Microfluidic Lab-on-a-Chip for Studies of Cell Migration under Spatial Confinement
Abstract
1. Introduction
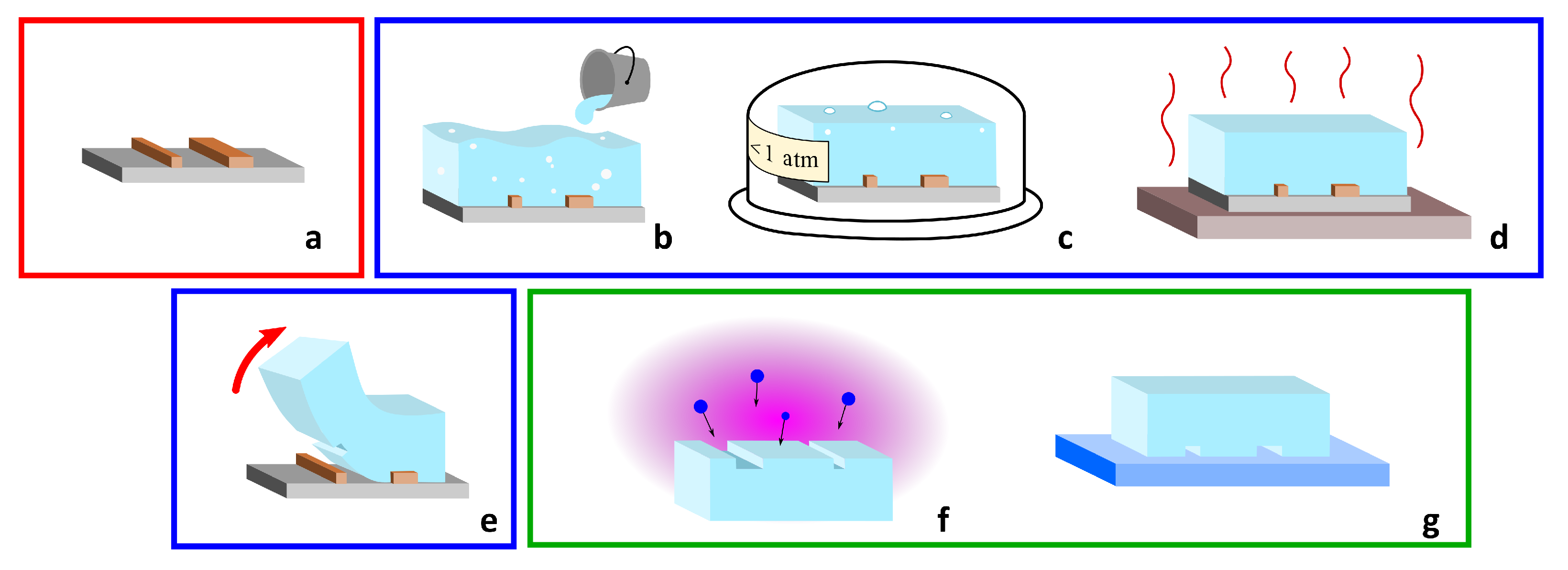
2. PDMS-Based Microfluidic Devices
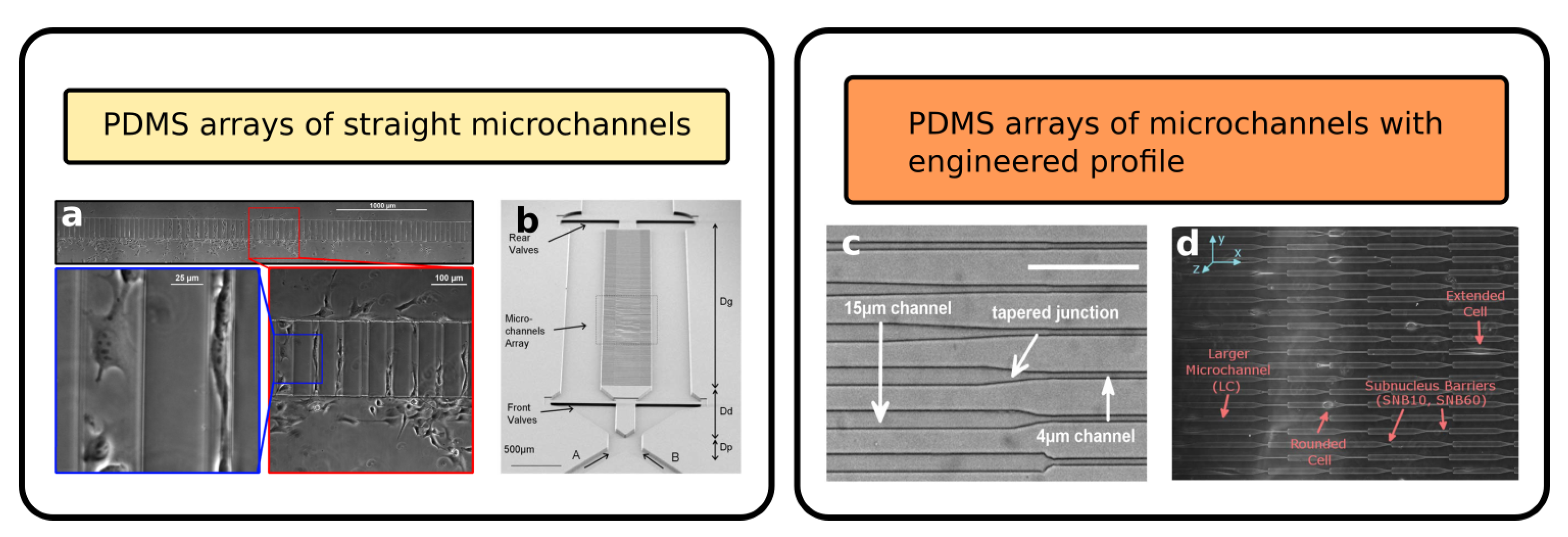
2.1. Arrays of Straight Channel as Cell Migration Assays
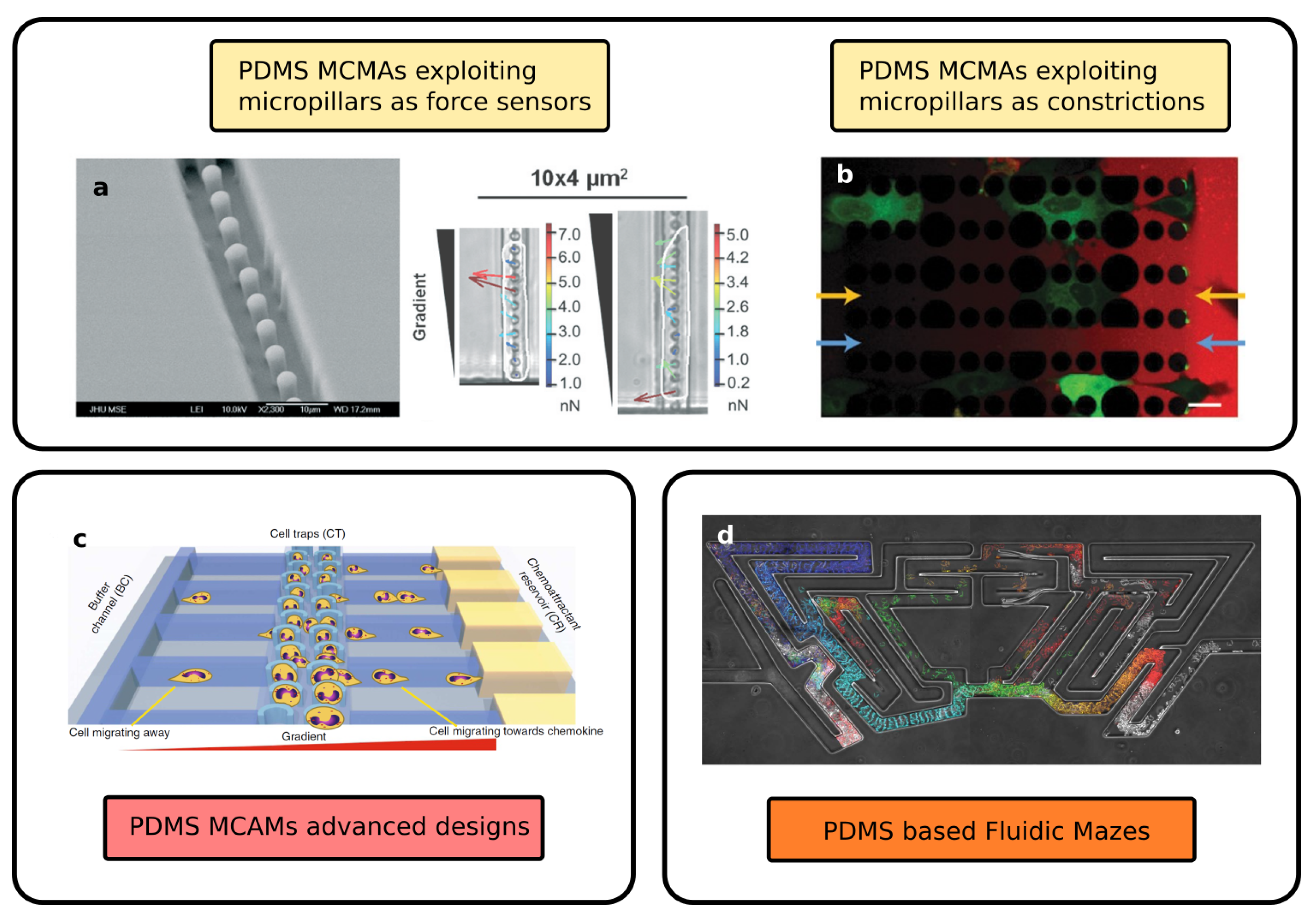
2.2. Advanced Geometries of Fluidic Networks
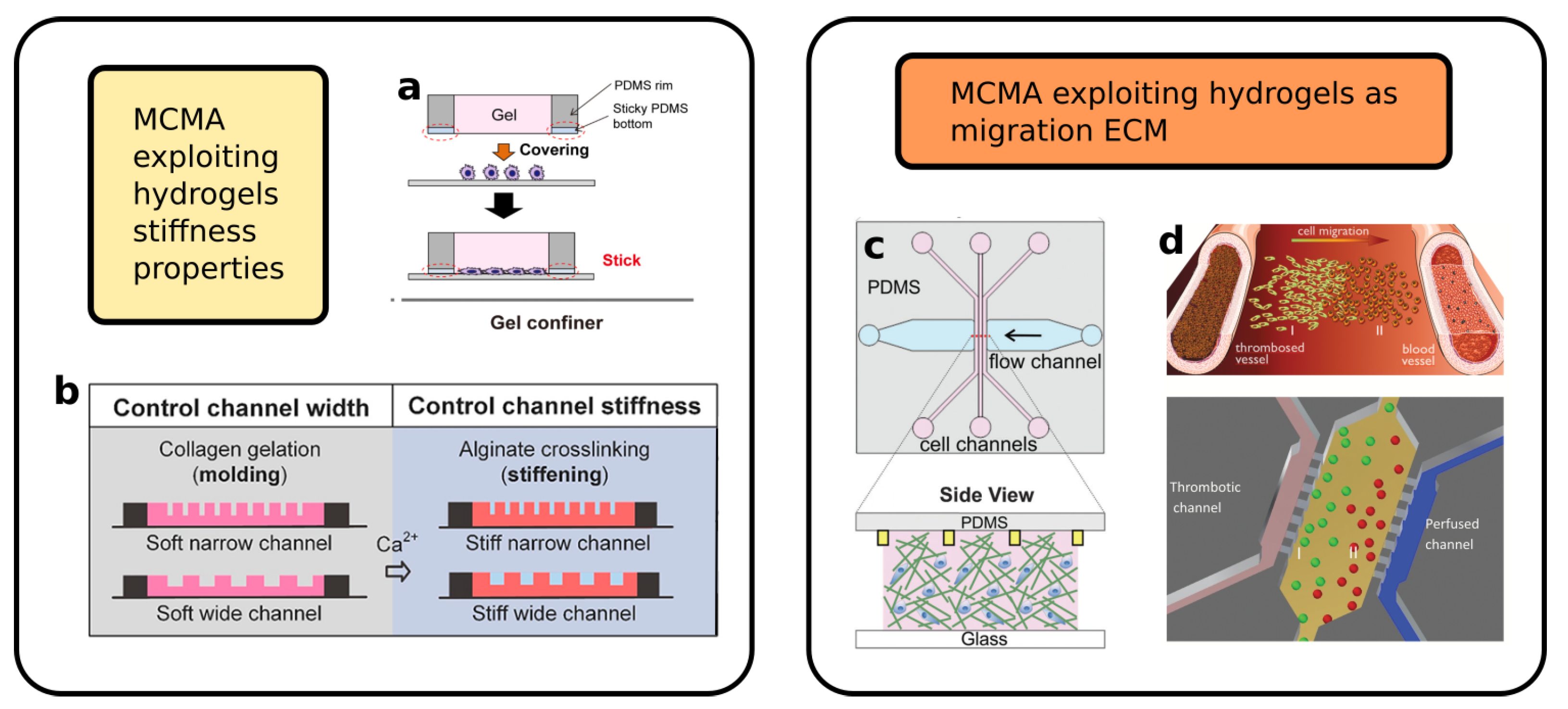
3. Hydrogels-Based Microfluidic Devices
4. Microfluidic Devices for Migration Assays Realized by Femtosecond Laser Micromachining
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| EMT | Epithelial-to-Mesenchymal Transition |
| ECM | Extra-cellular Matrix |
| MCMA | Microfluidic Cell Migration Assay |
| FLM | Femtosecond Laser Micromachining |
| TPP | Two-Photon Polymerization |
| FLICE | Femtosecond Laser Irradiation followed by Chemical Etching |
| PDMS | Polydimethylsiloxane |
References
- Paul, C.D.; Mistriotis, P.; Konstantopoulos, K. Cancer cell motility: Lessons from migration in confined spaces. Nat. Rev. Cancer 2016, 17, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Iwatsuki, M.; Mimori, K.; Yokobori, T.; Ishi, H.; Beppu, T.; Nakamori, S.; Baba, H.; Mori, M. Epithelial-mesenchymal transition in cancer development and its clinical significance. Cancer Sci. 2010, 101, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Gomez, S.J.; Maziveyi, M.; Alahari, S.K. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol. Cancer 2016, 15, 18. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef]
- Walker, C.; Mojares, E.; del Río Hernández, A. Role of Extracellular Matrix in Development and Cancer Progression. Int. J. Mol. Sci. 2018, 19, 3028. [Google Scholar] [CrossRef]
- Doolin, M.T.; Moriarty, R.A.; Stroka, K.M. Mechanosensing of mechanical confinement by mesenchymal-like cells. Front. Physiol. 2020, 11, 365. [Google Scholar] [CrossRef]
- Samuel, M.S.; Lopez, J.I.; McGhee, E.J.; Croft, D.R.; Strachan, D.; Timpson, P.; Munro, J.; Schröder, E.; Zhou, J.; Brunton, V.G.; et al. Actomyosin-Mediated Cellular Tension Drives Increased Tissue Stiffness and β-Catenin Activation to Induce Epidermal Hyperplasia and Tumor Growth. Cancer Cell 2011, 19, 776–791. [Google Scholar] [CrossRef]
- Nasrollahi, S.; Pathak, A. Topographic confinement of epithelial clusters induces epithelial-to-mesenchymal transition in compliant matrices. Sci. Rep. 2016, 6, 18831. [Google Scholar] [CrossRef]
- Lancaster, O.M.; Le Berre, M.; Dimitracopoulos, A.; Bonazzi, D.; Zlotek-Zlotkiewicz, E.; Picone, R.; Duke, T.; Piel, M.; Baum, B. Mitotic rounding alters cell geometry to ensure efficient bipolar spindle formation. Dev. Cell 2013, 25, 270–283. [Google Scholar] [CrossRef]
- Moriarty, R.A.; Stroka, K.M. Physical confinement alters sarcoma cell cycle progression and division. Cell Cycle 2018, 17, 2360–2373. [Google Scholar] [CrossRef] [PubMed]
- McGregor, A.L.; Hsia, C.R.; Lammerding, J. Squish and squeeze—The nucleus as a physical barrier during migration in confined environments. Curr. Opin. Cell Biol. 2016, 40, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Boucher, Y.; Baxter, L.T.; Jain, R.K. Interstitial pressure gradients in tissue-isolated and subcutaneous tumors: Implications for therapy. Cancer Res. 1990, 50, 4478–4484. [Google Scholar]
- Pickup, M.W.; Mouw, J.K.; Weaver, V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014, 15, 1243–1253. [Google Scholar] [CrossRef] [PubMed]
- Poltavets, V.; Kochetkova, M.; Pitson, S.M.; Samuel, M.S. The Role of the Extracellular Matrix and Its Molecular and Cellular Regulators in Cancer Cell Plasticity. Front. Oncol. 2018, 8, 431. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef]
- Weigelin, B.; Bakker, G.J.; Friedl, P. Intravital third harmonic generation microscopy of collective melanoma cell invasion. IntraVital 2012, 1, 32–43. [Google Scholar] [CrossRef]
- Alexander, S.; Weigelin, B.; Winkler, F.; Friedl, P. Preclinical intravital microscopy of the tumour-stroma interface: Invasion, metastasis, and therapy response. Curr. Opin. Cell Biol. 2013, 25, 659–671. [Google Scholar] [CrossRef]
- Ballestrem, C.; Wehrle-Haller, B.; Hinz, B.; Imhof, B.A. Actin-dependent Lamellipodia Formation and Microtubule-dependent Tail Retraction Control-directed Cell Migration. Mol. Biol. Cell 2000, 11, 2999–3012. [Google Scholar] [CrossRef]
- Mattila, P.K.; Lappalainen, P. Filopodia: Molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 2008, 9, 446–454. [Google Scholar] [CrossRef]
- Shankar, J.; Messenberg, A.; Chan, J.; Underhill, T.M.; Foster, L.J.; Nabi, I.R. Pseudopodial Actin Dynamics Control Epithelial-Mesenchymal Transition in Metastatic Cancer Cells. Cancer Res. 2010, 70, 3780–3790. [Google Scholar] [CrossRef]
- Friedl, P.; Wolf, K. Plasticity of cell migration: A multiscale tuning model. J. Cell Biol. 2009, 188, 11–19. [Google Scholar] [CrossRef]
- Talkenberger, K.; Cavalcanti-Adam, E.A.; Voss-Böhme, A.; Deutsch, A. Amoeboid-mesenchymal migration plasticity promotes invasion only in complex heterogeneous microenvironments. Sci. Rep. 2017, 7, 9237. [Google Scholar] [CrossRef]
- Wolf, K.; Mazo, I.; Leung, H.; Engelke, K.; Von Andrian, U.H.; Deryugina, E.I.; Strongin, A.Y.; Bröcker, E.B.; Friedl, P. Compensation mechanism in tumor cell migration: Mesenchymal–amoeboid transition after blocking of pericellular proteolysis. J. Cell Biol. 2003, 160, 267–277. [Google Scholar] [CrossRef]
- Graziani, V.; Rodriguez-Hernandez, I.; Maiques, O.; Sanz-Moreno, V. The amoeboid state as part of the epithelial-to-mesenchymal transition programme. Trends Cell Biol. 2022, 32, 228–242. [Google Scholar] [CrossRef]
- Gaertner, F.; Reis-Rodrigues, P.; de Vries, I.; Hons, M.; Aguilera, J.; Riedl, M.; Leithner, A.; Tasciyan, S.; Kopf, A.; Merrin, J.; et al. WASp triggers mechanosensitive actin patches to facilitate immune cell migration in dense tissues. Dev. Cell 2022, 57, 47–62.e9. [Google Scholar] [CrossRef]
- Alexander, S.; Koehl, G.E.; Hirschberg, M.; Geissler, E.K.; Friedl, P. Dynamic imaging of cancer growth and invasion: A modified skin-fold chamber model. Histochem. Cell Biol. 2008, 130, 1147–1154. [Google Scholar] [CrossRef]
- Friedl, P.; Alexander, S. Cancer Invasion and the Microenvironment: Plasticity and Reciprocity. Cell 2011, 147, 992–1009. [Google Scholar] [CrossRef]
- Ficorella, C.; Vázquez, R.M.; Heine, P.; Lepera, E.; Cao, J.; Warmt, E.; Osellame, R.; Käs, J.A. Normal epithelial and triple-negative breast cancer cells show the same invasion potential in rigid spatial confinement. New J. Phys. 2019, 21, 083016. [Google Scholar] [CrossRef]
- Ficorella, C.; Eichholz, H.M.; Sala, F.; Vázquez, R.M.; Osellame, R.; Käs, J.A. Intermediate filaments ensure resiliency of single carcinoma cells, while active contractility of the actin cortex determines their invasive potential. New J. Phys. 2021, 23, 083028. [Google Scholar] [CrossRef]
- Buxboim, A.; Swift, J.; Irianto, J.; Spinler, K.R.; Dingal, P.D.P.; Athirasala, A.; Kao, Y.R.C.; Cho, S.; Harada, T.; Shin, J.W.; et al. Matrix elasticity regulates lamin-A, C phosphorylation and turnover with feedback to actomyosin. Curr. Biol. 2014, 24, 1909–1917. [Google Scholar] [CrossRef]
- Cao, X.; Moeendarbary, E.; Isermann, P.; Davidson, P.M.; Wang, X.; Chen, M.B.; Burkart, A.K.; Lammerding, J.; Kamm, R.D.; Shenoy, V.B. A Chemomechanical Model for Nuclear Morphology and Stresses during Cell Transendothelial Migration. Biophys. J. 2016, 111, 1541–1552. [Google Scholar] [CrossRef]
- Denais, C.M.; Gilbert, R.M.; Isermann, P.; McGregor, A.L.; te Lindert, M.; Weigelin, B.; Davidson, P.M.; Friedl, P.; Wolf, K.; Lammerding, J. Nuclear envelope rupture and repair during cancer cell migration. Science 2016, 352, 353–358. [Google Scholar] [CrossRef]
- Hatch, E.M.; Hetzer, M.W. Nuclear envelope rupture is induced by actin-based nucleus confinement. J. Cell Biol. 2016, 215, 27–36. [Google Scholar] [CrossRef]
- Khatau, S.B.; Bloom, R.J.; Bajpai, S.; Razafsky, D.; Zang, S.; Giri, A.; Wu, P.H.; Marchand, J.; Celedon, A.; Hale, C.M.; et al. The distinct roles of the nucleus and nucleus-cytoskeleton connections in three-dimensional cell migration. Sci. Rep. 2012, 2, 488. [Google Scholar] [CrossRef]
- Raab, M.; Gentili, M.; de Belly, H.; Thiam, H.R.; Vargas, P.; Jimenez, A.J.; Lautenschlaeger, F.; Voituriez, R.; Lennon-Duménil, A.M.; Manel, N.; et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 2016, 352, 359–362. [Google Scholar] [CrossRef]
- Hulkower, K.I.; Herber, R.L. Cell Migration and Invasion Assays as Tools for Drug Discovery. Pharmaceutics 2011, 3, 107–124. [Google Scholar] [CrossRef]
- Friedl, P.; Hegerfeldt, Y.; Tusch, M. Collective cell migration in morphogenesis and cancer. Int. J. Dev. Biol. 2004, 48, 441–449. [Google Scholar] [CrossRef]
- Poujade, M.; Grasland-Mongrain, E.; Hertzog, A.; Jouanneau, J.; Chavrier, P.; Ladoux, B.; Buguin, A.; Silberzan, P. Collective migration of an epithelial monolayer in response to a model wound. Proc. Natl. Acad. Sci. USA 2007, 104, 15988–15993. [Google Scholar] [CrossRef]
- Boyden, S. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. J. Exp. Med. 1962, 115, 453–466. [Google Scholar] [CrossRef]
- Huang, Y.; Agrawal, B.; Sun, D.; Kuo, J.S.; Williams, J.C. Microfluidics-based devices: New tools for studying cancer and cancer stem cell migration. Biomicrofluidics 2011, 5, 013412. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.A.; Yin, T.I.; Reyes, D.; Urban, G.A. Microfluidic chip with integrated electrical cell-impedance sensing for monitoring single cancer cell migration in three-dimensional matrixes. Anal. Chem. 2013, 85, 11068–11076. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Vazquez, M. A Gal-MμS Device to Evaluate Cell Migratory Response to Combined Galvano-Chemotactic Fields. Biosensors 2017, 7, 54. [Google Scholar] [CrossRef] [PubMed]
- Nie, F.; Yamada, M.; Kobayashi, J.; Yamato, M.; Kikuchi, A.; Okano, T. On-chip cell migration assay using microfluidic channels. Biomaterials 2007, 28, 4017–4022. [Google Scholar] [CrossRef]
- Ilina, O.; Gritsenko, P.G.; Syga, S.; Lippoldt, J.; Porta, C.A.M.L.; Chepizhko, O.; Grosser, S.; Vullings, M.; Bakker, G.J.; Starruß, J.; et al. Cell–cell adhesion and 3D matrix confinement determine jamming transitions in breast cancer invasion. Nat. Cell Biol. 2020, 22, 1103–1115. [Google Scholar] [CrossRef]
- Liu, Y.J.; Le Berre, M.; Lautenschlaeger, F.; Maiuri, P.; Callan-Jones, A.; Heuzé, M.; Takaki, T.; Voituriez, R.; Piel, M. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell 2015, 160, 659–672. [Google Scholar] [CrossRef]
- Rolli, C.G.; Seufferlein, T.; Kemkemer, R.; Spatz, J.P. Impact of tumor cell cytoskeleton organization on invasiveness and migration: A microchannel-based approach. PLoS ONE 2010, 5, e8726. [Google Scholar] [CrossRef]
- Fu, Y.; Chin, L.K.; Bourouina, T.; Liu, A.Q.; VanDongen, A.M.J. Nuclear deformation during breast cancer cell transmigration. Lab Chip 2012, 12, 3774. [Google Scholar] [CrossRef]
- Tong, Z.; Balzer, E.M.; Dallas, M.R.; Hung, W.C.; Stebe, K.J.; Konstantopoulos, K. Chemotaxis of cell populations through confined spaces at single-cell resolution. PLoS ONE 2012, 7, e29211. [Google Scholar] [CrossRef]
- Irimia, D.; Charras, G.; Agrawal, N.; Mitchison, T.; Toner, M. Polar stimulation and constrained cell migration in microfluidic channels. Lab Chip 2007, 7, 1783. [Google Scholar] [CrossRef]
- Spuul, P.; Chi, P.Y.; Billottet, C.; Chou, C.F.; Génot, E. Microfluidic devices for the study of actin cytoskeleton in constricted environments: Evidence for podosome formation in endothelial cells exposed to a confined slit. Methods 2016, 94, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Cui, F.; Wen, Q.; Zhou, H.S. Effect of vimentin on cell migration in collagen-coated microchannels: A mimetic physiological confined environment. Biomicrofluidics 2021, 15, 034105. [Google Scholar] [CrossRef]
- Mak, M.; Reinhart-King, C.A.; Erickson, D. Microfabricated physical spatial gradients for investigating cell migration and invasion dynamics. PLoS ONE 2011, 6, e20825. [Google Scholar] [CrossRef] [PubMed]
- Mak, M.; Reinhart-King, C.A.; Erickson, D. Elucidating mechanical transition effects of invading cancer cells with a subnucleus-scaled microfluidic serial dimensional modulation device. Lab Chip 2013, 13, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Wang, R.; Chen, S.; Luo, T.; Chow, Y.T.; Sun, D. Microfluidic platform for probing cancer cells migration property under periodic mechanical confinement. Biomicrofluidics 2018, 12, 024118. [Google Scholar] [CrossRef]
- Raman, P.S.; Paul, C.D.; Stroka, K.M.; Konstantopoulos, K. Probing cell traction forces in confined microenvironments. Lab Chip 2013, 13, 4599. [Google Scholar] [CrossRef]
- Boneschansker, L.; Yan, J.; Wong, E.; Briscoe, D.M.; Irimia, D. Microfluidic platform for the quantitative analysis of leukocyte migration signatures. Nat. Commun. 2014, 5, 4787. [Google Scholar] [CrossRef]
- Doolin, M.T.; Stroka, K.M. Integration of Mesenchymal Stem Cells into a Novel Micropillar Confinement Assay. Tissue Eng. Part C Methods 2019, 25, 662–676. [Google Scholar] [CrossRef]
- Davidson, P.M.; Sliz, J.; Isermann, P.; Denais, C.; Lammerding, J. Design of a microfluidic device to quantify dynamic intra-nuclear deformation during cell migration through confining environments. Integr. Biol. 2015, 7, 1534–1546. [Google Scholar] [CrossRef]
- Tweedy, L.; Thomason, P.A.; Paschke, P.I.; Martin, K.; Machesky, L.M.; Zagnoni, M.; Insall, R.H. Seeing around corners: Cells solve mazes and respond at a distance using attractant breakdown. Science 2020, 369, eaay9792. [Google Scholar] [CrossRef]
- Belotti, Y.; McGloin, D.; Weijer, C.J. Analysis of barotactic and chemotactic guidance cues on directional decision-making of Dictyostelium discoideum cells in confined environments. Proc. Natl. Acad. Sci. USA 2020, 117, 25553–25559. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.Y.; Heilman, S.; Wasserman, M.; Archer, S.; Shuler, M.L.; Wu, M. A hydrogel-based microfluidic device for the studies of directed cell migration. Lab Chip 2007, 7, 763. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Kwon, J.E.; Cho, Y.K. Dendritic Cell Migration Is Tuned by Mechanical Stiffness of the Confining Space. Cells 2021, 10, 3362. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Cheng, B.; Yang, Y.; Liu, H.; Huang, G.; Han, L.; Li, F.; Xu, F. Microchannel Stiffness and Confinement Jointly Induce the Mesenchymal-Amoeboid Transition of Cancer Cell Migration. Nano Lett. 2019, 19, 5949–5958. [Google Scholar] [CrossRef]
- Huang, Y.L.; kuan Tung, C.; Zheng, A.; Kim, B.J.; Wu, M. Interstitial flows promote amoeboid over mesenchymal motility of breast cancer cells revealed by a three dimensional microfluidic model. Integr. Biol. 2015, 7, 1402–1411. [Google Scholar] [CrossRef]
- Anguiano, M.; Morales, X.; Castilla, C.; Pena, A.R.; Ederra, C.; Martínez, M.; Ariz, M.; Esparza, M.; Amaveda, H.; Mora, M.; et al. The use of mixed collagen-Matrigel matrices of increasing complexity recapitulates the biphasic role of cell adhesion in cancer cell migration: ECM sensing, remodeling and forces at the leading edge of cancer invasion. PLoS ONE 2020, 15, e0220019. [Google Scholar] [CrossRef]
- Ayuso, J.M.; Monge, R.; Martínez-González, A.; Virumbrales-Muñoz, M.; Llamazares, G.A.; Berganzo, J.; Hernández-Laín, A.; Santolaria, J.; Doblaré, M.; Hubert, C.; et al. Glioblastoma on a microfluidic chip: Generating pseudopalisades and enhancing aggressiveness through blood vessel obstruction events. Neuro-Oncology 2017, 19, 503–513. [Google Scholar] [CrossRef]
- Truong, D.; Puleo, J.; Llave, A.; Mouneimne, G.; Kamm, R.D.; Nikkhah, M. Breast Cancer Cell Invasion into a Three Dimensional Tumor-Stroma Microenvironment. Sci. Rep. 2016, 6, 34094. [Google Scholar] [CrossRef]
- Trappmann, B.; Baker, B.M.; Polacheck, W.J.; Choi, C.K.; Burdick, J.A.; Chen, C.S. Matrix degradability controls multicellularity of 3D cell migration. Nat. Commun. 2017, 8, 371. [Google Scholar] [CrossRef]
- Sima, F.; Kawano, H.; Miyawaki, A.; Kelemen, L.; Ormos, P.; Wu, D.; Xu, J.; Midorikawa, K.; Sugioka, K. 3D Biomimetic Chips for Cancer Cell Migration in Nanometer-Sized Spaces Using “Ship-in-a-Bottle” Femtosecond Laser Processing. ACS Appl. Bio Mater. 2018, 1, 1667–1676. [Google Scholar] [CrossRef]
- Sima, F.; Kawano, H.; Hirano, M.; Miyawaki, A.; Obata, K.; Serien, D.; Sugioka, K. Mimicking intravasation–extravasation with a 3D glass nanofluidic model for the chemotaxis-free migration of cancer cells in confined spaces. Adv. Mater. Technol. 2020, 5, 2000484. [Google Scholar] [CrossRef]
- Tayalia, P.; Mendonca, C.R.; Baldacchini, T.; Mooney, D.J.; Mazur, E. 3D cell-migration studies using two-photon engineered polymer scaffolds. Adv. Mater. 2008, 20, 4494–4498. [Google Scholar] [CrossRef]
- Olsen, M.H.; Hjortø, G.M.; Hansen, M.; Met, Ö.; Svane, I.M.; Larsen, N.B. In-chip fabrication of free-form 3D constructs for directed cell migration analysis. Lab Chip 2013, 13, 4800. [Google Scholar] [CrossRef]
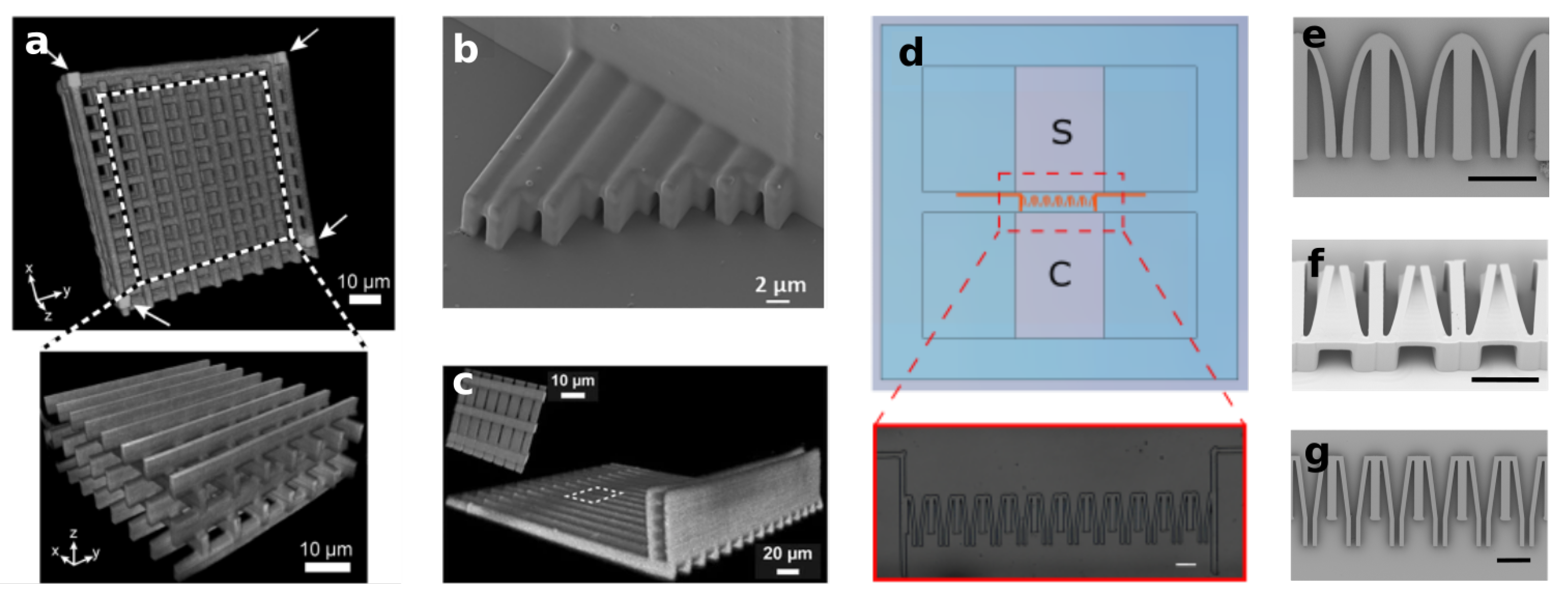
- Sala, F.; Ficorella, C.; Vázquez, R.M.; Eichholz, H.M.; Käs, J.A.; Osellame, R. Rapid Prototyping of 3D Biochips for Cell Motility Studies Using Two-Photon Polymerization. Front. Bioeng. Biotechnol. 2021, 9, 664094. [Google Scholar] [CrossRef] [PubMed]
- Sia, S.K.; Whitesides, G.M. Microfluidic devices fabricated in poly (dimethylsiloxane) for biological studies. Electrophoresis 2003, 24, 3563–3576. [Google Scholar] [CrossRef]
- M, K.R.; Chakraborty, S. PDMS microfluidics: A mini review. J. Appl. Polym. Sci. 2020, 137, 48958. [Google Scholar] [CrossRef]
- Banik, S.; Uchil, A.; Kalsang, T.; Chakrabarty, S.; Ali, M.A.; Srisungsitthisunti, P.; Mahato, K.K.; Surdo, S.; Mazumder, N. The revolution of PDMS microfluidics in cellular biology. Crit. Rev. Biotechnol. 2022, 1–19. [Google Scholar] [CrossRef]
- Zhang, M.; Wu, J.; Wang, L.; Xiao, K.; Wen, W. A simple method for fabricating multi-layer PDMS structures for 3D microfluidic chips. Lab Chip 2010, 10, 1199. [Google Scholar] [CrossRef]
- Hwang, Y.; Candler, R.N. Non-planar PDMS microfluidic channels and actuators: A review. Lab Chip 2017, 17, 3948–3959. [Google Scholar] [CrossRef]
- Park, S.-m.; Huh, Y.S.; Craighead, H.G.; Erickson, D. A method for nanofluidic device prototyping using elastomeric collapse. Proc. Natl. Acad. Sci. USA 2009, 106, 15549–15554. [Google Scholar] [CrossRef]
- Kim, M.; Ha, D.; Kim, T. Cracking-assisted photolithography for mixed-scale patterning and nanofluidic applications. Nat. Commun. 2015, 6, 6247. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Li, D. Fabrication of polydimethylsiloxane (PDMS) nanofluidic chips with controllable channel size and spacing. Lab Chip 2016, 16, 3767–3776. [Google Scholar] [CrossRef] [PubMed]
- Berthier, E.; Young, E.W.; Beebe, D. Engineers are from PDMS-land, Biologists are from Polystyrenia. Lab Chip 2012, 12, 1224–1237. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.P.; Salieb-Beugelaar, G.B.; Hunziker, P. PDMS with designer functionalities—Properties, modifications strategies, and applications. Prog. Polym. Sci. 2018, 83, 97–134. [Google Scholar] [CrossRef]
- Zhang, X.; Li, L.; Luo, C. Gel integration for microfluidic applications. Lab Chip 2016, 16, 1757–1776. [Google Scholar] [CrossRef] [PubMed]
- Bahram, M.; Mohseni, N.; Moghtader, M. An Introduction to Hydrogels and Some Recent Applications. In Emerging Concepts in Analysis and Applications of Hydrogels; InTech: London, UK, 2016. [Google Scholar] [CrossRef]
- Ling, Y.; Rubin, J.; Deng, Y.; Huang, C.; Demirci, U.; Karp, J.M.; Khademhosseini, A. A cell-laden microfluidic hydrogel. Lab Chip 2007, 7, 756. [Google Scholar] [CrossRef]
- Chung, S.; Sudo, R.; Vickerman, V.; Zervantonakis, I.K.; Kamm, R.D. Microfluidic Platforms for Studies of Angiogenesis, Cell Migration, and Cell–Cell Interactions. Ann. Biomed. Eng. 2010, 38, 1164–1177. [Google Scholar] [CrossRef]
- Osellame, R.; Hoekstra, H.J.; Cerullo, G.; Pollnau, M. Femtosecond laser microstructuring: An enabling tool for optofluidic lab-on-chips. Laser Photonics Rev. 2011, 5, 442–463. [Google Scholar] [CrossRef]
- Liao, Y.; Ju, Y.; Zhang, L.; He, F.; Zhang, Q.; Shen, Y.; Chen, D.; Cheng, Y.; Xu, Z.; Sugioka, K.; et al. Three-dimensional microfluidic channel with arbitrary length and configuration fabricated inside glass by femtosecond laser direct writing. Opt. Lett. 2010, 35, 3225–3227. [Google Scholar] [CrossRef]
- Maruo, S.; Nakamura, O.; Kawata, S. Three-dimensional microfabrication with two-photon-absorbed photopolymerization. Opt. Lett. 1997, 22, 132–134. [Google Scholar] [CrossRef]
- Cheng, D.; Jayne, R.K.; Tamborini, A.; Eyckmans, J.; White, A.E.; Chen, C.S. Studies of 3D directed cell migration enabled by direct laser writing of curved wave topography. Biofabrication 2019, 11, 021001. [Google Scholar] [CrossRef] [PubMed]
- Hjortø, G.M.; Olsen, M.H.; Svane, I.M.; Larsen, N.B. Confinement dependent chemotaxis in two-photon polymerized linear migration constructs with highly definable concentration gradients. Biomed. Microdevices 2015, 17, 30. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gökaltun, A.; Kang, Y.B.; Yarmush, M.L.; Usta, O.B.; Asatekin, A. Simple Surface Modification of Poly(dimethylsiloxane) via Surface Segregating Smart Polymers for Biomicrofluidics. Sci. Rep. 2019, 9, 7377. [Google Scholar] [CrossRef] [PubMed]
| References | Details | Constriction Characteristics | Schematic |
|---|---|---|---|
| PDMS—Straight channels array | |||
| Rolli et al. [47] 1 Fu et al. [48] 1 Tong et al. [49] 2 Irimia et al. [50] 1 Spuul et al. [51] 1 Zhou et al. [52] 2 Mishra and Vazquez [43] | Chemotaxis analysis, comparison of migration behaviors depending on channels dimensions or chemical stimuli | Constant cross-section. Channels characteristic dimension from 50 to m | 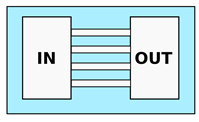 |
| PDMS—Microchannels with engineered profile | |||
| Mak et al. [53] Mak et al. [54] Ma et al. [55] Raman et al. [56] 2 Boneschansker et al. [57] | Study of migration strategies depending on the local 3D channel geometry, such as tapering or height modulation; Integration of cell traps or bendable micropillars as cell force probes | Variable cross-section. Width varying form 50 to m. Height varying from 15 to m | 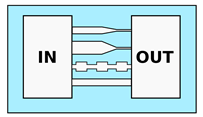 |
| PDMS—Micropillars | |||
| Doolin and Stroka [58] 2,3 Davidson et al. [59] 1 | Use of pillar arrays as ECM; Analysis of 2D cell motility depending on environment geometry; Study of cell migration through sub-nuclear dimension pores | Variable cross-section and 2D profile. Width varying from 50 to m | 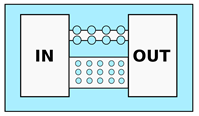 |
| PDMS—Fluidic Maze | |||
| Tweedy et al. [60] Belotti et al. [61] | Study of cell decision making during migration and cellular environment probing capacity (e.g., fluidic resistance or self-induced chemical gradient) | Constant single channel cross-section. Bifurcations, corners and widenings. Channels dimension from 5 to m | 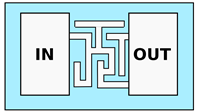 |
| Hydrogels—Microchannels | |||
| Cheng et al. [62] 1 Choi et al. [63] 1 Wang et al. [64] | Chemotaxis analysis, comparison of migration behaviors depending on channels dimensions or chemical stimuli; Possibility to modify mechanical properties of the channels, such as their stiffness | Constant cross-section. Channels dimension from 14 to m | 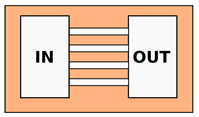 |
| Hydrogels—Migration matrix | |||
| Huang et al. [65] Anguiano et al. [66] Ayuso et al. [67] Truong et al. [68] Trappmann et al. [69] 4 | Use of hydrogel matrix as ECM, mimicking biological tissues in terms of porosity and stiffness. Possibility to embed the cells directly inside the matrix | No opened channels, cells migrate through the hydrogel. Possible presence of voids or pores with micrometric dimension. Mechanical stiffness ranges from few tens of Pa to tens of kPa (e.g., 18 kPa) | 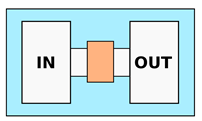 |
| FLM—Glass-based devices | |||
| Sima et al. [70] 2 Sima et al. [71] 2 | Microchannels with arbitrary cross-section realized in the bulk glass substrate. | Variable cross-section. Width varying from 5 to m | 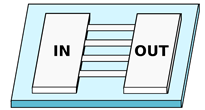 |
| FLM—Two-photon polymerization devices | |||
| Tayalia et al. [72] Olsen et al. [73] 2 Ficorella et al. [29] 1,2,3,4 Sala et al. [74] 1,2,3,4 | Polymeric 3D structures working as micrometer spatial constrains fabricated inside wider microfluidic channels. Possibility to arbitrary adjust the target geometry, from scaffolds or woodpiles to microchannels with arbitrary cross-section | Scaffold-like structure with porous size from m to m. Channel with variable cross-section, from m to m | 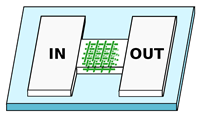 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sala, F.; Ficorella, C.; Osellame, R.; Käs, J.A.; Martínez Vázquez, R. Microfluidic Lab-on-a-Chip for Studies of Cell Migration under Spatial Confinement. Biosensors 2022, 12, 604. https://doi.org/10.3390/bios12080604
Sala F, Ficorella C, Osellame R, Käs JA, Martínez Vázquez R. Microfluidic Lab-on-a-Chip for Studies of Cell Migration under Spatial Confinement. Biosensors. 2022; 12(8):604. https://doi.org/10.3390/bios12080604
Chicago/Turabian StyleSala, Federico, Carlotta Ficorella, Roberto Osellame, Josef A. Käs, and Rebeca Martínez Vázquez. 2022. "Microfluidic Lab-on-a-Chip for Studies of Cell Migration under Spatial Confinement" Biosensors 12, no. 8: 604. https://doi.org/10.3390/bios12080604
APA StyleSala, F., Ficorella, C., Osellame, R., Käs, J. A., & Martínez Vázquez, R. (2022). Microfluidic Lab-on-a-Chip for Studies of Cell Migration under Spatial Confinement. Biosensors, 12(8), 604. https://doi.org/10.3390/bios12080604







