurner, C.; Klötzer, B.; Schwarz, S.; Bernardi, J.; Niaei, A.; Penner, S. Tailoring the metal-perovskite interface for promotional steering of the catalytic NO reduction by CO in the presence of H2O on Pd-lanthanum iron manganite composites. Appl. Catal. B Environ. 2022, 307, 121160. [Google Scholar] [CrossRef]
- Chen, J.; Yao, B.; Li, C.; Shi, G. An improved Hummers method for eco-friendly synthesis of graphene oxide. Carbon 2013, 64, 225–229. [Google Scholar] [CrossRef]
- Berg, K.E.; Clark, K.M.; Li, X.; Carter, E.M.; Volckens, J.; Henry, C.S. High-throughput, semi-automated dithiothreitol (DTT) assays for oxidative potential of fine particulate matter. Atmos. Environ. 2020, 222, 17132. [Google Scholar] [CrossRef]
- Sánchez-Paniagua, M.; Palenzuela-Batista, S.; Manzanares-Palenzuela, C.; López-Ruiz, B. Electrochemical genosensor for Klotho detection based on aliphatic and aromatic thiols self-assembled monolayers. Talanta 2020, 212, 120735. [Google Scholar] [CrossRef]
- Sohrabi, H.; Majidi, M.R.; Nami, F.; Asadpour-Zeynali, K.; Khataee, A.; Mokhtarzadeh, A. A novel engineered label-free Zn-based MOF/CMC/AuNPs electrochemical genosensor for highly sensitive determination of Haemophilus Influenzae in human plasma samples. Microchim. Acta 2021, 188, 1–16. [Google Scholar] [CrossRef]
- Claramunt, S.; Varea, A.; López-Díaz, D.; Velázquez, M.M.; Cornet, A.; Cirera, A. The importance of interbands on the interpretation of the Raman spectrum of graphene oxide. J. Phys. Chem. C 2015, 119, 10123–10129. [Google Scholar] [CrossRef]
- Inoue, J.; Okamoto, S.; Ishihara, S.; Koshibae, W.; Kawamura, Y.; Maekawa, S. Raman scattering by orbital waves in perovskite LaMnO3. Phys. B Condens. Matter 1997, 237, 51–53. [Google Scholar] [CrossRef]
- Hunter, N.; Azam, N.; Zobeiri, H.; Van Velson, N.; Mahjouri-Samani, M.; Wang, X. Interface Thermal Resistance between Monolayer WSe2 and SiO2: Raman Probing with Consideration of Optical–Acoustic Phonon Nonequilibrium. Adv. Mater. Interfaces 2022, 9, 2102059. [Google Scholar] [CrossRef]
- Zhang, P.; Li, Z.; Wang, H.; Zhuo, Y.; Yuan, R.; Chai, Y. DNA nanomachine-based regenerated sensing platform: A novel electrochemiluminescence resonance energy transfer strategy for ultra-high sensitive detection of microRNA from cancer cells. Nanoscale 2017, 9, 2310–2316. [Google Scholar] [CrossRef]
- Hao, K.; He, Y.; Lu, H.; Pu, S.; Zhang, Y.; Dong, H.; Zhang, X. High-sensitive surface plasmon resonance microRNA biosensor based on streptavidin functionalized gold nanorods-assisted signal amplification. Anal. Chim. Acta 2017, 954, 114–120. [Google Scholar] [CrossRef]
- Lu, L.; Tu, D.; Liu, Y.; Zhou, S.; Zheng, W.; Chen, X. Ultrasensitive detection of cancer biomarker microRNA by amplification of fluorescence of lanthanide nanoprobes. Nano Res. 2018, 11, 264–273. [Google Scholar] [CrossRef]
- Zhu, Y.; Qiu, D.; Yang, G.; Wang, M.; Zhang, Q.; Wang, P.; Ming, H.; Zhang, D.; Yu, Y.; Zou, G.; et al. Selective and sensitive detection of MiRNA-21 based on gold-nanorod functionalized polydiacetylene microtube waveguide. Biosens. Bioelectron. 2016, 85, 198–204. [Google Scholar] [CrossRef]
- Hosseinzadeh, E.; Ravan, H.; Mohammadi, A.; Pourghadamyari, H. Colorimetric detection of miRNA-21 by DNAzyme-coupled branched DNA constructs. Talanta 2020, 216, 120913. [Google Scholar] [CrossRef]
- Zhang, J.; Tang, L.; Yu, Q.; Qiu, W.; Li, K.; Cheng, L.; Zhang, T.; Qian, L.; Zhang, X.; Liu, G. Gold-platinum nanoflowers as colored and catalytic labels for ultrasensitive lateral flow MicroRNA-21 assay. Sens. Actuators B Chem. 2021, 344, 130325. [Google Scholar] [CrossRef]
- Pothipor, C.; Jakmunee, J.; Bamrungsap, S.; Ounnunkad, K. An electrochemical biosensor for simultaneous detection of breast cancer clinically related microRNAs based on a gold nanoparticles/graphene quantum dots/graphene oxide film. Analyst 2021, 146, 4000–4009. [Google Scholar] [CrossRef]
- Wang, J.; Lu, Z.; Tang, H.; Wu, L.; Wang, Z.; Wu, M.; Yi, X.; Wang, J. Multiplexed electrochemical detection of MiRNAs from sera of glioma patients at different stages via the novel conjugates of conducting magnetic microbeads and Diblock oligonucleotide-modified gold nanoparticles. Anal. Chem. 2017, 89, 10834–10840. [Google Scholar] [CrossRef]
- Xu, S.; Chang, Y.; Wu, Z.; Li, Y.; Yuan, R.; Chai, Y. One DNA circle capture probe with multiple target recognition domains for simultaneous electrochemical detection of miRNA-21 and miRNA-155. Biosens. Bioelectron. 2020, 149, 111848. [Google Scholar] [CrossRef]
- Lin, X.; Jiang, J.; Wang, J.; Xia, J.; Wang, R.; Diao, G. Competitive host-guest recognition initiated by DNAzyme-cleavage cycling for novel ratiometric electrochemical assay of miRNA-21. Sens. Actuators B Chem. 2021, 333, 129556. [Google Scholar] [CrossRef]
- Yang, K.; Li, J.; de la Chapelle, M.L.; Huang, G.; Wang, Y.; Zhang, J.; Xu, D.; Yao, J.; Yang, X.; Fu, W. A terahertz metamaterial biosensor for sensitive detection of microRNAs based on gold-nanoparticles and strand displacement amplification. Biosens. Bioelectron. 2021, 175, 112874. [Google Scholar] [CrossRef]



| Oligonucleotide | Sequences |
|---|---|
| Capture probe | 5′-SH-(CH20)6-UCAACAUCAGUCUGAUAAGCUA-3′ |
| miRNA-21 | 5′-UAGCUUAUCAGACUGAUGUUGA-3′ |
| Mismatch1 | 5′-UAGCUUAUCACACUGAUGUUGA-3′ |
| Mismatch2 | 5′-UAGCUUAUAAGACUAAUGUUGA-3′ |
| Mismatch3 | 5′-UUGCUUAUCGGACUGAUCUUGA-3′ |
| miRNA-126 | 5′-UCGUACCGUGAGUAAUAAUGCG-3′ |
| Methods | Modifier Agents | Linear Range | LOD | References |
|---|---|---|---|---|
| ECL a | GCE/Au NPs/DNA tweezer/report DNA | 0.02–150 pM | 6.3 fM | [62] |
| SPR b | Au/MB/miRNA/StreGNRs | 0.1–100 pM | 45 fM | [63] |
| Fluorescence | Au nanorod functionalized polydiacetylene microtube waveguide | 50–1000 fM | 10 fM | [64] |
| EC c | miRNA-21/DNA-Au NPs@MoS2 | 0.01–1000 | 0.78 fM | [65] |
| Colorimetric | - | 10 pM–1 nM | 1 pM | [66] |
| LFNAB d | AuPt NF-based | 2–5000 pM | 0.3 pM | [67] |
| Electrochemical | AuNPs/GQDs/GO | 0.001–1000 pM | 0.04 fM | [68] |
| Electrochemical | AuNPMMBs/MAuE | 5–600 fM | 0.20 fM | [69] |
| Electrochemical | TDN/depAu/GCE | 10 µM–10 nM | 18.9 aM | [70] |
| Ratiometric electrochemistry | Mg2+-dependent DNAzyme-cleavage cycling | 10 fM–0.1 nM | 1.8 fM | [71] |
| THz e spectroscopy | THz metamaterials | 1 fM–10 pM | 14.54 aM | [72] |
| Electrochemical genosensor | Perovskite-graphene oxide | 10 fM–100 nM | 2.94 fM | The present study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahbazi-Derakhshi, P.; Mahmoudi, E.; Majidi, M.M.; Sohrabi, H.; Amini, M.; Majidi, M.R.; Niaei, A.; Shaykh-Baygloo, N.; Mokhtarzadeh, A. An Ultrasensitive miRNA-Based Genosensor for Detection of MicroRNA 21 in Gastric Cancer Cells Based on Functional Signal Amplifier and Synthesized Perovskite-Graphene Oxide and AuNPs. Biosensors 2023, 13, 172. https://doi.org/10.3390/bios13020172
Shahbazi-Derakhshi P, Mahmoudi E, Majidi MM, Sohrabi H, Amini M, Majidi MR, Niaei A, Shaykh-Baygloo N, Mokhtarzadeh A. An Ultrasensitive miRNA-Based Genosensor for Detection of MicroRNA 21 in Gastric Cancer Cells Based on Functional Signal Amplifier and Synthesized Perovskite-Graphene Oxide and AuNPs. Biosensors. 2023; 13(2):172. https://doi.org/10.3390/bios13020172
Chicago/Turabian StyleShahbazi-Derakhshi, Payam, Elham Mahmoudi, Mir Mostafa Majidi, Hessamaddin Sohrabi, Mohammad Amini, Mir Reza Majidi, Aligholi Niaei, Nima Shaykh-Baygloo, and Ahad Mokhtarzadeh. 2023. "An Ultrasensitive miRNA-Based Genosensor for Detection of MicroRNA 21 in Gastric Cancer Cells Based on Functional Signal Amplifier and Synthesized Perovskite-Graphene Oxide and AuNPs" Biosensors 13, no. 2: 172. https://doi.org/10.3390/bios13020172
APA StyleShahbazi-Derakhshi, P., Mahmoudi, E., Majidi, M. M., Sohrabi, H., Amini, M., Majidi, M. R., Niaei, A., Shaykh-Baygloo, N., & Mokhtarzadeh, A. (2023). An Ultrasensitive miRNA-Based Genosensor for Detection of MicroRNA 21 in Gastric Cancer Cells Based on Functional Signal Amplifier and Synthesized Perovskite-Graphene Oxide and AuNPs. Biosensors, 13(2), 172. https://doi.org/10.3390/bios13020172





