A Simple and Universal Nucleic Acid Assay Platform Based on Personal Glucose Meter Using SARS-CoV-2 N Gene as the Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Instruments
2.2. Preparation of Probe1−Invertase Conjugates
2.3. Procedures of PGM-NAAP
2.4. Detection of Target Nucleic Acid in Genomic DNA Using PGM-NAAP
2.5. Detection of Target Nucleic Acid in Serum and Saliva Samples Using PGM-NAAP
2.6. Statistical Data Analysis
3. Results and Discussion
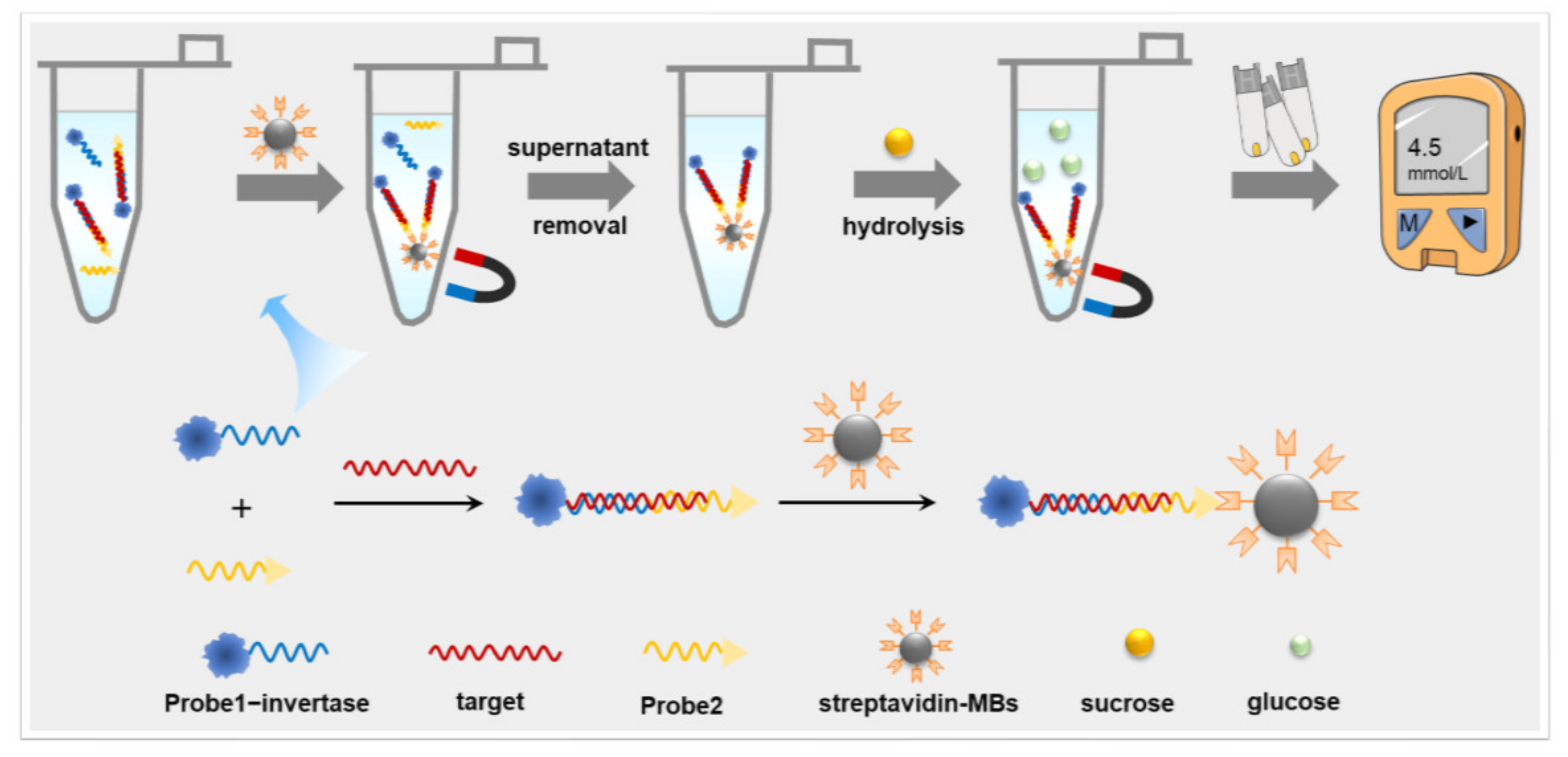
3.1. Principle of PGM-NAAP
3.2. Design and Selection of Sequences Targeted for the N-Gene of SARS-CoV-2
3.3. Optimization of Experimental Conditions
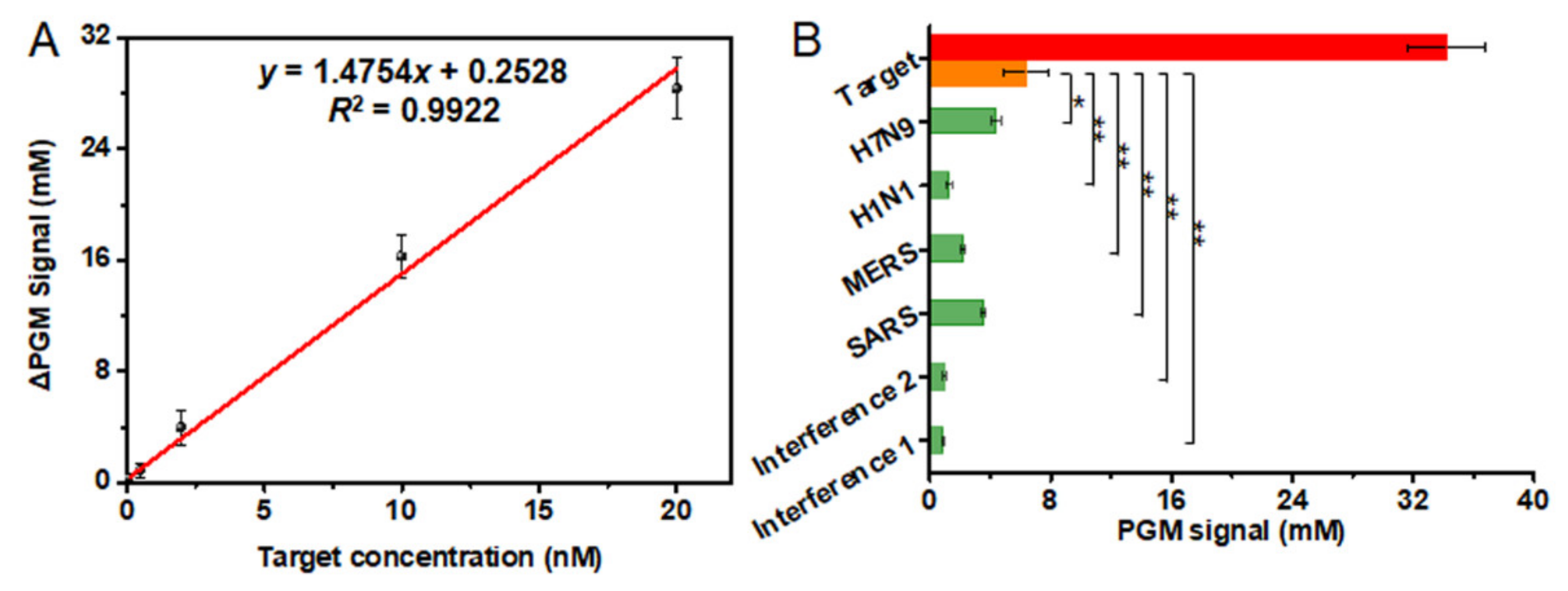
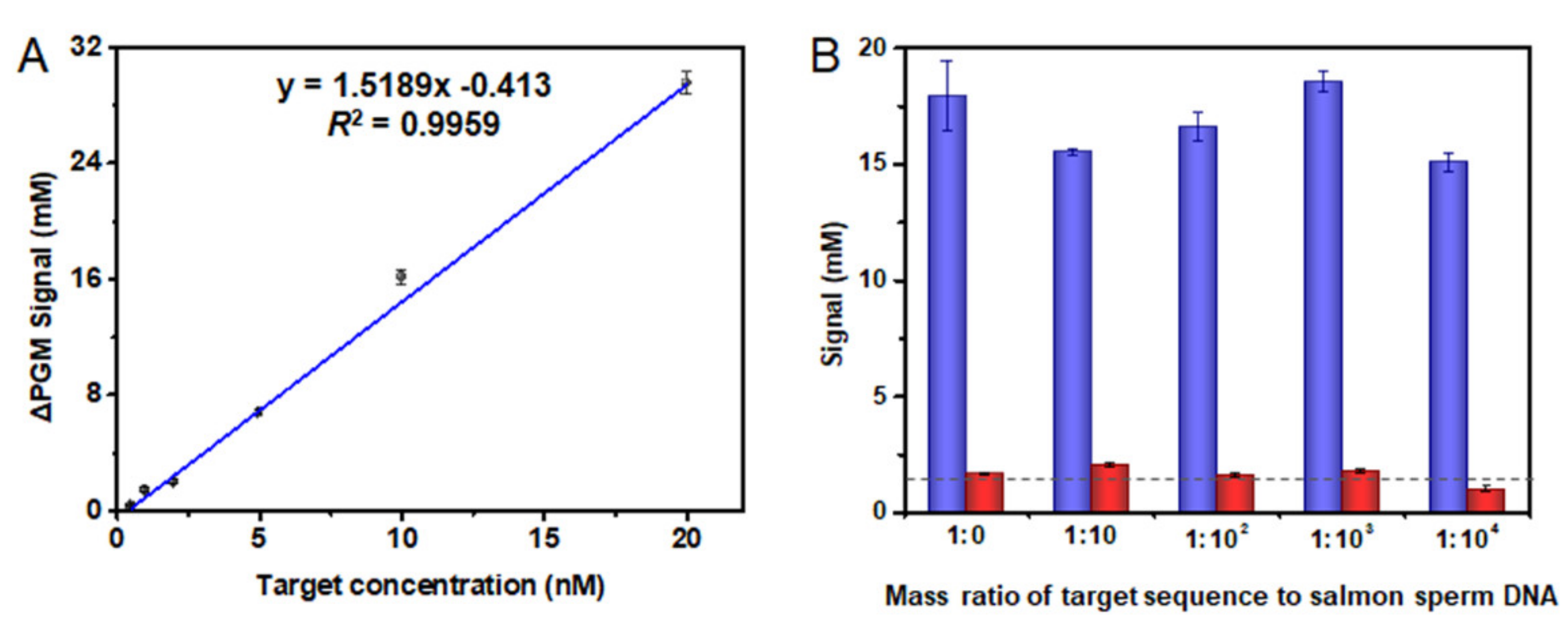
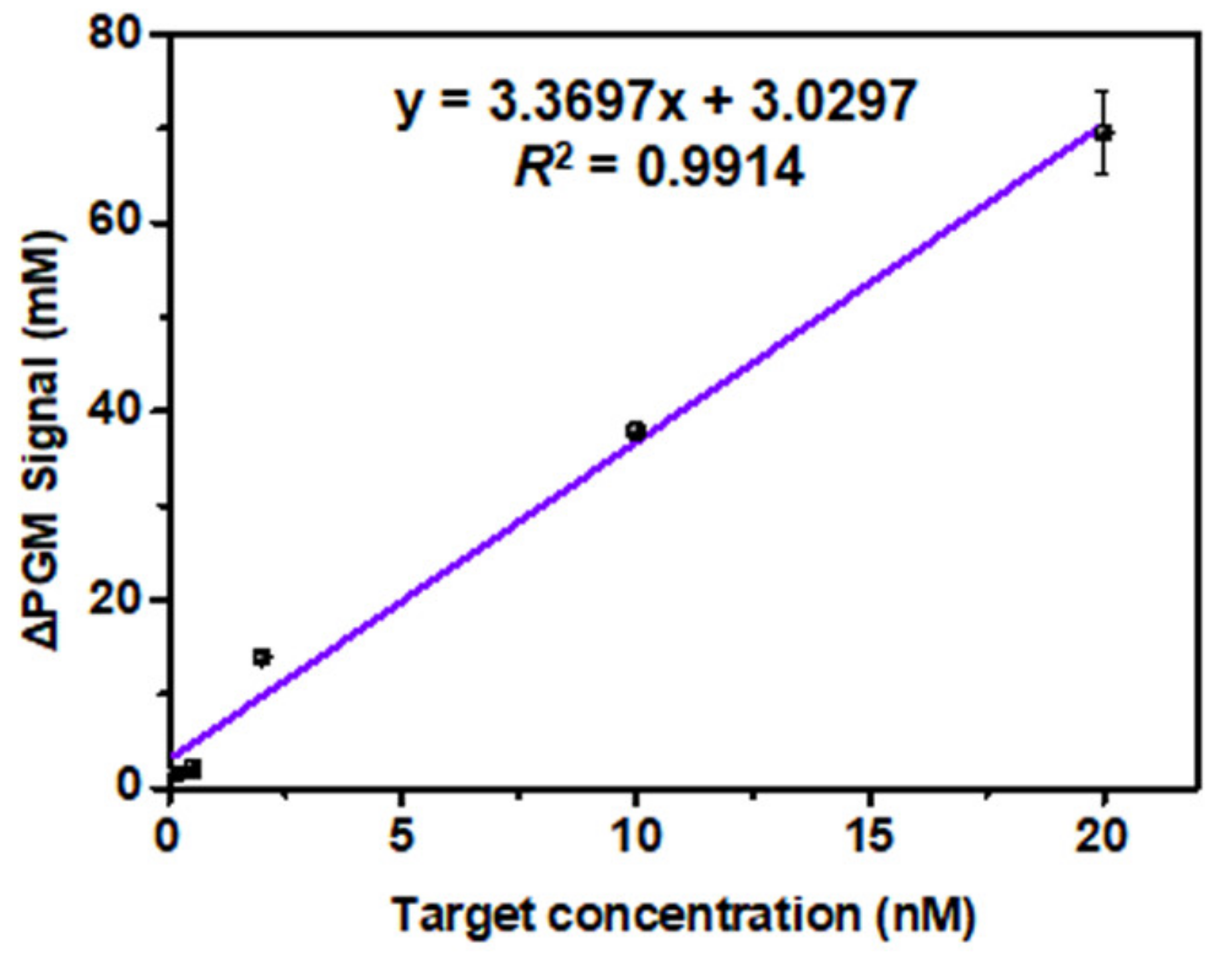
3.4. Analytical Characteristics of the PGM-NAAP
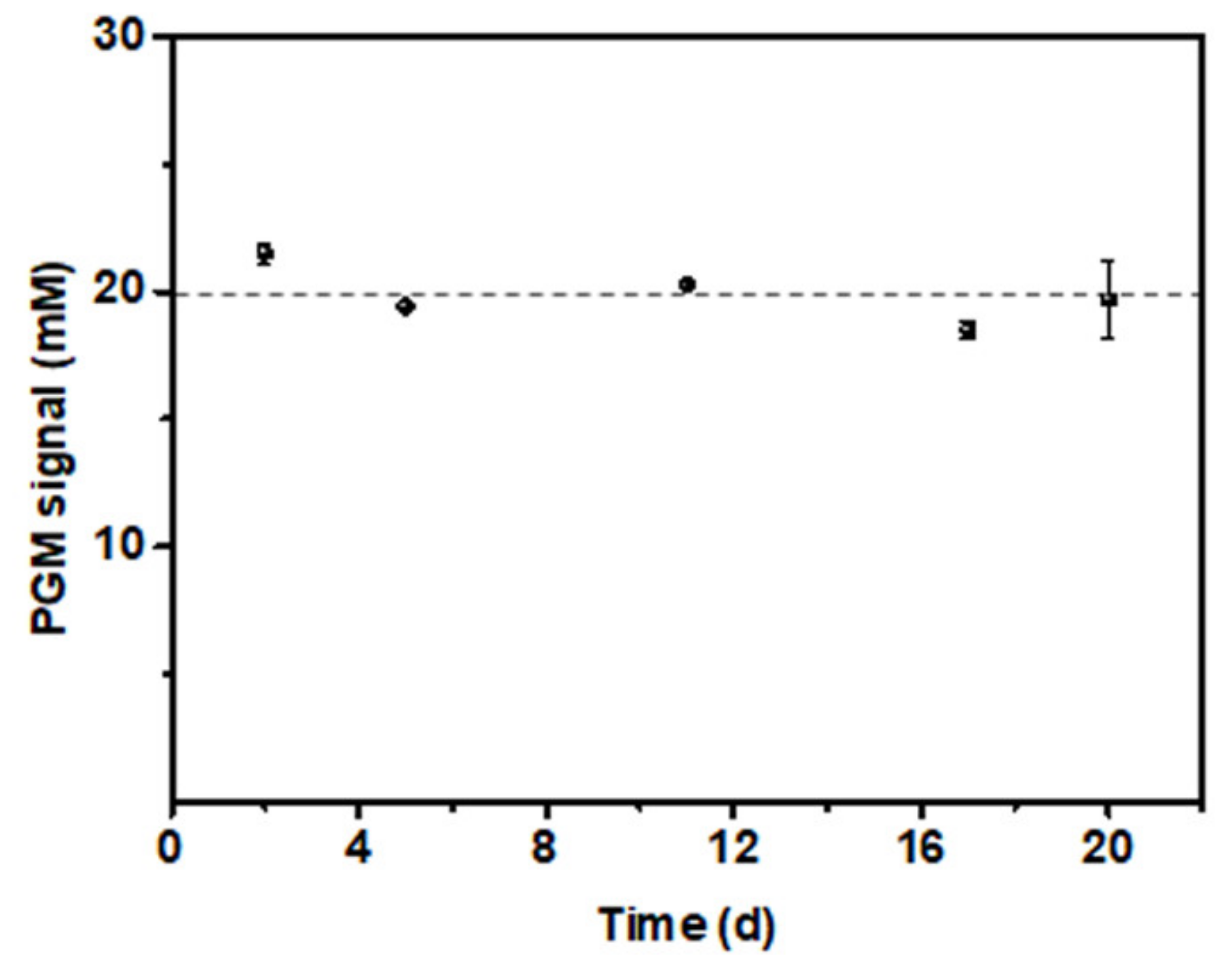
3.5. Stability Test of the PGM-NAAP
3.6. Detection of SARS-CoV-2 Related Nucleic Acid in Biological Fluids
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ravi, N.; Cortade, D.L.; Ng, E.; Wang, S.X. Diagnostics for SARS-CoV-2 detection: A comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020, 165, 112454. [Google Scholar] [CrossRef] [PubMed]
- Buitrago-Garcia, D.; Egli-Gany, D.; Counotte, M.J.; Hossmann, S.; Imeri, H.; Ipekci, A.M.; Salanti, G.; Low, N. Occurrence and transmission potential of asymptomatic and presymptomatic SARS-CoV-2 infections: A living systematic review and meta-analysis. PLoS Med. 2020, 17, e1003346. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Rabenau, H.F.; Adams, O.; Oberle, D.; Funk, M.B.; Keller-Stanislawski, B.; Timm, J.; Drosten, C.; Ciesek, S. SARS-CoV-2 asymptomatic and symptomatic patients and risk for transfusion transmission. Transfusion 2020, 60, 1119–1122. [Google Scholar] [CrossRef] [PubMed]
- Helmy, Y.A.; Fawzy, M.; Elaswad, A.; Sobieh, A.; Kenney, S.P.; Shehata, A.A. The COVID-19 Pandemic: A Comprehensive Review of Taxonomy, Genetics, Epidemiology, Diagnosis, Treatment, and Control. J. Clin. Med. 2020, 9, 1225. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y. The impact of the hypoxia-VEGF-vascular permeability on COVID-19-infected patients. Exploration 2021, 1, 20210051. [Google Scholar] [CrossRef]
- Singh, N.K.; Ray, P.; Carlin, A.F.; Magallanes, C.; Morgan, S.C.; Laurent, L.C.; Aronoff-Spencer, E.S.; Hall, D.A. Hitting the diagnostic sweet spot: Point-of-care SARS-CoV-2 salivary antigen testing with an off-the-shelf glucometer. Biosens. Bioelectron. 2021, 180, 113111. [Google Scholar] [CrossRef] [PubMed]
- Yuce, M.; Filiztekin, E.; Ozkaya, K.G. COVID-19 diagnosis -A review of current methods. Biosens. Bioelectron. 2021, 172, 112752. [Google Scholar] [CrossRef]
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C.W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14, 3822–3835. [Google Scholar] [CrossRef]
- Avivi-Mintz, S.; Lustig, Y.; Indenbaum, V.; Schwartz, E.; Danielli, A. Highly Sensitive and Specific SARS-CoV-2 Serological Assay Using a Magnetic Modulation Biosensing System. Biosensors 2022, 12, 7. [Google Scholar] [CrossRef]
- Grant, B.D.; Anderson, C.E.; Williford, J.R.; Alonzo, L.F.; Glukhova, V.A.; Boyle, D.S.; Weigl, B.H.; Nichols, K.P. SARS-CoV-2 Coronavirus Nucleocapsid Antigen-Detecting Half-Strip Lateral Flow Assay Toward the Development of Point of Care Tests Using Commercially Available Reagents. Anal. Chem. 2020, 92, 11305–11309. [Google Scholar] [CrossRef]
- Idili, A.; Parolo, C.; Alvarez-Diduk, R.; Merkoci, A. Rapid and Efficient Detection of the SARS-CoV-2 Spike Protein Using an Electrochemical Aptamer-Based Sensor. ACS Sens. 2021, 6, 3093–3101. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Ju, C.H.; Han, C.; Shi, R.; Chen, X.H.; Duan, D.M.; Yan, J.H.; Yan, X.Y. Nanozyme chemiluminescence paper test for rapid and sensitive detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 2021, 173, 112817. [Google Scholar] [CrossRef] [PubMed]
- Torrente-Rodriguez, R.M.; Lukas, H.; Tu, J.B.; Min, J.H.; Yang, Y.R.; Xu, C.H.; Rossiter, H.B.; Gao, W. SARS-CoV-2 RapidPlex: A Graphene-Based Multiplexed Telemedicine Platform for Rapid and Low-Cost COVID-19 Diagnosis and Monitoring. Matter 2020, 3, 1981–1998. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Irudayaraj, J. Paper-Based Test for Rapid On-Site Screening of SARS-CoV-2 in Clinical Samples. Biosensors 2021, 11, 488. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Zhou, H.S. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020, 165, 112349. [Google Scholar] [CrossRef] [PubMed]
- Ji, T.; Liu, Z.; Wang, G.; Guo, X.; Khan, S.A.; Lai, C.; Chen, H.; Huang, S.; Xia, S.; Chen, B.; et al. Detection of COVID-19: A review of the current literature and future perspectives. Biosens. Bioelectron. 2020, 166, 112455. [Google Scholar] [CrossRef]
- Zhao, H.; Liu, F.; Xie, W.; Zhou, T.-C.; OuYang, J.; Jin, L.; Li, H.; Zhao, C.-Y.; Zhang, L.; Wei, J.; et al. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sens. Actuators B Chem. 2021, 327, 128899. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.D.; Yu, G.X.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef]
- Ding, X.; Yin, K.; Li, Z.; Lalla, R.V.; Ballesteros, E.; Sfeir, M.M.; Liu, C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020, 11, 4711. [Google Scholar] [CrossRef]
- Huang, D.; Shi, Z.; Qian, J.; Bi, K.; Fang, M.; Xu, Z. A CRISPR-Cas12a-derived biosensor enabling portable personal glucose meter readout for quantitative detection of SARS-CoV-2. Biotechnol. Bioeng. 2021, 118, 1587–1596. [Google Scholar] [CrossRef]
- Moitra, P.; Alafeef, M.; Dighe, K.; Frieman, M.B.; Pan, D. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14, 7617–7627. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Shen, Y.; Wei, Y.; Yang, C.; Ge, X.; Wang, A.; Li, C.; Wan, Y.; Li, J. Botulinum toxin as an ultrasensitive reporter for bacterial and SARS-CoV-2 nucleic acid diagnostics. Biosens. Bioelectron. 2021, 176, 112953. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, K.G.; Neves Estrela, P.F.; Mendes, G.d.M.; dos Santos, C.A.; Silveira-Lacerda, E.d.P.; Mendes Duarte, G.R. Rapid molecular diagnostics of COVID-19 by RT-LAMP in a centrifugal polystyrene-toner based microdevice with end-point visual detection. Analyst 2021, 146, 1178–1187. [Google Scholar] [CrossRef] [PubMed]
- Nawattanapaiboon, K.; Pasomsub, E.; Prombun, P.; Wongbunmak, A.; Jenjitwanich, A.; Mahasupachai, P.; Vetcho, P.; Chayrach, C.; Manatjaroenlap, N.; Samphaongern, C.; et al. Colorimetric reverse transcription loop-mediated isothermal amplification (RT-LAMP) as a visual diagnostic platform for the detection of the emerging coronavirus SARS-CoV-2. Analyst 2021, 146, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Qian, C.Y.; Pang, Y.A.; Li, M.M.; Yang, Y.; Ma, H.J.; Zhao, M.Y.; Qian, F.; Yu, H.; Liu, Z.P.; et al. opvCRISPR: One-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens. Bioelectron. 2021, 172, 112766. [Google Scholar] [CrossRef] [PubMed]
- Xiong, E.; Jiang, L.; Tian, T.; Hu, M.; Yue, H.; Huang, M.; Lin, W.; Jiang, Y.; Zhu, D.; Zhou, X. Simultaneous Dual-Gene Diagnosis of SARS-CoV-2 Based on CRISPR/Cas9-Mediated Lateral Flow Assay. Angew. Chem. Int. Ed. 2021, 60, 5307–5315. [Google Scholar] [CrossRef]
- Gu, C.; Lan, T.; Shi, H.; Lu, Y. Portable Detection of Melamine in Milk Using a Personal Glucose Meter Based on an In Vitro Selected Structure-Switching Aptamer. Anal. Chem. 2015, 87, 7676–7682. [Google Scholar] [CrossRef]
- Xiang, Y.; Lan, T.; Lu, Y. Using the widely available blood glucose meter to monitor insulin and HbA1c. J. Diabetes Sci. Technol. 2014, 8, 855–858. [Google Scholar] [CrossRef]
- Xiang, Y.; Lu, Y. Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat. Chem. 2011, 3, 697–703. [Google Scholar] [CrossRef]
- Xiang, Y.; Lu, Y. An invasive DNA approach toward a general method for portable quantification of metal ions using a personal glucose meter. Chem. Commun. 2013, 49, 585–587. [Google Scholar] [CrossRef]
- Xiao, W.; Gao, Y.; Zhang, Y.; Li, J.; Liu, Z.; Nie, J.; Li, J. Enhanced 3D paper-based devices with a personal glucose meter for highly sensitive and portable biosensing of silver ion. Biosens. Bioelectron. 2019, 137, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Gong, J.; Rong, P.; Liu, C.; Chen, J. A portable and quantitative biosensor for cadmium detection using glucometer as the point-of-use device. Talanta 2019, 198, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.S.; Shi, D.M.; Zhu, S.M.; Wang, B.J.; Zhang, X.J.; Wang, G.F. Portable Aptasensor of Aflatoxin B1 in Bread Based on a Personal Glucose Meter and DNA Walking Machine. ACS Sensors 2018, 3, 1368–1375. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Cui, X.K.; Chivukula, V.; Iyer, S.S. Detection of Enzymes, Viruses, and Bacteria Using Glucose Meters. Anal. Chem. 2018, 90, 11589–11598. [Google Scholar] [CrossRef]
- Huang, H.; Zhao, G.; Dou, W. Portable and quantitative point-of-care monitoring of Escherichia coli O157:H7 using a personal glucose meter based on immunochromatographic assay. Biosens. Bioelectron. 2018, 107, 266–271. [Google Scholar] [CrossRef]
- Jang, Y.-J.; Lee, K.-H.; Yoo, T.H.; Kim, D.-M. Interfacing a Personal Glucose Meter with Cell-Free Protein Synthesis for Rapid Analysis of Amino Acids. Anal. Chem. 2019, 91, 2531–2535. [Google Scholar] [CrossRef]
- Lin, B.Q.; Liu, D.; Yan, J.M.; Qiao, Z.; Zhong, Y.X.; Yan, J.W.; Zhu, Z.; Ji, T.H.; Yang, C.J. Enzyme-Encapsulated Liposome-Linked Immunosorbent Assay Enabling Sensitive Personal Glucose Meter Readout for Portable Detection of Disease Biomarkers. ACS Appl. Mater. Interfaces 2016, 8, 6890–6897. [Google Scholar] [CrossRef]
- Wu, T.T.; Yang, Y.M.; Cao, Y.; Song, Y.C.; Xu, L.P.; Zhang, X.J.; Wang, S.T. Bioinspired DNA-Inorganic Hybrid Nanoflowers Combined with a Personal Glucose Meter for Onsite Detection of miRNA. ACS Appl. Mater. Interfaces 2018, 10, 42050–42057. [Google Scholar] [CrossRef]
- Xue, Q.W.; Kong, Y.C.; Wang, H.S.; Jiang, W. Liposome-encoded magnetic beads initiated by padlock exponential rolling circle amplification for portable and accurate quantification of microRNAs. Chem. Commun. 2017, 53, 10772–10775. [Google Scholar] [CrossRef]
- Zhang, J.J.; Lu, Y. Biocomputing for Portable, Resettable, and Quantitative Point-of-Care Diagnostics: Making the Glucose Meter a Logic-Gate Responsive Device for Measuring Many Clinically Relevant Targets. Angew. Chem. Int. Edit. 2018, 57, 9702–9706. [Google Scholar] [CrossRef]
- Amalfitano, E.; Karlikow, M.; Norouzi, M.; Jaenes, K.; Cicek, S.; Masum, F.; Mousavi, P.S.; Guo, Y.; Tang, L.; Sydor, A.; et al. A glucose meter interface for point-of-care gene circuit-based diagnostics. Nat. Commun. 2021, 12, 724. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Hu, Y.S.; He, Y.; Lan, T.; Zhang, J.J. Translating daily COVID-19 screening into a simple glucose test: A proof of concept study. Chem. Sci. 2021, 12, 9022–9030. [Google Scholar] [CrossRef] [PubMed]
- Toldra, A.; Alcaraz, C.; Diogene, J.; O’Sullivan, C.K.; Campas, M. Detection of Ostreopsis cf. ovata in environmental samples using an electrochemical DNA-based biosensor. Sci. Total Environ. 2019, 689, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Gaiani, G.; Cucchi, F.; Toldra, A.; Andree, K.B.; Rey, M.; Tsumuraya, T.; O’Sullivan, C.K.; Diogene, J.; Campas, M. Electrochemical biosensor for the dual detection of Gambierdiscus australes and Gambierdiscus excentricus in field samples. First report of G. excentricus in the Balearic Islands. Sci. Total Environ. 2022, 806, 150915. [Google Scholar] [CrossRef]
- Xiong, H.W.; Ye, X.; Li, Y.; Wang, L.J.; Zhang, J.; Fang, X.E.; Kong, J.L. Rapid Differential Diagnosis of Seven Human Respiratory Coronaviruses Based on Centrifugal Microfluidic Nucleic Acid Assay. Anal. Chem. 2020, 92, 14297–14302. [Google Scholar] [CrossRef]
- Wang, Y.X.; Zhang, Y.; Chen, J.B.; Wang, M.J.; Zhang, T.; Luo, W.X.; Li, Y.L.; Wu, Y.P.; Zeng, B.; Zhang, K.X.; et al. Detection of SARS-CoV-2 and Its Mutated Variants via CRISPR-Cas13-Based Transcription Amplification. Anal. Chem. 2021, 93, 3393–3402. [Google Scholar] [CrossRef]
- Pang, B.; Xu, J.; Liu, Y.; Peng, H.; Feng, W.; Cao, Y.; Wu, J.; Xiao, H.; Pabbaraju, K.; Tipples, G.; et al. Isothermal Amplification and Ambient Visualization in a Single Tube for the Detection of SARS-CoV-2 Using Loop-Mediated Amplification and CRISPR Technology. Anal. Chem. 2020, 92, 16204–16212. [Google Scholar] [CrossRef]
- Fu, X.; Sun, J.; Ye, Y.; Zhang, Y.; Sun, X. A rapid and ultrasensitive dual detection platform based on Cas12a for simultaneous detection of virulence and resistance genes of drug-resistant Salmonella. Biosens. Bioelectron. 2022, 195, 113682. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, T.; Pan, R.; Wen, Y.; Xu, J.; Zhang, L.; He, S.; Liang, G. A Simple and Universal Nucleic Acid Assay Platform Based on Personal Glucose Meter Using SARS-CoV-2 N Gene as the Model. Biosensors 2022, 12, 249. https://doi.org/10.3390/bios12040249
Li T, Pan R, Wen Y, Xu J, Zhang L, He S, Liang G. A Simple and Universal Nucleic Acid Assay Platform Based on Personal Glucose Meter Using SARS-CoV-2 N Gene as the Model. Biosensors. 2022; 12(4):249. https://doi.org/10.3390/bios12040249
Chicago/Turabian StyleLi, Tian, Rui Pan, Yuhan Wen, Jiaqi Xu, Liping Zhang, Suna He, and Gaofeng Liang. 2022. "A Simple and Universal Nucleic Acid Assay Platform Based on Personal Glucose Meter Using SARS-CoV-2 N Gene as the Model" Biosensors 12, no. 4: 249. https://doi.org/10.3390/bios12040249
APA StyleLi, T., Pan, R., Wen, Y., Xu, J., Zhang, L., He, S., & Liang, G. (2022). A Simple and Universal Nucleic Acid Assay Platform Based on Personal Glucose Meter Using SARS-CoV-2 N Gene as the Model. Biosensors, 12(4), 249. https://doi.org/10.3390/bios12040249



