Facile Label-Free Electrochemical DNA Biosensor for Detection of Osteosarcoma-Related Survivin Gene
Abstract
1. Introduction
2. Experimental Sections
2.1. Reagents and Apparatus
2.2. Interaction of CB with dsDNA in Solution
2.3. UV-vis Spectroscopy Experiment
2.4. Fabrication of Electrochemical DNA Biosensor for Survivin Gene Detection
3. Results and Discussion
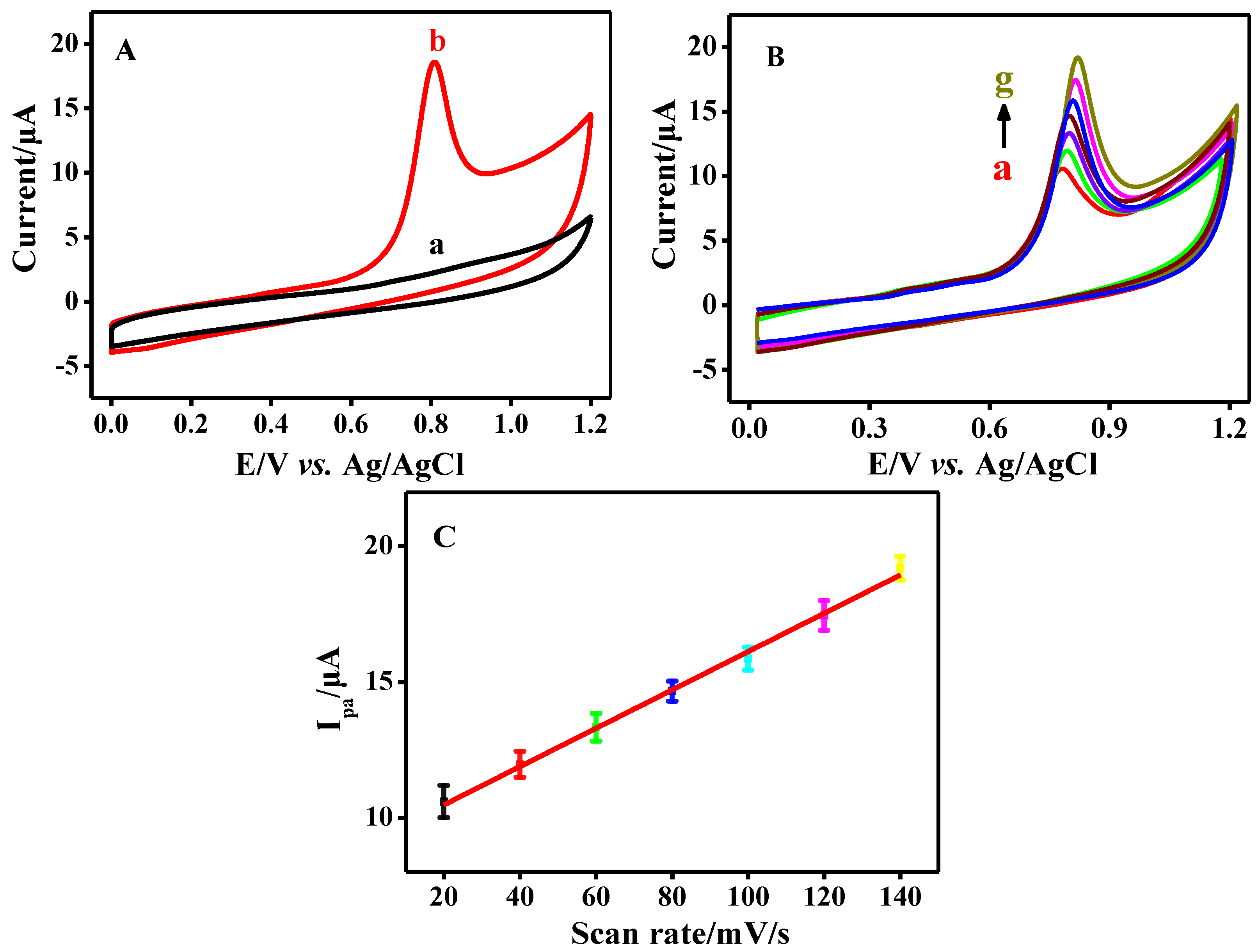
3.1. Electrochemical Behavior of CB
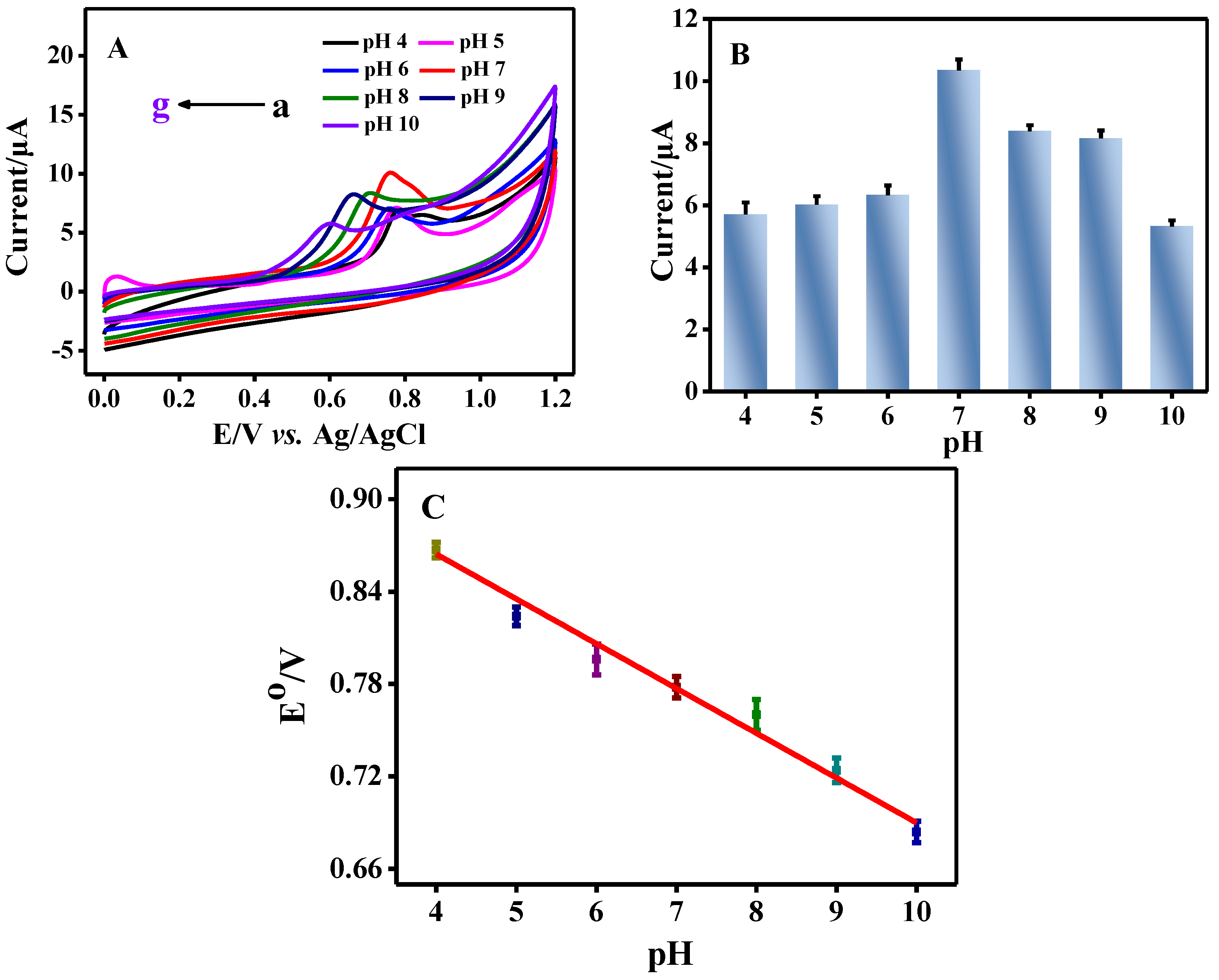
3.2. The Interaction between CB and Salmon Sperm dsDNA
3.2.1. Electrochemical Investigation on the Interaction between CB and Salmon Sperm dsDNA
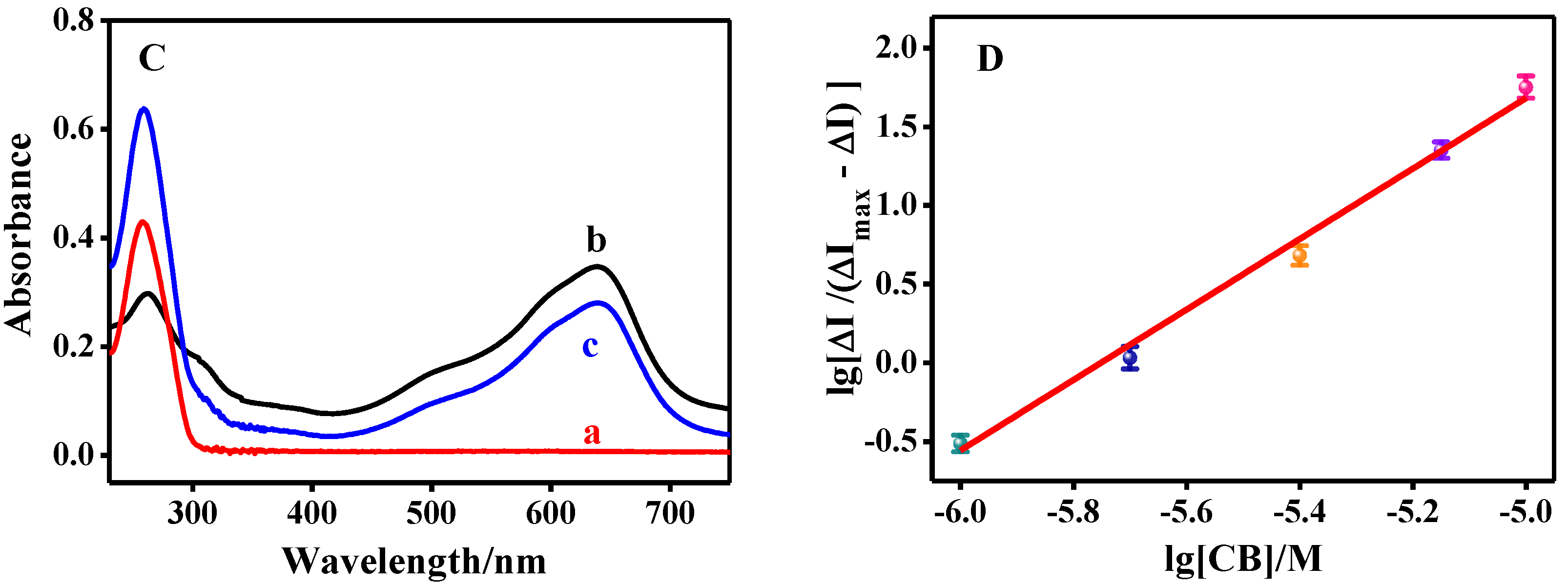
3.2.2. UV–vis Spectroscopic Study on the Interaction between CB and Salmon Sperm dsDNA
3.3. The Equilibrium Constant between CB and Salmon Sperm dsDNA
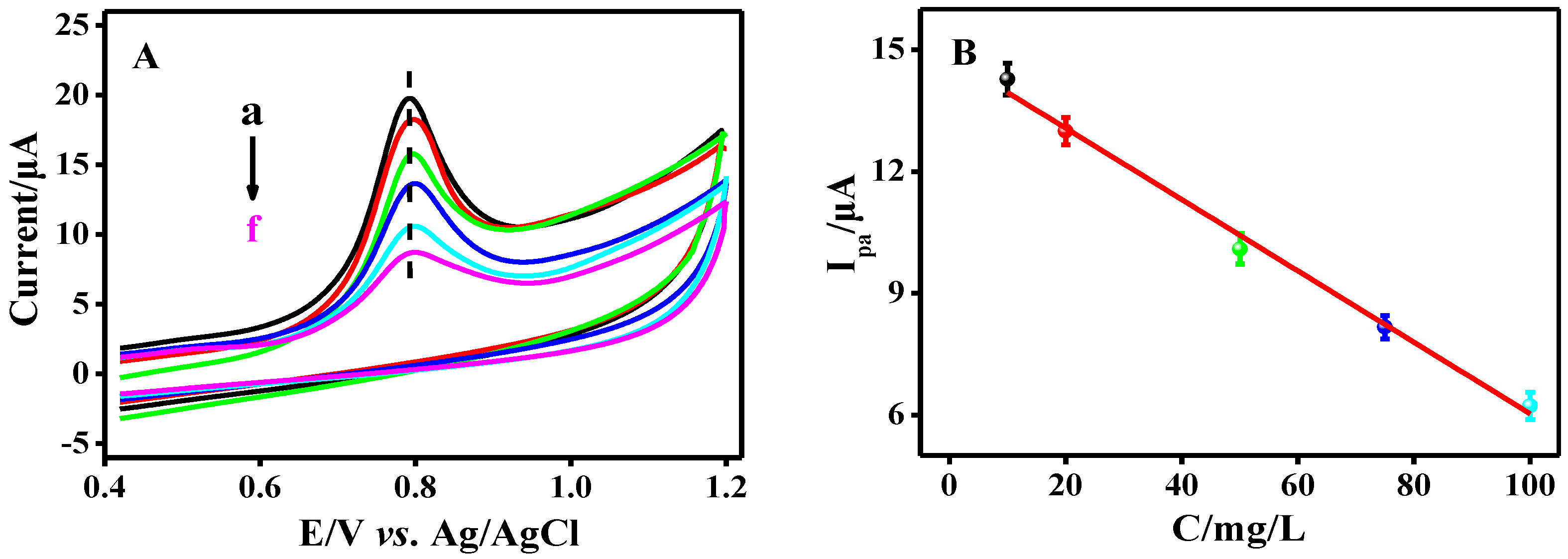
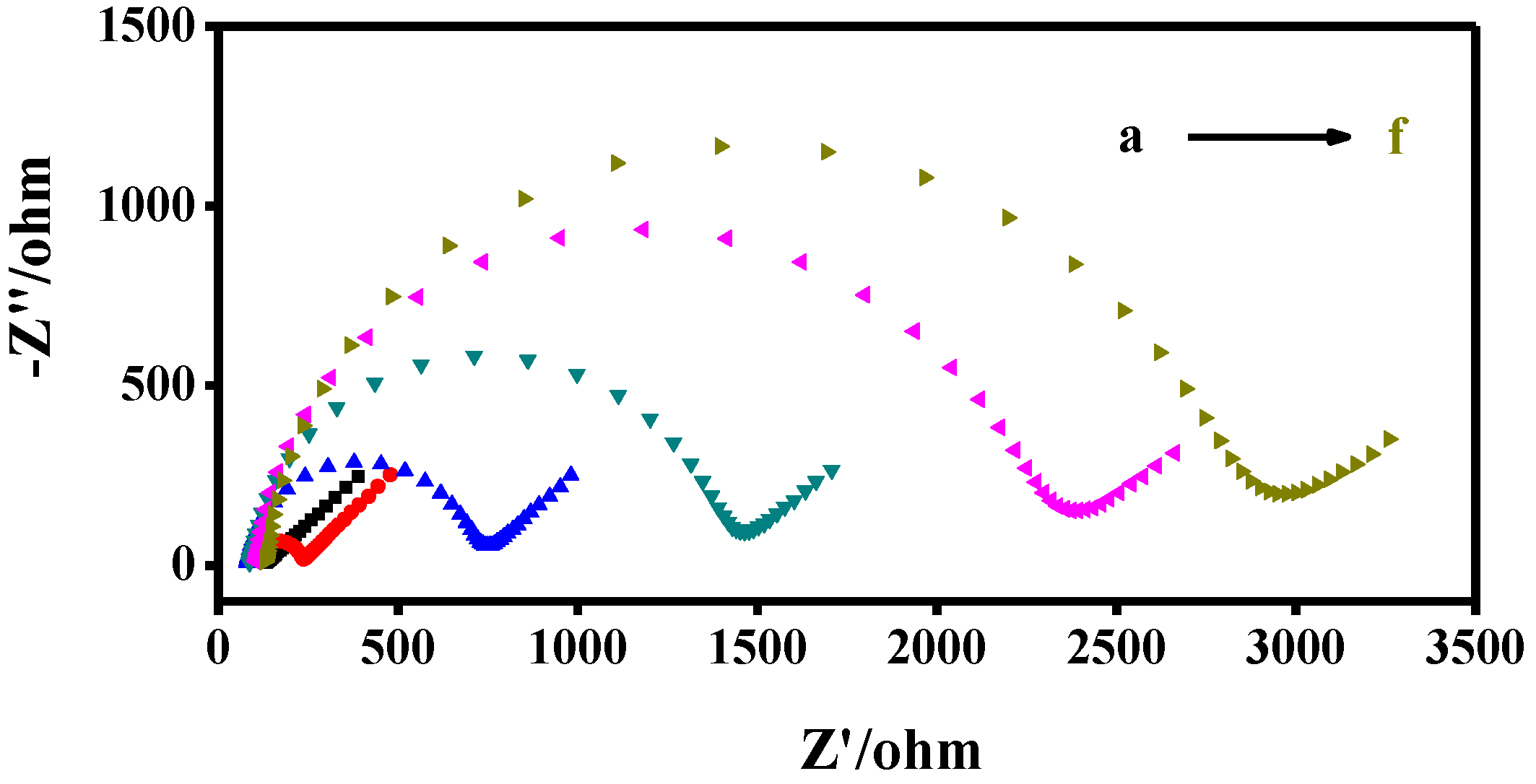
3.4. Electrochemical Characterization of Modified GCE Electrodes
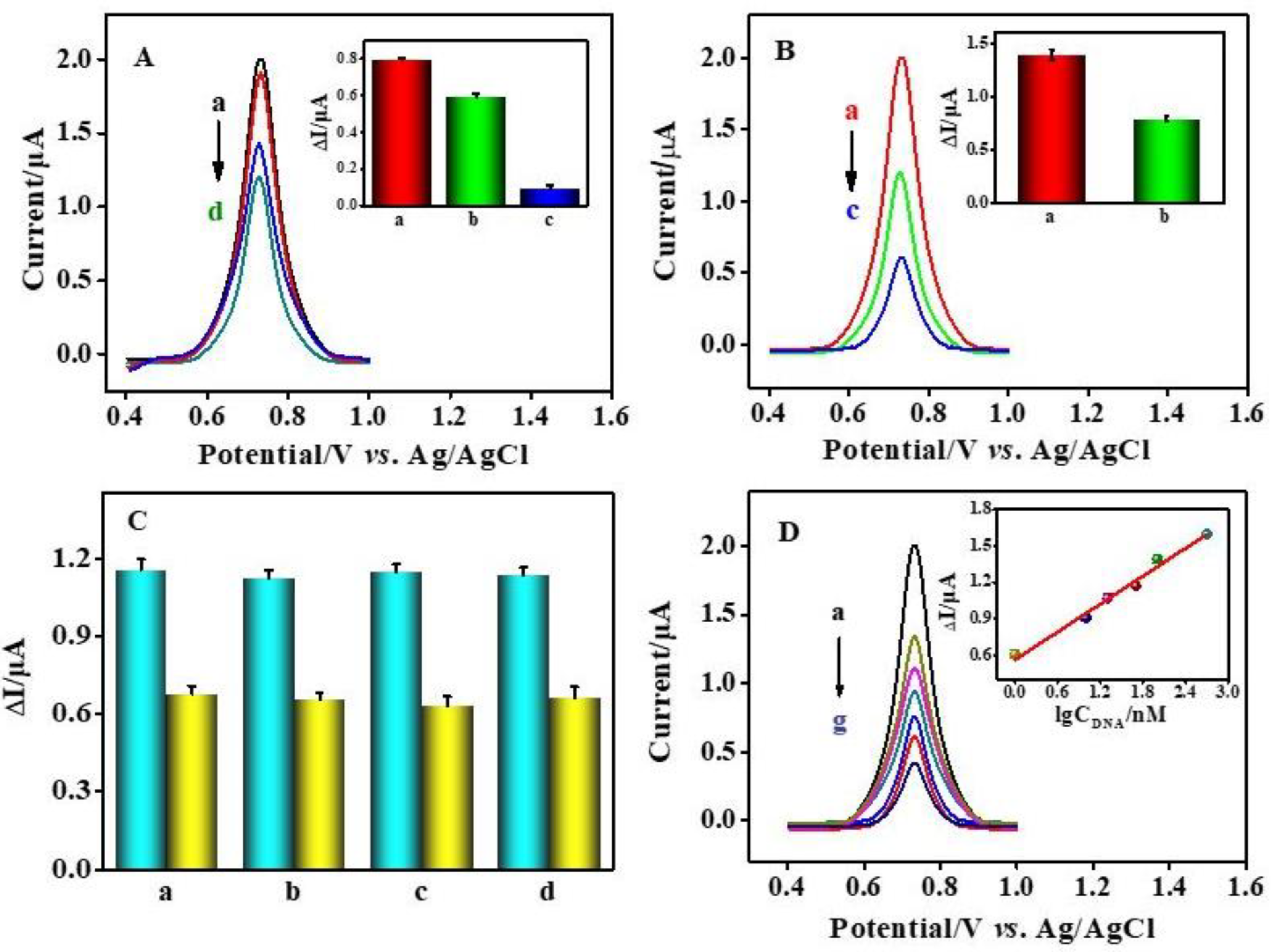
3.5. Performance of the Designed Biosensor for Osteosarcoma-Related Survivin Gene Detection
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Meyers, P.A.; Gorlick, R. Osteosarcoma. Pediatr. Clin. N. Am. 1997, 44, 973–989. [Google Scholar] [CrossRef]
- Ottaviani, G.; Jaffe, N. The epidemiology of osteosarcoma. Cancer Treat. Res. 2009, 152, 3–13. [Google Scholar] [PubMed]
- Moreno, F.; Cacciavillano, W.; Cipolla, M.; Coirini, M.; Streitenberger, P.; Martí, J.L.; Palladino, M.; Morici, M.; Onoratelli, M.; Drago, G.; et al. Childhood osteosarcoma: Incidence and survival in Argentina. Report from the National Pediatric Cancer Registry, ROHA Network 2000–2013. Pediatr. Blood Cancer 2017, 64, e26533. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Da, M.; Zhuang, Z.; Wu, W.; Wu, Z.; Wu, Y.; Shen, H. Effects of Survivin on cell proliferation and apoptosis in MG-63 cells in vitro. Cell Biol. Int. 2009, 33, 119–124. [Google Scholar] [CrossRef]
- Zhong, G.; Liu, A.; Xu, X.; Sun, Z.; Chen, J.; Wang, K.; Liu, Q.; Lin, X.; Lin, J. Detection of femtomolar level osteosarcoma-related gene via a chronocoulometric DNA biosensor based on nanostructure gold electrode. Int. J. Nanomed. 2012, 7, 527–536. [Google Scholar] [CrossRef][Green Version]
- Soleimanpour, E.; Babaei, E. Survivin as a Potential Target for Cancer Therapy. Asian Pac. J. Cancer Prev. 2015, 16, 6187–6191. [Google Scholar] [CrossRef]
- Porichis, F.; Hart, M.G.; Griesbeck, M.; Everett, H.L.; Hassan, M.; Baxter, A.; Lindqvist, M.; Miller, S.M.; Soghoian, D.Z.; Kavanagh, D.G.; et al. High-throughput detection of miRNAs and gene-specific mRNA at the single-cell level by flow cytometry. Nat. Commun. 2014, 5, 5641. [Google Scholar] [CrossRef]
- Hamidi-Asl, E.; Palchetti, I.; Hasheminejad, E.; Mascini, M. A review on the electrochemical biosensors for determination of microRNAs. Talanta 2013, 115, 74–83. [Google Scholar] [CrossRef]
- Yin, G.; Fu, B.; Xu, B.; Han, J.; Xue, Y.; Chen, H.; Zhang, X.; Wang, G. Identification of osteosarcoma by microRNA-coupled nuclease digestion on interdigitated electrode sensor. Biotechnol. Appl. Biochem. 2021, 69, 1094–1100. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, J.; Zhuang, Q.; Chen, J.; Lin, X. Electrochemical Studies of the Interaction of 2-Nitroacridone with DNA and Determination of DNA. Electroanalysis 2007, 19, 1765–1772. [Google Scholar] [CrossRef]
- Deng, C.; Xia, Y.; Xiao, C.; Nie, Z.; Yang, M.; Si, S. Electrochemical oxidation of purine and pyrimidine bases based on the boron-doped nanotubes modified electrode. Biosens. Bioelectron. 2012, 31, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Millan, K.M.; Mikkelsen, S.R. Sequence-selective biosensor for DNA based on electroactive hybridization indicators. Anal. Chem. 1993, 65, 2317–2323. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Wu, J.; Ju, H. Electrochemical sensing of heavy metal ions with inorganic, organic and bio-materials. Biosens. Bioelectron. 2015, 63, 276–286. [Google Scholar] [CrossRef]
- Drummond, T.G.; Hill, M.G.; Barton, J.K. Electrochemical DNA sensors. Nat. Biotechnol. 2003, 21, 1192–1199. [Google Scholar] [CrossRef]
- Peng, H.; Soeller, C.; Vigar, N.; Kilmartin, P.A.; Cannell, M.B.; Bowmaker, G.A.; Cooney, R.P.; Travas-Sejdic, J. Label-free electrochemical DNA sensor based on functionalised conducting copolymer. Biosens. Bioelectron. 2005, 20, 1821–1828. [Google Scholar] [CrossRef]
- Marrazza, G.; Chianella, I.; Mascini, M. Disposable DNA electrochemical sensor for hybridization detection. Biosens. Bioelectron. 1999, 14, 43–51. [Google Scholar] [CrossRef]
- Tansil, N.C.; Xie, H.; Xie, F.; Gao, Z. Direct Detection of DNA with an Electrocatalytic Threading Intercalator. Anal. Chem. 2003, 77, 126–134. [Google Scholar] [CrossRef]
- Noorbakhsh, A.; Salimi, A. Amperometric detection of hydrogen peroxide at nano-nickel oxide/thionine and celestine blue nanocomposite-modified glassy carbon electrodes. Electrochim. Acta 2009, 54, 6312–6321. [Google Scholar] [CrossRef]
- Hamidi-Asl, E.; Raoof, J.B.; Meibodi, A.S.; Bazgir, Z.H. Celestine blue as a new indicator in electrochemical DNA biosensors. Sci. China Chem. 2016, 59, 128–134. [Google Scholar] [CrossRef]
- Sun, Z.; Ma, Z.; Zhang, W.; Wang, X.; Fan, C.; Li, G. Electrochemical investigations of baicalin and DNA-baicalin interactions. Anal. Bioanal. Chem. 2004, 379, 283–286. [Google Scholar] [CrossRef]
- Nazari-Vanani, R.; Sattarahmady, N.; Yadegari, H.; Delshadi, N.; Hatam, G.; Heli, H. Electrochemical quantitation of Leishmania infantum based on detection of its kDNA genome and transduction of non-spherical gold nanoparticles. Anal. Chim. Acta 2018, 1041, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Stobiecka, M.; Chalupa, A.; Dworakowska, B. Piezometric biosensors for anti-apoptotic protein survivin based on buried positive-potential barrier and immobilized monoclonal antibodies. Biosens. Bioelectron. 2016, 84, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Stobiecka, M.; Chalupa, A. Modulation of Plasmon-Enhanced Resonance Energy Transfer to Gold Nanoparticles by Protein Survivin Channeled-Shell Gating. J. Phys. Chem. B 2015, 119, 13227–13235. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, K.; Krazinski, B.E.; Kowalczyk, A.E.; Dworakowska, B.; Jakiela, S.; Stobiecka, M. Hairpin–Hairpin Molecular Beacon Interactions for Detection of Survivin mRNA in Malignant SW480 Cells. ACS Appl. Mater. Interfaces 2018, 10, 17028–17039. [Google Scholar] [CrossRef]
- Khan, S.; Jutzy, J.M.S.; Aspe, J.R.; McGregor, D.W.; Neidigh, J.W.; Wall, N.R. Survivin is released from cancer cells via exosomes. Apoptosis 2010, 16, 1–12. [Google Scholar] [CrossRef]
- Konopka, K.; Spain, C.; Yen, A.; Overlid, N.; Gebremedhin, S.; Duzgunes, N. Correlation between the levels of survivin and survivin promoter-driven gene expression in cancer and non-cancer cells. Cell. Mol. Biol. Lett. 2009, 14, 70–89. [Google Scholar] [CrossRef]


| Synthetic Oligonucleotides | Sequence (5′→3′) |
|---|---|
| Capture probe (CP, S1) | NH2-TTGGAGGGCTGCGCCTGCACCC |
| Target DNA (S2) | GGGTGCAGGCGCAGCCCTCCAA |
| Single-base mismatched target DNA (S3) | GGGTGCAAGCGCAGCCCTCCAA |
| Non-complementary target DNA (S4) | AATTGTACGTCATTCGATCCTA |
| Partial BCR sequence (S5) | GAAGGGCACTACCCATCT |
| Partial ABL sequence (S6) | AGGCTGGTCCTGAACTCT |
| Strategy | Readout | Detection Limit | References |
|---|---|---|---|
| AuQC/AHT/AbSurvivin | EQCM biosensor | 1.87 ± 0.7 nM in lysate solution; 1.7 ± 0.8 nM in 10 mM PBS | [22] |
| AuNP@Cit/Survivin | gated RET a | 0.240 ± 0.007 nM (synthetic solutions) | [23] |
| Hairpin-hairpin molecular beacon | fluorescence | 26 nM | [24] |
| Eletrophoresis | WB b | not specified | [25] |
| ELISA c | colorimetry | 5.8 ± 2.3 to 24.3 ± 2.9 ng suvivin/mg proteins in cancer cells lysates (35 ± 14 to 148 ± 18 nM) d | [26] |
| CB/Survivin DNA | label-free electrochemical DNA biosensor | 0.046 nM | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Zhong, Y.; Ye, J.-X.; Lei, Y.; Liu, A.-L. Facile Label-Free Electrochemical DNA Biosensor for Detection of Osteosarcoma-Related Survivin Gene. Biosensors 2022, 12, 747. https://doi.org/10.3390/bios12090747
Chen Y, Zhong Y, Ye J-X, Lei Y, Liu A-L. Facile Label-Free Electrochemical DNA Biosensor for Detection of Osteosarcoma-Related Survivin Gene. Biosensors. 2022; 12(9):747. https://doi.org/10.3390/bios12090747
Chicago/Turabian StyleChen, Yao, Yu Zhong, Ji-Xing Ye, Yun Lei, and Ai-Lin Liu. 2022. "Facile Label-Free Electrochemical DNA Biosensor for Detection of Osteosarcoma-Related Survivin Gene" Biosensors 12, no. 9: 747. https://doi.org/10.3390/bios12090747
APA StyleChen, Y., Zhong, Y., Ye, J.-X., Lei, Y., & Liu, A.-L. (2022). Facile Label-Free Electrochemical DNA Biosensor for Detection of Osteosarcoma-Related Survivin Gene. Biosensors, 12(9), 747. https://doi.org/10.3390/bios12090747




