Genetically Modified Soybean Detection Using a Biosensor Electrode with a Self-Assembled Monolayer of Gold Nanoparticles
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Experimental Process
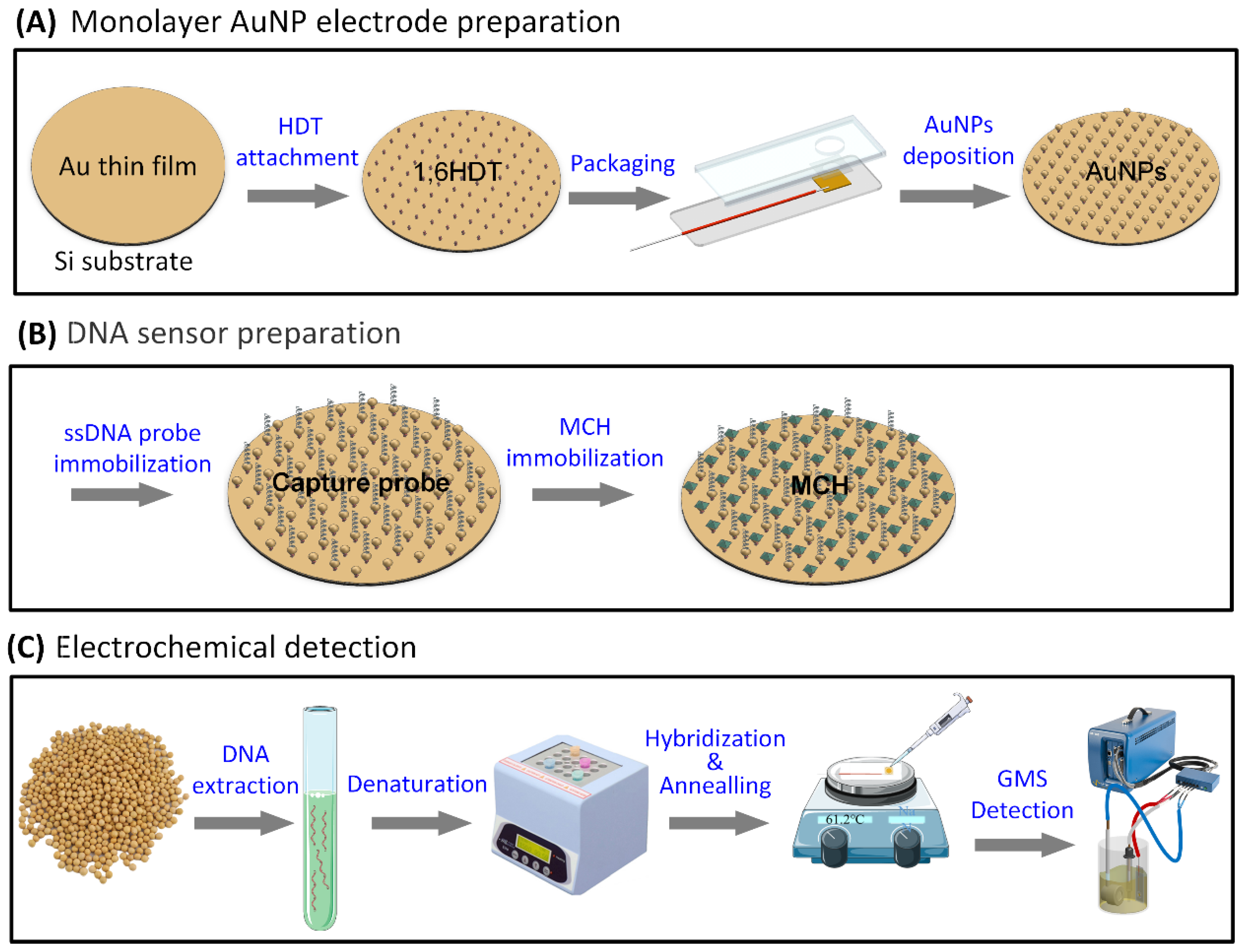
2.2.1. Monolayer AuNPs Electrode Preparation
2.2.2. DNA Sensor Preparation
2.2.3. Electrochemical Detection
- (1)
- Target dsDNA preparation
- (2)
- Detection of genetically modified soybean
2.3. Target dsDNA PCR Amplification and Gel Electrophoresis Analysis
2.4. The Quantification of dsDNA
3. Results and Discussion
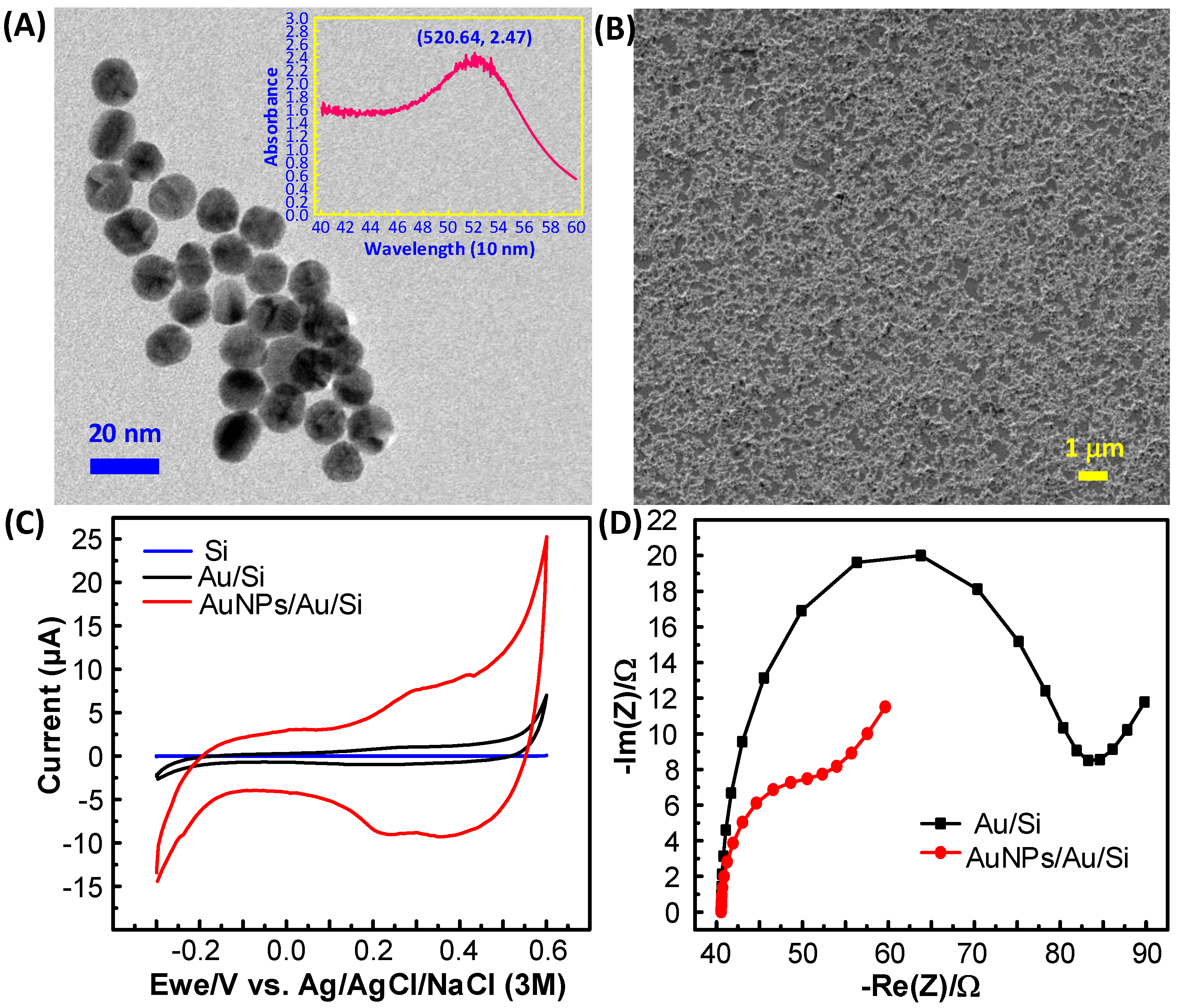
3.1. Characterization of the Monolayer AuNPs Electrode
3.2. Characterization of the Extracted Target dsDNA
3.3. GMS Detection
- (1)
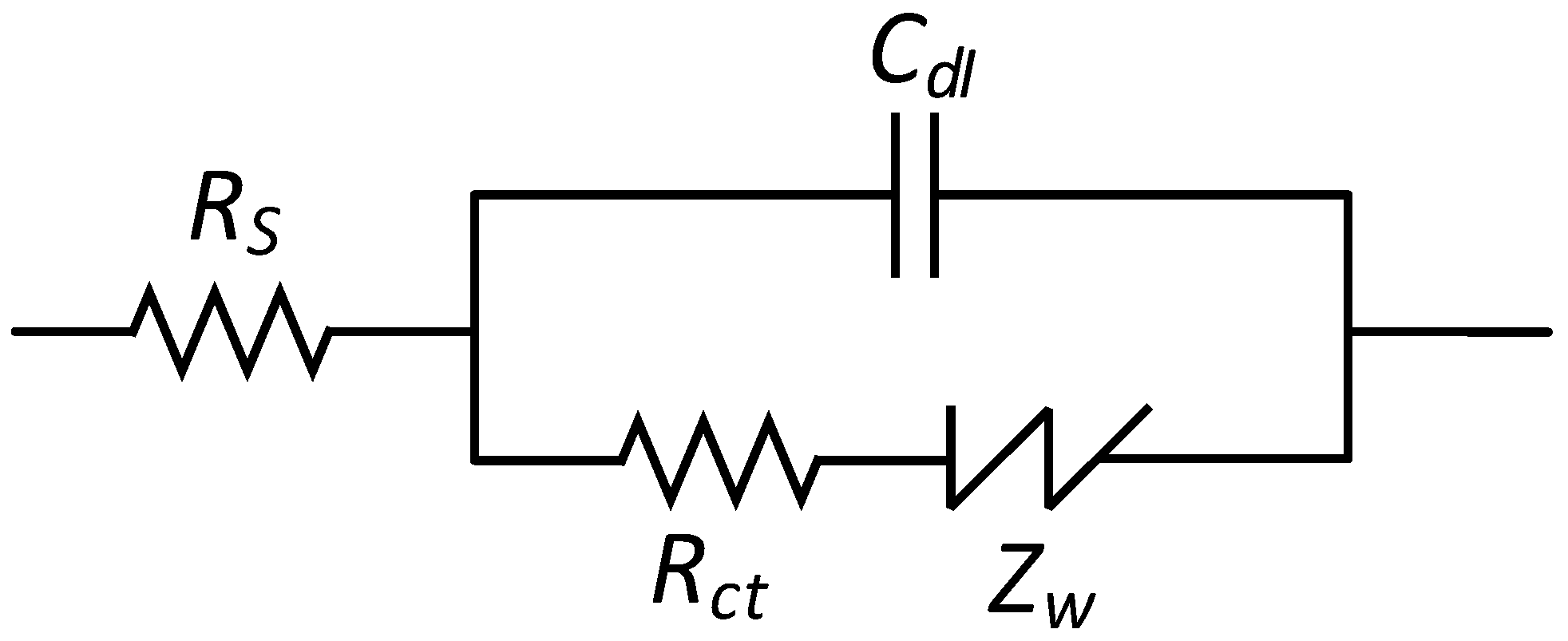
- EIS characteristics of the sensing electrode
- (2)
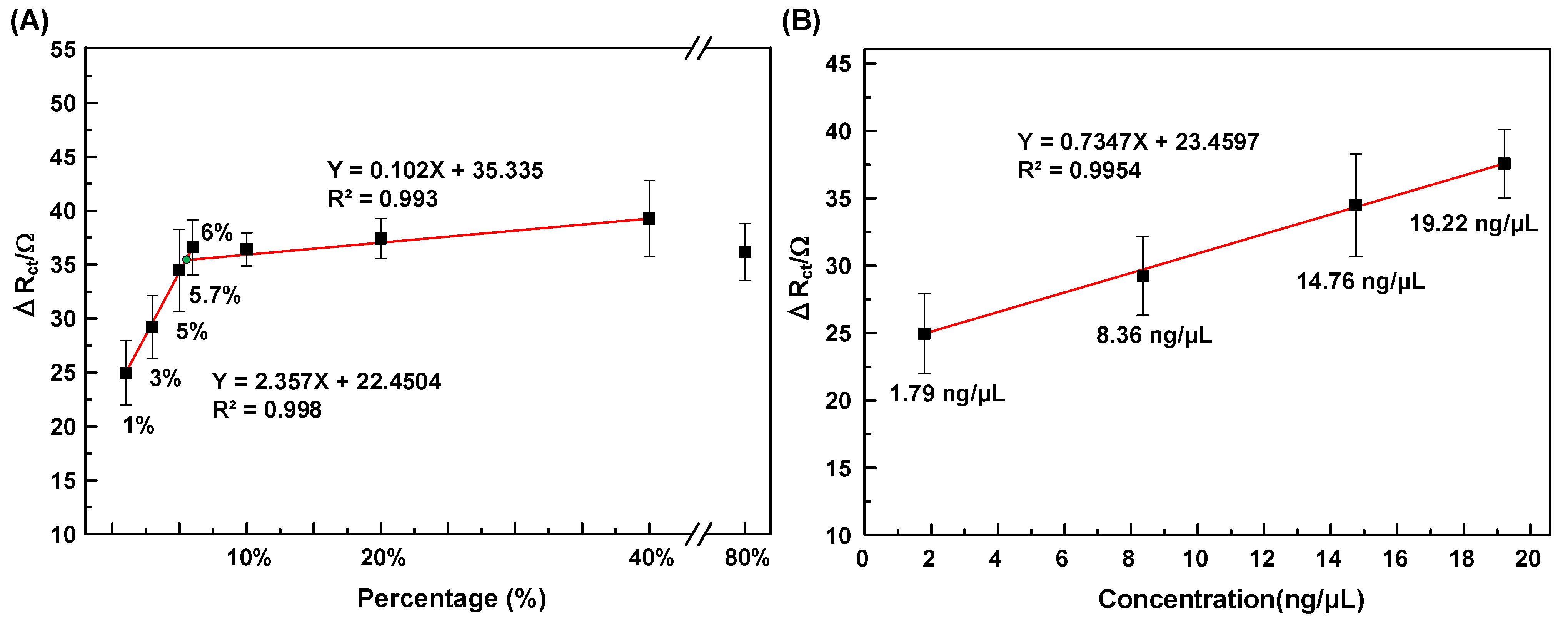
- Calibration curve for GMS detection
- (3)
- Performance comparisons
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Key, S.; Ma, J.K.-C.; Drake, P.M. Genetically modified plants and human health. J. R. Soc. Med. 2008, 101, 290–298. [Google Scholar] [CrossRef] [PubMed]
- James, C. 20th Anniversary (1996 to 2015) of the Global Commercialization of Biotech Crops and Biotech Crop Highlights in 2015; ISAAA Brief No. 51; ISAAA: Ithaca, NY, USA, 2015. [Google Scholar]
- Mohajer, M.B.; Ardekani, A.M. Genetically modified foods and social concerns. Avicenna J. Med. Biotechnol. 2011, 3, 109–117. [Google Scholar]
- Lucht, J.M. Public Acceptance of Plant Biotechnology and GM Crops. Viruses 2015, 7, 4254–4281. [Google Scholar] [CrossRef]
- Ponti, L. Transgenic Crops and Sustainable Agriculture in the European Context. Bull. Sci. Technol. Soc. 2005, 25, 289–305. [Google Scholar] [CrossRef]
- Dona, A.; Arvanitoyannis, I.S. Health Risks of Genetically Modified Foods. Crit. Rev. Food Sci. Nutr. 2009, 49, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Ujj, O. European and American views on genetically modified foods. New Atlantis 2016, 49, 77–92. [Google Scholar]
- Kamle, M.; Kumar, P.J.; Patra, J.K.; Bajpai, V.K. Current perspectives on genetically modified crops and detection methods. 3 Biotech 2017, 7, 219. [Google Scholar] [CrossRef]
- Qian, C.; Wang, R.; Wu, H.; Ping, J.; Wu, J. Recent advances in emerging DNA-based methods for genetically modified organisms (GMOs) rapid detection. TrAC Trends Anal. Chem. 2018, 109, 19–31. [Google Scholar] [CrossRef]
- Akiyama, H.; Makiyama, D.; Nakamura, K.; Sasaki, N.; Minegishi, Y.; Mano, J.; Kitta, K.; Ozeki, Y.; Teshima, R. A Novel Detection System for the Genetically Modified Canola (Brassica rapa) Line RT73. Anal. Chem. 2010, 82, 9909–9916. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zhang, M.; Ma, M.; Hai, H.; Li, J.; Shan, Y. A novel electrochemical DNA biosensor for transgenic soybean detection based on triple signal amplification. Anal. Chim. Acta 2019, 1078, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Xu, Y.; Huang, K.; Luo, Y.; Xu, W. Multiplex pyrosequencing quantitative detection combined with universal primer-multiplex-PCR for genetically modified organisms. Food Chem. 2020, 320, 126634. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhang, X.; Wu, B.; Qian, C.; Zhang, F.; Wang, L.; Ye, Z.; Wu, J. Rapid on-site detection of genetically modified soybean products by real-time loop-mediated isothermal amplification coupled with a designed portable amplifier. Food Chem. 2020, 323, 126819. [Google Scholar] [CrossRef] [PubMed]
- Takabatake, R.; Kagiya, Y.; Minegishi, Y.; Yeasmin, S.; Futo, S.; Noguchi, A.; Kondo, K.; Mano, J.; Kitta, K. Development and evaluation of rapid screening detection methods for genetically modified crops using loop-mediated isothermal amplification. Food Chem. 2018, 252, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Emslie, K.R.; Whaites, L.; Griffiths, K.R.; Murby, E.J. Sampling Plan and Test Protocol for the Semiquantitative Detection of Genetically Modified Canola (Brassica napus) Seed in Bulk Canola Seed. J. Agric. Food Chem. 2007, 55, 4414–4421. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.-J.; Cho, I.-H.; Kim, H.-S.; Jeon, J.-W.; Hwang, S.-Y.; Paek, S.-H. Development of a chemiluminometric immunosensor array for on-site monitoring of genetically modified organisms. Sens. Actuators B Chem. 2011, 155, 598–605. [Google Scholar] [CrossRef]
- Zeng, H.; Wang, J.; Jia, J.; Wu, G.; Yang, Q.; Liu, X.; Tang, X. Development of a lateral flow test strip for simultaneous detection of BT-Cry1Ab, BT-Cry1Ac and CP4 EPSPS proteins in genetically modified crops. Food Chem. 2020, 335, 127627. [Google Scholar] [CrossRef]
- Broeders, S.; Fraiture, M.-A.; Vandermassen, E.; Delvoye, M.; Barbau-Piednoir, E.; Lievens, A.; Roosens, N. New qualitative trait-specific SYBR®Green qPCR methods to expand the panel of GMO screening methods used in the CoSYPS. Eur. Food Res. Technol. 2015, 241, 275–287. [Google Scholar] [CrossRef][Green Version]
- Broeders, S.; Papazova, N.; Van den Bulcke, M.; Roosens, N. Development of a Molecular Platform for GMO Detection in Food and Feed on the Basis of “Combinatory qPCR” Technology; Hernandez-Rodriguez, P., Gome, A.P.R., Eds.; Polymerase Chain Reaction, InTech: Rijeka, Croatia, 2012; Volume 1, pp. 363–404. [Google Scholar]
- Salisu, I.B.; Shahid, A.A.; Yaqoob, A.; Ali, Q.; Bajwa, K.S.; Rao, A.Q.; Husnain, T. Molecular Approaches for High Throughput Detection and Quantification of Genetically Modified Crops: A Review. Front. Plant Sci. 2017, 8, 1670. [Google Scholar] [CrossRef] [PubMed]
- López, M.S.-P.; Manzanares-Palenzuela, C.L.; López-Ruiz, B. Biosensors for GMO Testing: Nearly 25 Years of Research. Crit. Rev. Anal. Chem. 2018, 48, 391–405. [Google Scholar] [CrossRef] [PubMed]
- Manzanares-Palenzuela, C.L.; Martín-Fernández, B.; Sánchez-Paniagua López, M.; López-Ruiz, B. Electrochemical genosensors as innovative tools for detection of genetically modified organisms. Trends Anal. Chem. 2015, 66, 19–31. [Google Scholar] [CrossRef]
- Wang, S.; Liu, Q.; Li, H.; Li, Y.; Hao, N.; Qian, J.; Zhu, W.; Wang, K. Fabrication of label-free electrochemical impedimetric DNA biosensor for detection of genetically modified soybean by recognizing CaMV 35S promoter. J. Electroanal. Chem. 2016, 782, 19–25. [Google Scholar] [CrossRef]
- Tam, P.D. Genetically modified organism (GMO) detection by biosensor based on SWCNT material. Curr. Appl. Phys. 2015, 15, 397–401. [Google Scholar] [CrossRef]
- Sun, W.; Zhang, Y.; Hu, A.; Lu, Y.; Shi, F.; Lei, B.; Sun, Z. Electrochemical DNA Biosensor Based on Partially Reduced Graphene Oxide Modified Carbon Ionic Liquid Electrode for the Detection of Transgenic Soybean A2704-12 Gene Sequence. Electroanalysis 2013, 25, 1417–1424. [Google Scholar] [CrossRef]
- Aghili, Z.; Nasirizadeh, N.; Divsalar, A.; Shoeibi, S.; Yaghmaei, P. A nanobiosensor composed of Exfoliated Graphene Oxide and Gold Nano-Urchins, for detection of GMO products. Biosens. Bioelectron. 2017, 95, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Manzanares-Palenzuela, C.; Martín-Clemente, J.; Lobo-Castañón, M.; López-Ruiz, B. Electrochemical detection of magnetically-entrapped DNA sequences from complex samples by multiplexed enzymatic labelling: Application to a transgenic food/feed quantitative survey. Talanta 2017, 164, 261–267. [Google Scholar] [CrossRef]
- Barroso, M.F.; Freitas, M.; Oliveira, M.B.P.; De-Los-Santos-Álvarez, N.; Lobo-Castañón, M.J.; Delerue-Matos, C. 3D-nanostructured Au electrodes for the event-specific detection of MON810 transgenic maize. Talanta 2015, 134, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.-C.; Chuang, M.-C.; Ho, J.-A.A. Electrochemical sensor for multiplex screening of genetically modified DNA: Identification of biotech crops by logic-based biomolecular analysis. Biosens. Bioelectron. 2013, 50, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Manzanares-Palenzuela, C.L.; Mafra, I.; Costa, J.; Barroso, M.F.; De-Los-Santos-Álvarez, N.; Delerue-Matos, C.; Oliveira, M.B.P.; Lobo-Castañón, M.J.; López-Ruiz, B. Electrochemical magnetoassay coupled to PCR as a quantitative approach to detect the soybean transgenic event GTS40-3-2 in foods. Sens. Actuators B Chem. 2016, 222, 1050–1057. [Google Scholar] [CrossRef]
- Bak, A.; Emerson, J.B. Cauliflower mosaic virus (CaMV) Biology, Management, and Relevance to GM Plant Detection for Sustainable Organic Agriculture. Front. Sustain. Food Syst. 2020, 4, 21. [Google Scholar] [CrossRef]
- Arugula, M.A.; Zhang, Y.; Simonian, A.L. Biosensors as 21st Century Technology for Detecting Genetically Modified Organisms in Food and Feed. Anal. Chem. 2013, 86, 119–129. [Google Scholar] [CrossRef]
- Mahmoud, A.M.; Tang, T.; Harrison, D.J.; Lee, W.E.; Jemere, A.B. A regenerating self-assembled gold nanoparticle-containing electrochemical impedance sensor. Biosens. Bioelectron. 2014, 56, 328–333. [Google Scholar] [CrossRef]
- Barsoukov, E.; Macdonald, J.R. Impedance Spectroscopy: Theory, Experiment, and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2005. [Google Scholar]
- Deng, Y.; Wu, J.; Tu, K.; Xu, H.; Ma, L.; Chen, J.; Wang, J. Preparation of DNA-based biosensor for electrochemically identification of transgenic soybean. Int. J. Electrochem. Sci. 2020, 15, 4556–4566. [Google Scholar] [CrossRef]
- Li, Y.; Sun, L.; Liu, Q.; Han, E.; Hao, N.; Zhang, L.; Wang, S.; Cai, J.; Wang, K. Photoelectrochemical CaMV35S biosensor for discriminating transgenic from non-transgenic soybean based on SiO2@CdTe quantum dots core-shell nanoparticles as signal indicators. Talanta 2016, 161, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Jamaluddin, R.Z.A.R.; Heng, L.Y.; Tan, L.L.; Chong, K.F. A Biosensor for Genetic Modified Soybean DNA Determination via Adsorption of Anthraquinone-2-sulphonic Acid in Reduced Graphene Oxide. Electroanalysis 2017, 30, 250–258. [Google Scholar] [CrossRef]


| Con. (%) | Measured Con. (ng/μL) | Real Con. (ng/μL) | Recov. Rate (%) | RSD (%) |
|---|---|---|---|---|
| 4% | 11.399 | 10.96 | 104.01 | 6.24 |
| 5.7% | 16.912 | 16.5 | 102.49 | 2.54 |
| Electrode | Detection Method | Sample Type | Linear Range | LOD | Refer. |
|---|---|---|---|---|---|
| AuNPs/MWCNT-rGONR | EIS (label-free) | PCR amplify. | 1 × 10−16–5 × 10−10 M (PCR) | 3.3 × 10−17 M (PCR) | [23] |
| ROG/CPE | DPV with Methylene blue indicator | PCR amplify. | 3 × 10−15–5 × 10−7 M (PCR) | 1.7 × 10−5 M (PCR) | [35] |
| AuNPs-rGO | PEC with (SiO2@CdTe QDs) indicator | PCR amplify. | 1 × 10−16–5 × 10−10 M (PCR) | 5 × 10−17 M (PCR) | [36] |
| Poly(nBA-NAS)-rGO | DPV with (AQMS) indicator | Real sample | 1 × 10−15–1 × 10−8 M (Synthetic) | 6.3 × 10−16 M (Synthetic) | [37] |
| Monolayer AuNPs | EIS (label-free) | Real sample | 1.792–1.922 × 101 ng/mL | 1.792 ng/mL | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chou, C.-C.; Lin, Y.-T.; Kuznetsova, I.; Wang, G.-J. Genetically Modified Soybean Detection Using a Biosensor Electrode with a Self-Assembled Monolayer of Gold Nanoparticles. Biosensors 2022, 12, 207. https://doi.org/10.3390/bios12040207
Chou C-C, Lin Y-T, Kuznetsova I, Wang G-J. Genetically Modified Soybean Detection Using a Biosensor Electrode with a Self-Assembled Monolayer of Gold Nanoparticles. Biosensors. 2022; 12(4):207. https://doi.org/10.3390/bios12040207
Chicago/Turabian StyleChou, Cheng-Chi, Ying-Ting Lin, Iren Kuznetsova, and Gou-Jen Wang. 2022. "Genetically Modified Soybean Detection Using a Biosensor Electrode with a Self-Assembled Monolayer of Gold Nanoparticles" Biosensors 12, no. 4: 207. https://doi.org/10.3390/bios12040207
APA StyleChou, C.-C., Lin, Y.-T., Kuznetsova, I., & Wang, G.-J. (2022). Genetically Modified Soybean Detection Using a Biosensor Electrode with a Self-Assembled Monolayer of Gold Nanoparticles. Biosensors, 12(4), 207. https://doi.org/10.3390/bios12040207







