Quantitative Imaging of Genetically Encoded Fluorescence Lifetime Biosensors
Abstract
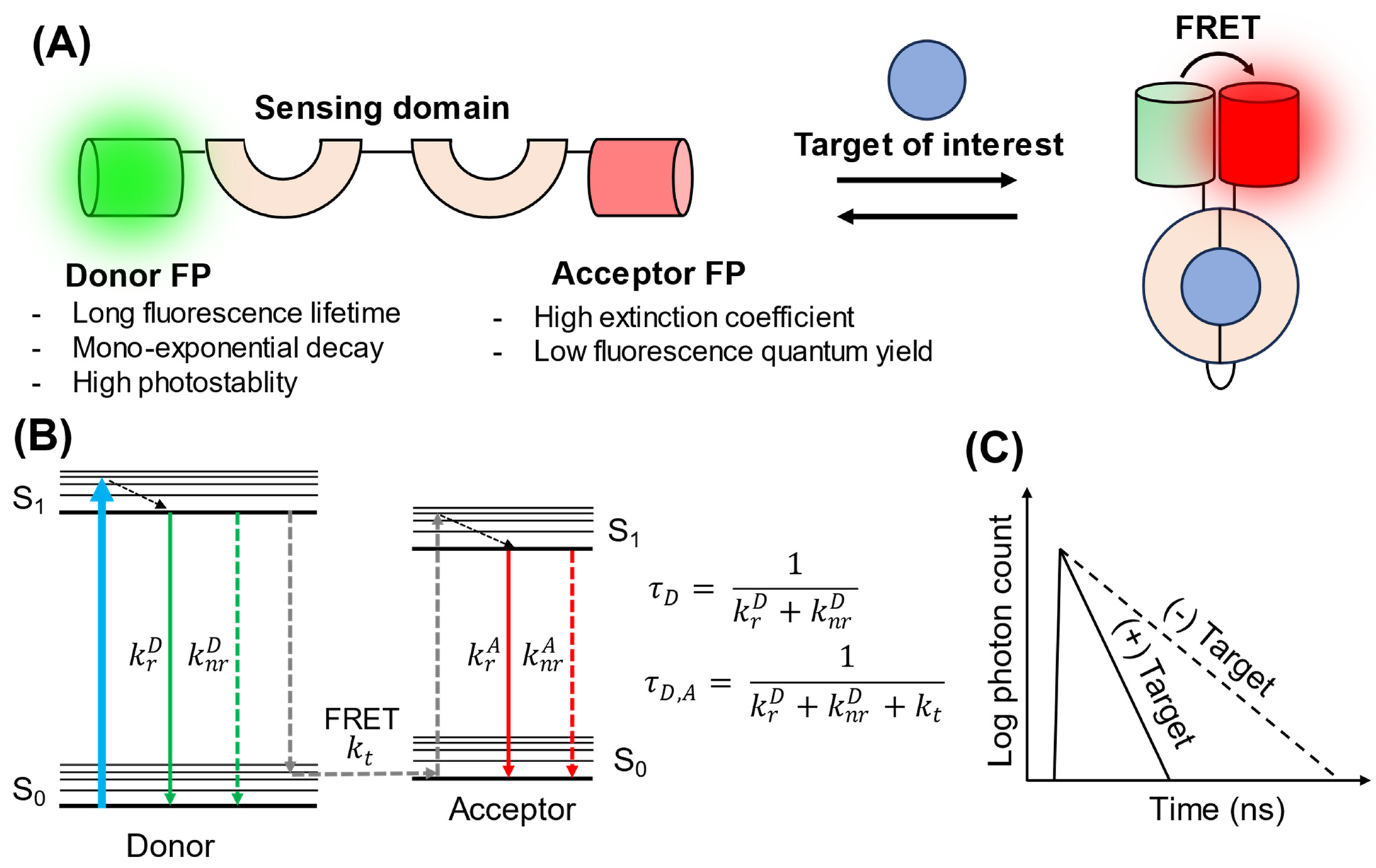
:1. Introduction
2. FRET–FLIM Biosensors
| Targets | Names | FRET Pairs | 2p-FLIM | 𝜏free (ns) | 𝜏bind (ns) | Δ𝜏(bind-free) (ns) | Ref. |
|---|---|---|---|---|---|---|---|
| Protein kinase A (PKA) | FLIM–AKAR | EGFP–cpsREACh | Yes | 1.85 | 1.70 | −0.15 | [29] |
| AKARet | sREAChet–EGFP | Yes | - | - | −0.21 | [30] | |
| AKAR5 | mEGFP–sREACh | Yes | - | - | 0.20 | [31] | |
| tAKARα | EGFP–cpsREACh | Yes | - | - | −0.26 | [32] | |
| Protein kinase B (PKB)/Akt | GFP–Akt–YFP | GFP–YFP | 2.30 | 1.70 | −0.60 | [33] | |
| Protein kinase C (PKC) | ITRACKα | mEGFP–mCherry | Yes | - | - | −0.22 | [19] |
| ITRACKβ | mEGFP–mCherry | Yes | - | - | −0.18 | [19] | |
| ITRACKγ | mEGFP–mCherry | Yes | - | - | −0.16 | [19] | |
| IDOCKSα | mEGFP–mCherry | Yes | - | - | −0.31 | [19] | |
| IDOCKSβ | mEGFP–mCherry | Yes | - | - | −0.23 | [19] | |
| IDOCKSγ | mEGFP–mCherry | Yes | - | - | −0.22 | [19] | |
| Aurora Kinase A | ShadowG–AURKA–mTQ2 | ShadowG–mTQ2 | - | - | 0.15 | [34] | |
| ShadowY–AURKA–mTQ2 | ShadowY–mTQ2 | - | - | 0.15 | [34] | ||
| Calcium/calmodulin-dependent kinase II (CaMKII) | Green–Camuiα | mEGFP–REACh | Yes | 1.67 | 2.08 | 0.41 | [20] |
| mRFP/GFP–Camui | mRFP–GFP | 1.82 | 2.13 | 0.31 | [35] | ||
| Camuiα–mRmC | mRuby2–mCherry_I202Y/T | - | - | 0.10 | [36] | ||
| Extracellular signal-regulated kinase (ERK) | EKARet | sREAChet–EGFP | Yes | - | - | −0.23 | [30] |
| cAMP | TEpacVV | mTQ–cp173Venus-Venus | 2.28 | 3.03 | 0.75 | [22] | |
| CEpacVV | mECFP–cp173Venus-Venus | 1.64 | 2.02 | 0.38 | [22] | ||
| EpacSH189 | mTQ2–tdDark-cp173Venus | 1.93 | 3.41 | 1.48 | [17] | ||
| Ca2+ | TN-L15 | CFP–Citrine | 2.36 | 1.9 | −0.46 | [37] | |
| mTFP–TnC-Cit | mTFP1–Citrine | 2.51 | 2.18 | −0.33 | [37] | ||
| NAD+ | ChemoD–NAD | ShadowG–HaloTag7 | 2.21 | 3.37 | 1.16 | [38] | |
| pH | pH–Lemon | mTQ2–EYFP | Yes | 3.69 (pH 4.03) | 2.48 (pH 7.01) | −1.21 | [39] |
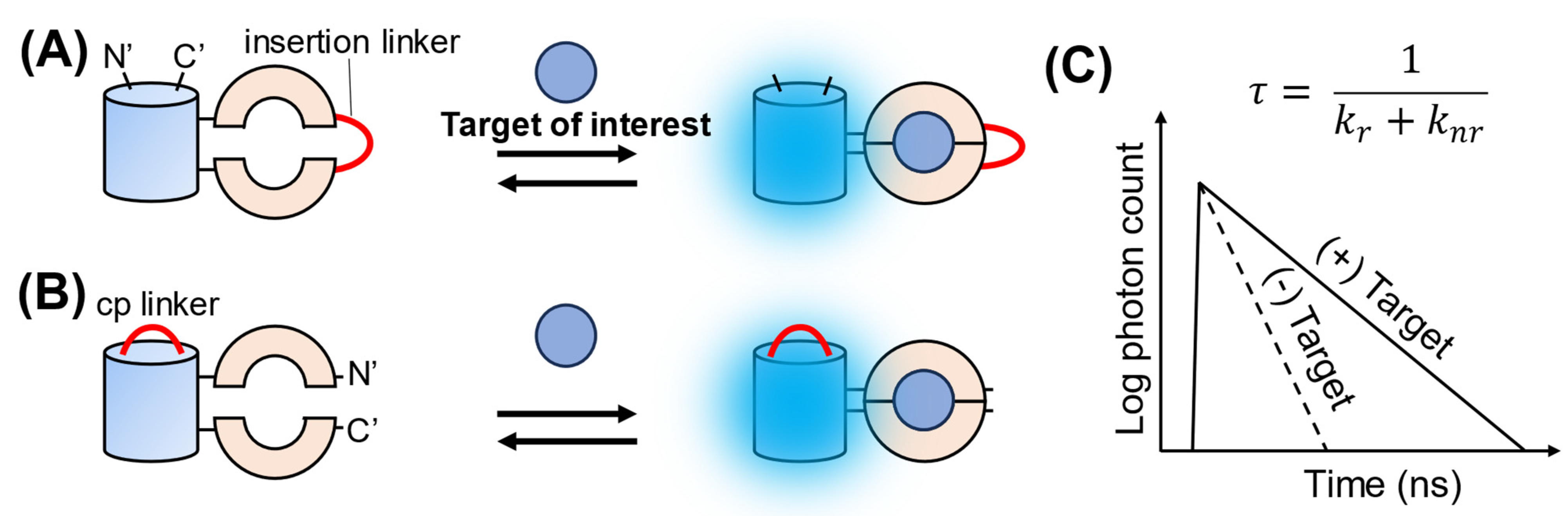
3. Single-FP-Based FLIM Biosensors
4. Challenges and Limitations
5. Future Perspectives and Conclusion
5.1. Strengthening in the Design and Screening Methods
5.2. Potential Applications and Impact on Biological Research
5.3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Chalfie, M.; Tu, Y.; Euskirchen, G.; Ward, W.W.; Prasher, D.C. Green Fluorescent Protein as a Marker for Gene Expression. Science 1994, 263, 802–805. [Google Scholar] [CrossRef]
- Miyawaki, A.; Llopis, J.; Heim, R.; McCaffery, J.M.; Adams, J.A.; Ikura, M.; Tsien, R.Y. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 1997, 388, 882–887. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Araki, S.; Wu, J.; Teramoto, T.; Chang, Y.F.; Nakano, M.; Abdelfattah, A.S.; Fujiwara, M.; Ishihara, T.; Nagai, T.; et al. An expanded palette of genetically encoded Ca2+ indicators. Science 2011, 333, 1888–1891. [Google Scholar] [CrossRef]
- Arai, S.; Kriszt, R.; Harada, K.; Looi, L.-S.; Matsuda, S.; Wongso, D.; Suo, S.; Ishiura, S.; Tseng, Y.-H.; Raghunath, M.; et al. RGB-Color Intensiometric Indicators to Visualize Spatiotemporal Dynamics of ATP in Single Cells. Angew. Chem. Int. Ed. 2018, 57, 10873–10878. [Google Scholar] [CrossRef] [PubMed]
- Jing, M.; Li, Y.; Zeng, J.; Huang, P.; Skirzewski, M.; Kljakic, O.; Peng, W.; Qian, T.; Tan, K.; Zou, J.; et al. An optimized acetylcholine sensor for monitoring in vivo cholinergic activity. Nat. Methods 2020, 17, 1139–1146. [Google Scholar] [CrossRef]
- Evans, S.W.; Shi, D.-Q.; Chavarha, M.; Plitt, M.H.; Taxidis, J.; Madruga, B.; Fan, J.L.; Hwang, F.-J.; van Keulen, S.C.; Suomivuori, C.-M.; et al. A positively tuned voltage indicator for extended electrical recordings in the brain. Nat. Methods 2023, 20, 1104–1113. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Wazawa, T.; Sakamoto, J.; Vu, C.Q.; Nakano, M.; Kamei, Y.; Nagai, T. Intracellular Heat Transfer and Thermal Property Revealed by Kilohertz Temperature Imaging with a Genetically Encoded Nanothermometer. Nano Lett. 2022, 22, 5698–5707. [Google Scholar] [CrossRef]
- Boersma, A.J.; Zuhorn, I.S.; Poolman, B. A sensor for quantification of macromolecular crowding in living cells. Nat. Methods 2015, 12, 227–229. [Google Scholar] [CrossRef]
- Mo, G.C.H.; Posner, C.; Rodriguez, E.A.; Sun, T.; Zhang, J. A rationally enhanced red fluorescent protein expands the utility of FRET biosensors. Nat. Commun. 2020, 11, 1848. [Google Scholar] [CrossRef]
- Hashizume, R.; Fujii, H.; Mehta, S.; Ota, K.; Qian, Y.; Zhu, W.; Drobizhev, M.; Nasu, Y.; Zhang, J.; Bito, H.; et al. A genetically encoded far-red fluorescent calcium ion biosensor derived from a biliverdin-binding protein. Protein Sci. 2022, 31, e4440. [Google Scholar] [CrossRef]
- Prasher, D.C.; Eckenrode, V.K.; Ward, W.W.; Prendergast, F.G.; Cormier, M.J. Primary structure of the Aequorea victoria green-fluorescent protein. Gene 1992, 111, 229–233. [Google Scholar] [CrossRef]
- Kogure, T.; Karasawa, S.; Araki, T.; Saito, K.; Kinjo, M.; Miyawaki, A. A fluorescent variant of a protein from the stony coral Montipora facilitates dual-color single-laser fluorescence cross-correlation spectroscopy. Nat. Biotechnol. 2006, 24, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Tantama, M.; Hung, Y.P.; Yellen, G. Imaging Intracellular pH in Live Cells with a Genetically Encoded Red Fluorescent Protein Sensor. J. Am. Chem. Soc. 2011, 133, 10034–10037. [Google Scholar] [CrossRef] [PubMed]
- Berezin, M.Y.; Achilefu, S. Fluorescence Lifetime Measurements and Biological Imaging. Chem. Rev. 2010, 110, 2641–2684. [Google Scholar] [CrossRef] [PubMed]
- Rupsa, D.; Tiffany, M.H.; Joe, T.S.; Amani, A.G.; Melissa, C.S. Fluorescence lifetime imaging microscopy: Fundamentals and advances in instrumentation, analysis, and applications. J. Biomed. Opt. 2020, 25, 071203. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: Boston, MA, USA, 2006; p. XXVI, 954. [Google Scholar]
- Klarenbeek, J.; Goedhart, J.; van Batenburg, A.; Groenewald, D.; Jalink, K. Fourth-Generation Epac-Based FRET Sensors for cAMP Feature Exceptional Brightness, Photostability and Dynamic Range: Characterization of Dedicated Sensors for FLIM, for Ratiometry and with High Affinity. PLoS ONE 2015, 10, e0122513. [Google Scholar] [CrossRef]
- Harkes, R.; Kukk, O.; Mukherjee, S.; Klarenbeek, J.; van den Broek, B.; Jalink, K. Dynamic FRET-FLIM based screening of signal transduction pathways. Sci. Rep. 2021, 11, 20711. [Google Scholar] [CrossRef]
- Colgan, L.A.; Hu, M.; Misler, J.A.; Parra-Bueno, P.; Moran, C.M.; Leitges, M.; Yasuda, R. PKCα integrates spatiotemporally distinct Ca2+ and autocrine BDNF signaling to facilitate synaptic plasticity. Nat. Neurosci. 2018, 21, 1027–1037. [Google Scholar] [CrossRef]
- Lee, S.-J.R.; Escobedo-Lozoya, Y.; Szatmari, E.M.; Yasuda, R. Activation of CaMKII in single dendritic spines during long-term potentiation. Nature 2009, 458, 299–304. [Google Scholar] [CrossRef]
- Zhong, W.; Wu, M.; Chang, C.-W.; Merrick, K.A.; Merajver, S.D.; Mycek, M.-A. Picosecond-resolution fluorescence lifetime imaging microscopy: A useful tool for sensing molecular interactions in vivo via FRET. Opt. Express 2007, 15, 18220–18235. [Google Scholar] [CrossRef]
- Klarenbeek, J.B.; Goedhart, J.; Hink, M.A.; Gadella, T.W.J.; Jalink, K. A mTurquoise-Based cAMP Sensor for Both FLIM and Ratiometric Read-Out Has Improved Dynamic Range. PLoS ONE 2011, 6, e19170. [Google Scholar] [CrossRef]
- Sarkisyan, K.S.; Goryashchenko, A.S.; Lidsky, P.V.; Gorbachev, D.A.; Bozhanova, N.G.; Gorokhovatsky, A.Y.; Pereverzeva, A.R.; Ryumina, A.P.; Zherdeva, V.V.; Savitsky, A.P.; et al. Green Fluorescent Protein with Anionic Tryptophan-Based Chromophore and Long Fluorescence Lifetime. Biophys. J. 2015, 109, 380–389. [Google Scholar] [CrossRef]
- Ganesan, S.; Ameer-beg, S.M.; Ng, T.T.C.; Vojnovic, B.; Wouters, F.S. A dark yellow fluorescent protein (YFP)-based Resonance Energy-Accepting Chromoprotein (REACh) for Förster resonance energy transfer with GFP. Proc. Natl. Acad. Sci. USA 2006, 103, 4089–4094. [Google Scholar] [CrossRef]
- Murakoshi, H.; Shibata, A.C.E. ShadowY: A dark yellow fluorescent protein for FLIM-based FRET measurement. Sci. Rep. 2017, 7, 6791. [Google Scholar] [CrossRef]
- Pettikiriarachchi, A.; Gong, L.; Perugini, M.A.; Devenish, R.J.; Prescott, M. Ultramarine, a Chromoprotein Acceptor for Förster Resonance Energy Transfer. PLoS ONE 2012, 7, e41028. [Google Scholar] [CrossRef]
- van der Linden, F.H.; Mahlandt, E.K.; Arts, J.J.G.; Beumer, J.; Puschhof, J.; de Man, S.M.A.; Chertkova, A.O.; Ponsioen, B.; Clevers, H.; van Buul, J.D.; et al. A turquoise fluorescence lifetime-based biosensor for quantitative imaging of intracellular calcium. Nat. Commun. 2021, 12, 7159. [Google Scholar] [CrossRef] [PubMed]
- Shinoda, H.; Ma, Y.; Nakashima, R.; Sakurai, K.; Matsuda, T.; Nagai, T. Acid-Tolerant Monomeric GFP from Olindias formosa. Cell Chem. Biol. 2018, 25, 330–338.e337. [Google Scholar] [CrossRef]
- Chen, Y.; Saulnier, J.; Yellen, G.; Sabatini, B. A PKA activity sensor for quantitative analysis of endogenous GPCR signaling via 2-photon FRET-FLIM imaging. Front. Pharmacol. 2014, 5, 56. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Yasuda, R. Imaging ERK and PKA Activation in Single Dendritic Spines during Structural Plasticity. Neuron 2017, 93, 1315–1324.e1313. [Google Scholar] [CrossRef] [PubMed]
- Tillo, S.E.; Xiong, W.-H.; Takahashi, M.; Miao, S.; Andrade, A.L.; Fortin, D.A.; Yang, G.; Qin, M.; Smoody, B.F.; Stork, P.J.S.; et al. Liberated PKA Catalytic Subunits Associate with the Membrane via Myristoylation to Preferentially Phosphorylate Membrane Substrates. Cell Rep. 2017, 19, 617–629. [Google Scholar] [CrossRef]
- Ma, L.; Jongbloets, B.C.; Xiong, W.-H.; Melander, J.B.; Qin, M.; Lameyer, T.J.; Harrison, M.F.; Zemelman, B.V.; Mao, T.; Zhong, H. A Highly Sensitive A-Kinase Activity Reporter for Imaging Neuromodulatory Events in Awake Mice. Neuron 2018, 99, 665–679.e665. [Google Scholar] [CrossRef]
- Calleja, V.; Ameer-Beg, S.M.; Vojnovic, B.; Woscholski, R.; Downward, J.; Larijani, B. Monitoring conformational changes of proteins in cells by fluorescence lifetime imaging microscopy. Biochem. J. 2003, 372, 33–40. [Google Scholar] [CrossRef]
- Bertolin, G.; Sizaire, F.; Déméautis, C.; Chapuis, C.; Mérola, F.; Erard, M.; Tramier, M. Optimized FRET Pairs and Quantification Approaches To Detect the Activation of Aurora Kinase A at Mitosis. ACS Sens. 2019, 4, 2018–2027. [Google Scholar] [CrossRef]
- Kwok, S.; Lee, C.; Sánchez, S.A.; Hazlett, T.L.; Gratton, E.; Hayashi, Y. Genetically encoded probe for fluorescence lifetime imaging of CaMKII activity. Biochem. Biophys. Res. Commun. 2008, 369, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Nakahata, Y.; Nabekura, J.; Murakoshi, H. Dual observation of the ATP-evoked small GTPase activation and Ca2+ transient in astrocytes using a dark red fluorescent protein. Sci. Rep. 2016, 6, 39564. [Google Scholar] [CrossRef] [PubMed]
- Laine, R.; Stuckey, D.W.; Manning, H.; Warren, S.C.; Kennedy, G.; Carling, D.; Dunsby, C.; Sardini, A.; French, P.M.W. Fluorescence Lifetime Readouts of Troponin-C-Based Calcium FRET Sensors: A Quantitative Comparison of CFP and mTFP1 as Donor Fluorophores. PLoS ONE 2012, 7, e49200. [Google Scholar] [CrossRef] [PubMed]
- Hellweg, L.; Edenhofer, A.; Barck, L.; Huppertz, M.-C.; Frei, M.S.; Tarnawski, M.; Bergner, A.; Koch, B.; Johnsson, K.; Hiblot, J. A general method for the development of multicolor biosensors with large dynamic ranges. Nat. Chem. Biol. 2023, 19, 1147–1157. [Google Scholar] [CrossRef]
- Burgstaller, S.; Bischof, H.; Gensch, T.; Stryeck, S.; Gottschalk, B.; Ramadani-Muja, J.; Eroglu, E.; Rost, R.; Balfanz, S.; Baumann, A.; et al. pH-Lemon, a Fluorescent Protein-Based pH Reporter for Acidic Compartments. ACS Sens. 2019, 4, 883–891. [Google Scholar] [CrossRef]
- Arai, S.; Itoh, H.; Vu, C.Q.; Nakayama, M.; Oshima, M.; Morita, A.; Okamoto, K.; Okuda, S.; Teranishi, A.; Osawa, M.; et al. qMaLioffG: A single green fluorescent protein FLIM indicator enabling quantitative imaging of endogenous ATP. bioRxiv 2023. [Google Scholar] [CrossRef]
- Shimolina, L.; Potekhina, E.; Druzhkova, I.; Lukina, M.; Dudenkova, V.; Belousov, V.; Shcheslavskiy, V.; Zagaynova, E.; Shirmanova, M. Fluorescence lifetime-based pH mapping of tumors in vivo using genetically encoded sensor SypHerRed. Biophys. J. 2022, 121, 1156–1165. [Google Scholar] [CrossRef]
- Mongeon, R.; Venkatachalam, V.; Yellen, G. Cytosolic NADH-NAD+ Redox Visualized in Brain Slices by Two-Photon Fluorescence Lifetime Biosensor Imaging. Antioxid. Redox Signal. 2016, 25, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Díaz-García, C.M.; Mongeon, R.; Lahmann, C.; Koveal, D.; Zucker, H.; Yellen, G. Neuronal Stimulation Triggers Neuronal Glycolysis and Not Lactate Uptake. Cell Metab. 2017, 26, 361–374.e364. [Google Scholar] [CrossRef] [PubMed]
- Díaz-García, C.M.; Lahmann, C.; Martínez-François, J.R.; Li, B.; Koveal, D.; Nathwani, N.; Rahman, M.; Keller, J.P.; Marvin, J.S.; Looger, L.L.; et al. Quantitative in vivo imaging of neuronal glucose concentrations with a genetically encoded fluorescence lifetime sensor. J. Neurosci. Res. 2019, 97, 946–960. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Chen, P.; Tilden, E.; Aggarwal, S.; Oldenborg, A.; Chen, Y. Fluorescence lifetime enables high-resolution analysis of neuromodulator dynamics across time and animals. bioRxiv 2023. [Google Scholar] [CrossRef]
- Koveal, D.; Rosen, P.C.; Meyer, D.J.; Díaz-García, C.M.; Wang, Y.; Cai, L.-H.; Chou, P.J.; Weitz, D.A.; Yellen, G. A high-throughput multiparameter screen for accelerated development and optimization of soluble genetically encoded fluorescent biosensors. Nat. Commun. 2022, 13, 2919. [Google Scholar] [CrossRef]
- Li, L.; Cheng, Y.; Shen, S.; Zhou, J.; Wang, A.; Chen, G.; Xu, J.; Yang, Y.; Zhao, Y.; Zhang, S.; et al. Sensitive detection via the time-resolved fluorescence of circularly permuted yellow fluorescent protein biosensors. Sens. Actuators B Chem. 2020, 321, 128614. [Google Scholar] [CrossRef]
- Zhuo, Y.; Solntsev, K.M.; Reddish, F.; Tang, S.; Yang, J.J. Effect of Ca2+ on the Steady-State and Time-Resolved Emission Properties of the Genetically Encoded Fluorescent Sensor CatchER. J. Phys. Chem. B 2015, 119, 2103–2111. [Google Scholar] [CrossRef]
- Goryashchenko, A.S.; Pakhomov, A.A.; Ryabova, A.V.; Romanishkin, I.D.; Maksimov, E.G.; Orsa, A.N.; Serova, O.V.; Mozhaev, A.A.; Maksimova, M.A.; Martynov, V.I.; et al. FLIM-Based Intracellular and Extracellular pH Measurements Using Genetically Encoded pH Sensor. Biosensors 2021, 11, 340. [Google Scholar] [CrossRef]
- Bilan, D.S.; Pase, L.; Joosen, L.; Gorokhovatsky, A.Y.; Ermakova, Y.G.; Gadella, T.W.J.; Grabher, C.; Schultz, C.; Lukyanov, S.; Belousov, V.V. HyPer-3: A Genetically Encoded H2O2 Probe with Improved Performance for Ratiometric and Fluorescence Lifetime Imaging. ACS Chem. Biol. 2013, 8, 535–542. [Google Scholar] [CrossRef]
- Nasu, Y.; Shen, Y.; Kramer, L.; Campbell, R.E. Structure- and mechanism-guided design of single fluorescent protein-based biosensors. Nat. Chem. Biol. 2021, 17, 509–518. [Google Scholar] [CrossRef]
- Alvarez, L.; Widzgowski, B.; Ossato, G.; van den Broek, B.; Jalink, K.; Kuschel, L.; Roberti, M.J.; Hecht, F. SP8 FALCON: A novel concept in fluorescence lifetime imaging enabling video-rate confocal FLIM. Nat. Methods 2019, 20, 2–4. [Google Scholar]
- Carolyn, J.; Klaus, S. Refractive index sensing using Fluorescence Lifetime Imaging (FLIM). J. Phys. Conf. Ser. 2006, 45, 223–230. [Google Scholar] [CrossRef]
- Vu, C.Q.; Fukushima, S.-i.; Wazawa, T.; Nagai, T. A highly-sensitive genetically encoded temperature indicator exploiting a temperature-responsive elastin-like polypeptide. Sci. Rep. 2021, 11, 16519. [Google Scholar] [CrossRef] [PubMed]
- Sharick, J.T.; Walsh, C.M.; Sprackling, C.M.; Pasch, C.A.; Pham, D.L.; Esbona, K.; Choudhary, A.; Garcia-Valera, R.; Burkard, M.E.; McGregor, S.M.; et al. Metabolic Heterogeneity in Patient Tumor-Derived Organoids by Primary Site and Drug Treatment. Front. Oncol. 2020, 10, 553. [Google Scholar] [CrossRef]
- Vogel, S.S.; Thaler, C.; Blank, P.S.; Koushik, S.V. Chapter 10: Time-Resloved Fluorescence Anisotropy. In Flim Microscopy in Biology and Medicine; Periasamy, A., Clegg, R.M., Eds.; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Kreitz, J.; Friedrich, M.J.; Guru, A.; Lash, B.; Saito, M.; Macrae, R.K.; Zhang, F. Programmable protein delivery with a bacterial contractile injection system. Nature 2023, 616, 357–364. [Google Scholar] [CrossRef]
- Joron, K.; Viegas, J.O.; Haas-Neill, L.; Bier, S.; Drori, P.; Dvir, S.; Lim, P.S.L.; Rauscher, S.; Meshorer, E.; Lerner, E. Fluorescent protein lifetimes report densities and phases of nuclear condensates during embryonic stem-cell differentiation. Nat. Commun. 2023, 14, 4885. [Google Scholar] [CrossRef]
- Hoepker, A.C.; Wang, A.; Le Marois, A.; Suhling, K.; Yan, Y.; Marriott, G. Genetically encoded sensors of protein hydrodynamics and molecular proximity. Proc. Natl. Acad. Sci. USA 2015, 112, E2569–E2574. [Google Scholar] [CrossRef]
- Grimm, J.B.; Lavis, L.D. Caveat fluorophore: An insiders’ guide to small-molecule fluorescent labels. Nat. Methods 2022, 19, 149–158. [Google Scholar] [CrossRef]
- Deo, C.; Abdelfattah, A.S.; Bhargava, H.K.; Berro, A.J.; Falco, N.; Farrants, H.; Moeyaert, B.; Chupanova, M.; Lavis, L.D.; Schreiter, E.R. The HaloTag as a general scaffold for far-red tunable chemigenetic indicators. Nat. Chem. Biol. 2021, 17, 718–723. [Google Scholar] [CrossRef]
- Zhang, D.; Chen, Z.; Du, Z.; Bao, B.; Su, N.; Chen, X.; Ge, Y.; Lin, Q.; Yang, L.; Hua, Y.; et al. Design of a palette of SNAP-tag mimics of fluorescent proteins and their use as cell reporters. Cell Discov. 2023, 9, 56. [Google Scholar] [CrossRef]
- Farrants, H.; Shuai, Y.; Lemon, W.C.; Hernandez, C.M.; Yang, S.; Patel, R.; Qiao, G.; Frei, M.S.; Grimm, J.B.; Hanson, T.L.; et al. A modular chemigenetic calcium indicator enables in vivo functional imaging with near-infrared light. bioRxiv 2023. [Google Scholar] [CrossRef]
- Schaaf, T.M.; Li, A.; Grant, B.D.; Peterson, K.; Yuen, S.; Bawaskar, P.; Kleinboehl, E.; Li, J.; Thomas, D.D.; Gillispie, G.D. Red-Shifted FRET Biosensors for High-Throughput Fluorescence Lifetime Screening. Biosensors 2018, 8, 99. [Google Scholar] [CrossRef]
- Schaaf, T.M.; Peterson, K.C.; Grant, B.D.; Bawaskar, P.; Yuen, S.; Li, J.; Muretta, J.M.; Gillispie, G.D.; Thomas, D.D. High-Throughput Spectral and Lifetime-Based FRET Screening in Living Cells to Identify Small-Molecule Effectors of SERCA. SLAS Discov. 2017, 22, 262–273. [Google Scholar] [CrossRef]
- Rebbeck, R.; Ginsburg, K.S.; Ko, C.Y.; Fasoli, A.; Rusch, K.; Cai, G.F.; Dong, X.; Thomas, D.D.; Bers, D.M.; Cornea, R.L. Synergistic FRET assays for drug discovery targeting RyR2 channels. J. Mol. Cell. Cardiol. 2022, 168, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Guzmán, C.; Oetken-Lindholm, C.; Abankwa, D. Automated High-Throughput Fluorescence Lifetime Imaging Microscopy to Detect Protein–Protein Interactions. J. Lab. Autom. 2016, 21, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Liu, L.; Zou, P. Functional imaging-guided cell selection for evolving genetically encoded fluorescent indicators. Cell Rep. Methods 2023, 3, 100544. [Google Scholar] [CrossRef]
- Lo, C.H.; Vunnam, N.; Lewis, A.K.; Chiu, T.-L.; Brummel, B.E.; Schaaf, T.M.; Grant, B.D.; Bawaskar, P.; Thomas, D.D.; Sachs, J.N. An Innovative High-Throughput Screening Approach for Discovery of Small Molecules That Inhibit TNF Receptors. SLAS Discov. 2017, 22, 950–961. [Google Scholar] [CrossRef]
- Rebbeck, R.T.; Essawy, M.M.; Nitu, F.R.; Grant, B.D.; Gillispie, G.D.; Thomas, D.D.; Bers, D.M.; Cornea, R.L. High-Throughput Screens to Discover Small-Molecule Modulators of Ryanodine Receptor Calcium Release Channels. SLAS Discov. 2017, 22, 176–186. [Google Scholar] [CrossRef]


| Targets | Names | FP Reporters | 2p-FLIM | 𝜏free (ns) | 𝜏bind (ns) | Δ𝜏(bind-free) (ns) | Ref. |
|---|---|---|---|---|---|---|---|
| NAD+/NADH | Peredox | cpT–Sapphire | Yes | 2.63 | 1.87 | −0.76 | [42] |
| Glucose | iGlucoSnFR–TS | cpT–Sapphire | Yes | 1.40 | 1.78 | 0.38 | [44] |
| Lactate | LiLac | cp–mTQ2 | Yes | 3.00 | 1.80 | −1.20 | [46] |
| ATP | qMaLioffG | Citrine | 2.57 | 1.49 | −1.08 | [40] | |
| Acetylcholine | GRABACh3.0 | cpGFP | Yes | 3.34 | 3.51 | 0.17 | [45] |
| Histidine | FHisJ | cpYFP | 2.80 | 1.60 | −1.20 | [47] | |
| Ca2+ | Tq–Ca–FLITS | cp–mTQ2 | 1.40 | 2.78 | 1.38 | [27] | |
| CatchER | EGFP | 2.18 | 2.61 | 0.43 | [48] | ||
| RCaMP1h | cp–mRuby | Yes | - | - | 1.10 | [43] | |
| pH | pHRed | mKeima–A213S | Yes | 1.72 (pH 5) | 2.12 (pH 8) | 0.40 | [13] |
| SypHerRed | cp–mApple | Yes | 0.72 (pH 6.9) | 1.05 (pH 7.7) | 0.33 | [41] | |
| SypHer3s | cpYFP | Yes | 1.20 (pH 6.5) | 2.30 (pH 9.5) | 1.10 | [49] | |
| H2O2 | Hyper3 | cpGFP | 1.29 | 0.92 | −0.37 | [50] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vu, C.Q.; Arai, S. Quantitative Imaging of Genetically Encoded Fluorescence Lifetime Biosensors. Biosensors 2023, 13, 939. https://doi.org/10.3390/bios13100939
Vu CQ, Arai S. Quantitative Imaging of Genetically Encoded Fluorescence Lifetime Biosensors. Biosensors. 2023; 13(10):939. https://doi.org/10.3390/bios13100939
Chicago/Turabian StyleVu, Cong Quang, and Satoshi Arai. 2023. "Quantitative Imaging of Genetically Encoded Fluorescence Lifetime Biosensors" Biosensors 13, no. 10: 939. https://doi.org/10.3390/bios13100939
APA StyleVu, C. Q., & Arai, S. (2023). Quantitative Imaging of Genetically Encoded Fluorescence Lifetime Biosensors. Biosensors, 13(10), 939. https://doi.org/10.3390/bios13100939





