Bio-Impedance Spectroscopy of Retained Cells Using a Micro-Perforated Sensing Membrane Filtrating Whole Blood Samples under High Flowrate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Manufacturing Process
2.2. Design Parameters
2.3. Experimental Setup
2.4. Calibration and Control Protocol
2.5. Experimental Protocol
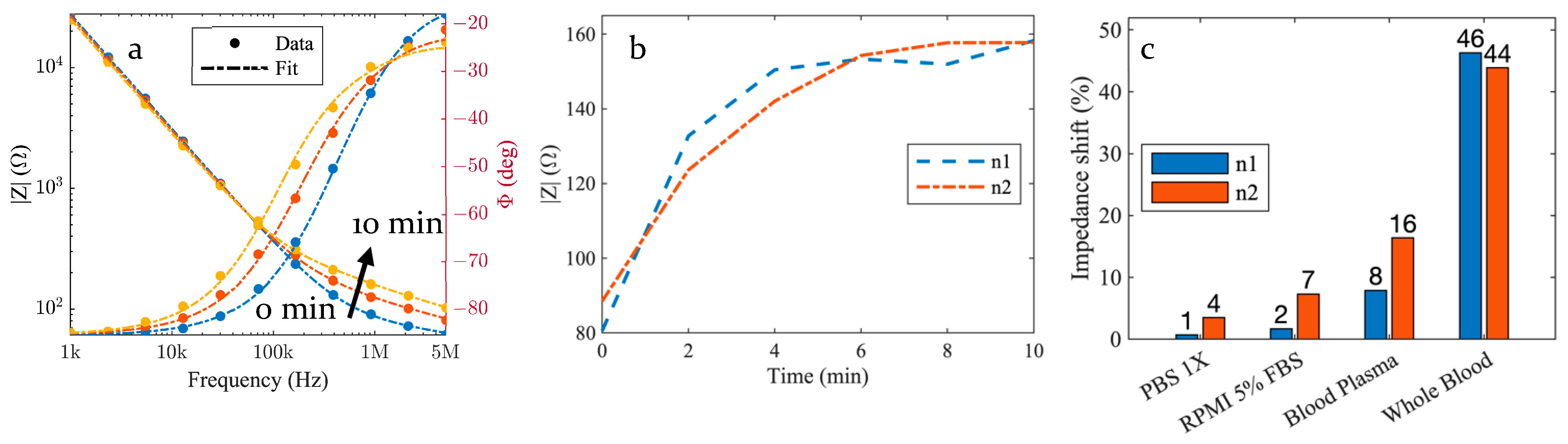
3. Results
3.1. Impedance Evolution
3.2. Control Samples
4. Discussion
4.1. Electrical Circuit Model
4.2. Interface Impedance Model
4.3. Medium Impedance Model and Dispersion
4.4. Impedance Spectra over Time
4.5. Capture and Detection Kinetics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hansson, O.; Edelmayer, R.M.; Boxer, A.L.; Carrillo, M.C.; Mielke, M.M.; Rabinovici, G.D.; Salloway, S.; Sperling, R.; Zetterberg, H.; Teunissen, C.E. The Alzheimer’s Association appropriate use recommendations for blood biomarkers in Alzheimer’s disease. Alzheimers Dement. 2022, 18, 2669–2686. [Google Scholar] [CrossRef] [PubMed]
- Shahjahan, M.; Islam, M.J.; Hossain, M.T.; Mishu, M.A.; Hasan, J.; Brown, C. Blood biomarkers as diagnostic tools: An overview of climate-driven stress responses in fish. Sci. Total Environ. 2022, 843, 156910. [Google Scholar] [CrossRef]
- Newcombe, V.F.J.; Ashton, N.J.; Posti, J.P.; Glocker, B.; Manktelow, A.; Chat, D.A.; Correia, M.M.; Williams, G.B.; Simrén, J.; Takala, R.S.K.; et al. Post-acute blood biomarkers and disease progression in traumatic brain injury. Brain 2022, 145, 2064–2076. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, G.L.; Hudson, I.L.; Hayes, K.; James, J. Prevention of transfusion-acquired cytomegalovirus infection in infants by blood filtration to remove leucocytes. Lancet 1989, 333, 1228–1231. [Google Scholar] [CrossRef] [PubMed]
- Tutino, V.M.; Fricano, S.; Chien, A.; Patel, T.R.; Monteiro, A.; Rai, H.H.; Dmytriw, A.A.; Chaves, L.D.; Waqas, M.; Levy, E.I.; et al. Gene expression profiles of ischemic stroke clots retrieved by mechanical thrombectomy are associated with disease etiology. J. NeuroInterv. Surg. 2023, 15, e33–e40. [Google Scholar] [CrossRef]
- Ronco, C.; Clark, W.R. Haemodialysis membranes. Nat. Rev. Nephrol. 2018, 14, 394–410. [Google Scholar] [CrossRef] [PubMed]
- Martín-Núñez, E.; Pérez-Castro, A.; Tagua, V.G.; Hernández-Carballo, C.; Ferri, C.; Pérez-Delgado, N.; Rodríguez-Ramos, S.; Cerro-López, P.; López-Castillo, Á.; Delgado-Molinos, A.; et al. Klotho expression in peripheral blood circulating cells is associated with vascular and systemic inflammation in atherosclerotic vascular disease. Sci. Rep. 2022, 12, 8422. [Google Scholar] [CrossRef]
- Liu, C.; Xiang, X.; Han, S.; Lim, H.Y.; Li, L.; Zhang, X.; Ma, Z.; Yang, L.; Guo, S.; Soo, R.; et al. Blood-based liquid biopsy: Insights into early detection and clinical management of lung cancer. Cancer Lett. 2022, 524, 91–102. [Google Scholar] [CrossRef]
- Ilie, M.; Hofman, V.; Long-Mira, E.; Selva, E.; Vignaud, J.-M.; Padovani, B.; Mouroux, J.; Marquette, C.-H.; Hofman, P. “Sentinel” Circulating Tumor Cells Allow Early Diagnosis of Lung Cancer in Patients with Chronic Obstructive Pulmonary Disease. PLoS ONE 2014, 9, e111597. [Google Scholar] [CrossRef] [PubMed]
- Yeo, D.; Bastian, A.; Strauss, H.; Saxena, P.; Grimison, P.; Rasko, J.E.J. Exploring the Clinical Utility of Pancreatic Cancer Circulating Tumor Cells. Int. J. Mol. Sci. 2022, 23, 1671. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Liquid biopsy and minimal residual disease—Latest advances and implications for cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Kiniwa, Y.; Nakamura, K.; Mikoshiba, A.; Ashida, A.; Akiyama, Y.; Morimoto, A.; Okuyama, R. Usefulness of monitoring circulating tumor cells as a therapeutic biomarker in melanoma with BRAF mutation. BMC Cancer 2021, 21, 287. [Google Scholar] [CrossRef] [PubMed]
- Jarvas, G.; Szerenyi, D.; Tovari, J.; Takacs, L.; Guttman, A. Modification of Hemodialysis Membranes for Efficient Circulating Tumor Cell Capture for Cancer Therapy. Molecules 2021, 26, 4845. [Google Scholar] [CrossRef] [PubMed]
- Tamminga, M.; Andree, K.C.; van den Bos, H.; Hiltermann, T.J.N.; Mentink, A.; Spierings, D.C.J.; Lansdorp, P.; Timens, W.; Schuuring, E.; Terstappen, L.W.M.M.; et al. Leukapheresis increases circulating tumour cell yield in non-small cell lung cancer, counts related to tumour response and survival. Br. J. Cancer 2022, 126, 409–418. [Google Scholar] [CrossRef]
- Hiller, J.G.; Perry, N.J.; Poulogiannis, G.; Riedel, B.; Sloan, E.K. Perioperative events influence cancer recurrence risk after surgery. Nat. Rev. Clin. Oncol. 2018, 15, 205–218. [Google Scholar] [CrossRef]
- Chemi, F.; Rothwell, D.G.; McGranahan, N.; Gulati, S.; Abbosh, C.; Pearce, S.P.; Zhou, C.; Wilson, G.A.; Jamal-Hanjani, M.; Birkbak, N.; et al. Pulmonary venous circulating tumor cell dissemination before tumor resection and disease relapse. Nat. Med. 2019, 25, 1534–1539. [Google Scholar] [CrossRef]
- Rand, P.W.; Austin, W.H.; Lacombe, E.; Barker, N. pH and blood viscosity. J. Appl. Physiol. 1968, 25, 550–559. [Google Scholar] [CrossRef]
- Pedro, B.G.; Marcôndes, D.W.C.; Bertemes-Filho, P. Analytical Model for Blood Glucose Detection Using Electrical Impedance Spectroscopy. Sensors 2020, 20, 6928. [Google Scholar] [CrossRef]
- Addabbo, T.; Fort, A.; Kapita, P.; Mugnaini, M.; Vignoli, V.; Barygina, V.; Becatti, M.; Fiorillo, C.; Taddei, N. A Compact System for Blood Impedance Measurements for ROS Evaluation. In Proceedings of the 2018 IEEE International Symposium on Medical Measurements and Applications (MeMeA), Rome, Italy, 11–13 June 2018; pp. 1–5. [Google Scholar] [CrossRef]
- Bavil, A.K.; Sticker, D.; Rothbauer, M.; Ertl, P.; Kim, J. A microparticle-labeled microfluidic impedance immunosensor array for enhancing sensitivity. Analyst 2021, 146, 3289–3298. [Google Scholar] [CrossRef]
- Pajkossy, T.; Jurczakowski, R. Electrochemical impedance spectroscopy in interfacial studies. Curr. Opin. Electrochem. 2017, 1, 53–58. [Google Scholar] [CrossRef]
- Wu, J. Understanding the Electric Double-Layer Structure, Capacitance, and Charging Dynamics. Chem. Rev. 2022, 122, 10821–10859. [Google Scholar] [CrossRef]
- Cole, K.S. Permeability and Imperbeability of Cell Membranes for Ions. Cold Spring Harb. Symp. Quant. Biol. 1940, 8, 110–122. [Google Scholar] [CrossRef]
- Gabriel, S.; Lau, R.W.; Gabriel, C. The dielectric properties of biological tissues: III. Parametric models for the dielectric spectrum of tissues. Phys. Med. Biol. 1996, 41, 2271–2293. [Google Scholar] [CrossRef]
- Fricke, H.; Morse, S. The Electric Resistance and Capacity of Bood for Frequencies Between 800 and 4½ Million Cycles. J. Gen. Physiol. 1925, 9, 153–167. [Google Scholar] [CrossRef]
- Foster, K.R.; Schwan, H.P. Dielectric properties of tissues and biological materials: A critical review. Crit. Rev. Biomed. Eng. 1989, 17, 25–104. [Google Scholar] [PubMed]
- Olthuis, W.; Streekstra, W.; Bergveld, P. Theoretical and experimental determination of cell constants of planar-interdigitated electrolyte conductivity sensors. Sens. Actuators B Chem. 1995, 24, 252–256. [Google Scholar] [CrossRef]
- Debye, P.J.W. Polar Molecules. In Dover Books Explaining Science and Mathematics; Dover: New York, NY, USA, 1960. [Google Scholar]
- Cole, K.S.; Cole, R.H. Dispersion and Absorption in Dielectrics I. Alternating Current Characteristics. J. Chem. Phys. 1941, 9, 341–351. [Google Scholar] [CrossRef]
- Hurt, W.D. Multiterm Debye Dispersion Relations for Permittivity of Muscle. IEEE Trans. Biomed. Eng. 1985, BME-32, 60–64. [Google Scholar] [CrossRef]
- Smith, K.J.; Jana, J.A.; Kaehr, A.; Purcell, E.; Opdycke, T.; Paoletti, C.; Cooling, L.; Thamm, D.H.; Hayes, D.F.; Nagrath, S. Inertial focusing of circulating tumor cells in whole blood at high flow rates using the microfluidic CTCKeyTM device for CTC enrichment. Lab. Chip 2021, 21, 3559–3572. [Google Scholar] [CrossRef]
- Lux, A.; Bott, H.; Malek, N.P.; Zengerle, R.; Maucher, T.; Hoffmann, J. Real-Time Detection of Tumor Cells during Capture on a Filter Element Significantly Enhancing Detection Rate. Biosensors 2021, 11, 312. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sagot, M.; Bou, E.; Bourrier, D.; Cerf, A.; Aubert, H.; Vieu, C. Bio-Impedance Spectroscopy of Retained Cells Using a Micro-Perforated Sensing Membrane Filtrating Whole Blood Samples under High Flowrate. Biosensors 2023, 13, 996. https://doi.org/10.3390/bios13120996
Sagot M, Bou E, Bourrier D, Cerf A, Aubert H, Vieu C. Bio-Impedance Spectroscopy of Retained Cells Using a Micro-Perforated Sensing Membrane Filtrating Whole Blood Samples under High Flowrate. Biosensors. 2023; 13(12):996. https://doi.org/10.3390/bios13120996
Chicago/Turabian StyleSagot, Matthieu, Elise Bou, David Bourrier, Aline Cerf, Hervé Aubert, and Christophe Vieu. 2023. "Bio-Impedance Spectroscopy of Retained Cells Using a Micro-Perforated Sensing Membrane Filtrating Whole Blood Samples under High Flowrate" Biosensors 13, no. 12: 996. https://doi.org/10.3390/bios13120996
APA StyleSagot, M., Bou, E., Bourrier, D., Cerf, A., Aubert, H., & Vieu, C. (2023). Bio-Impedance Spectroscopy of Retained Cells Using a Micro-Perforated Sensing Membrane Filtrating Whole Blood Samples under High Flowrate. Biosensors, 13(12), 996. https://doi.org/10.3390/bios13120996





