Cuff-Less Blood Pressure Prediction from ECG and PPG Signals Using Fourier Transformation and Amplitude Randomization Preprocessing for Context Aggregation Network Training
Abstract
:1. Introduction
2. Materials and Methods
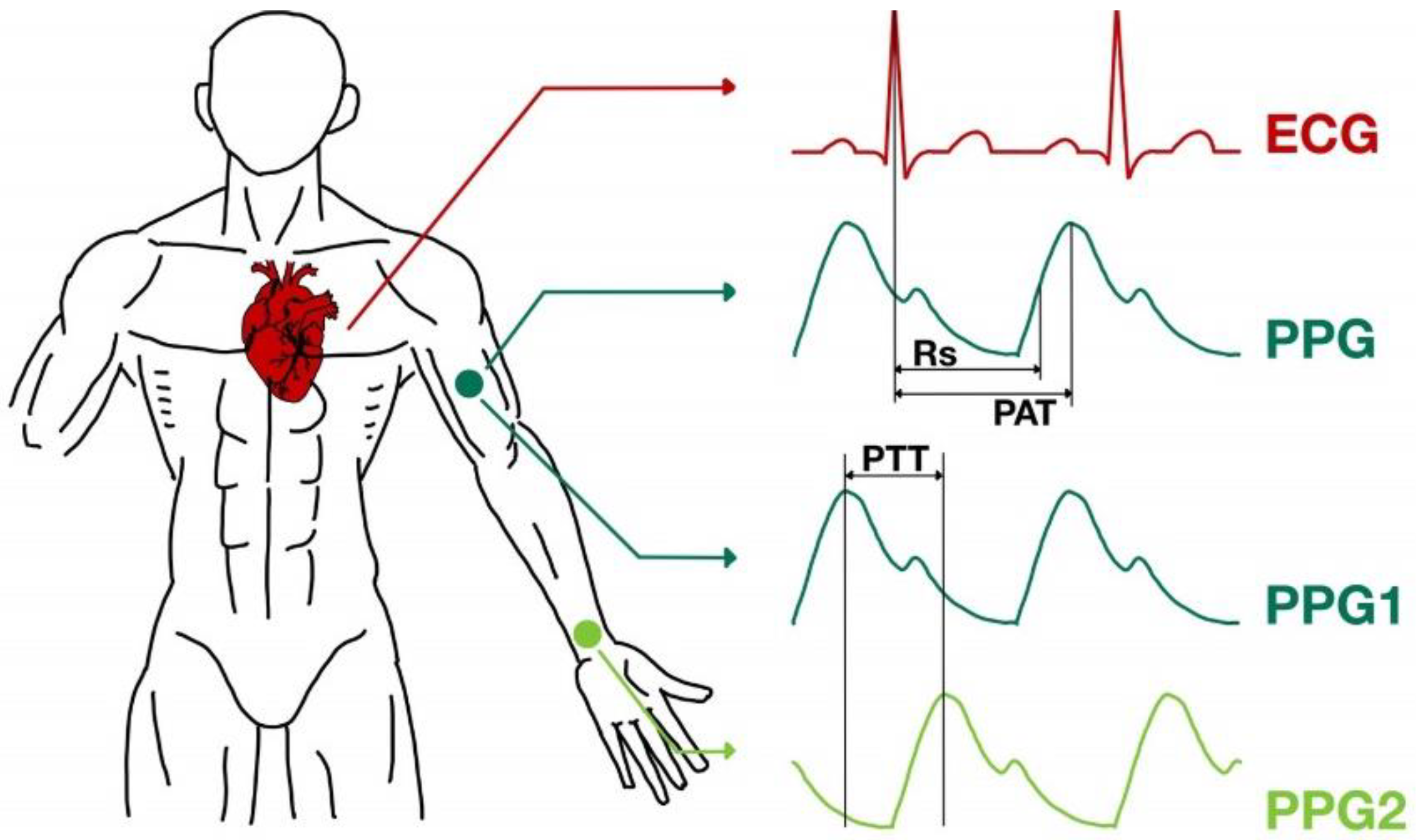
2.1. ECG and PPG Signal Database
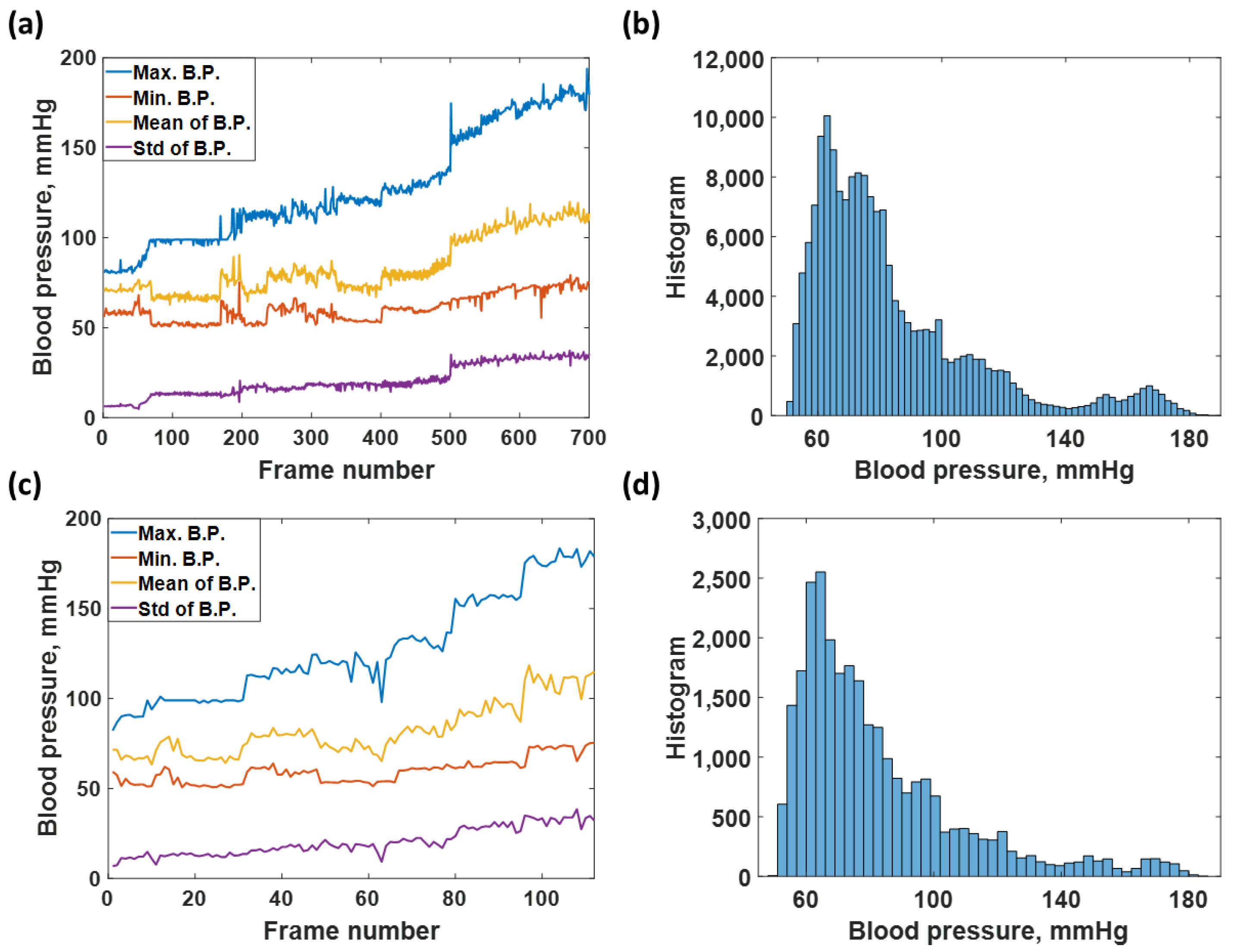
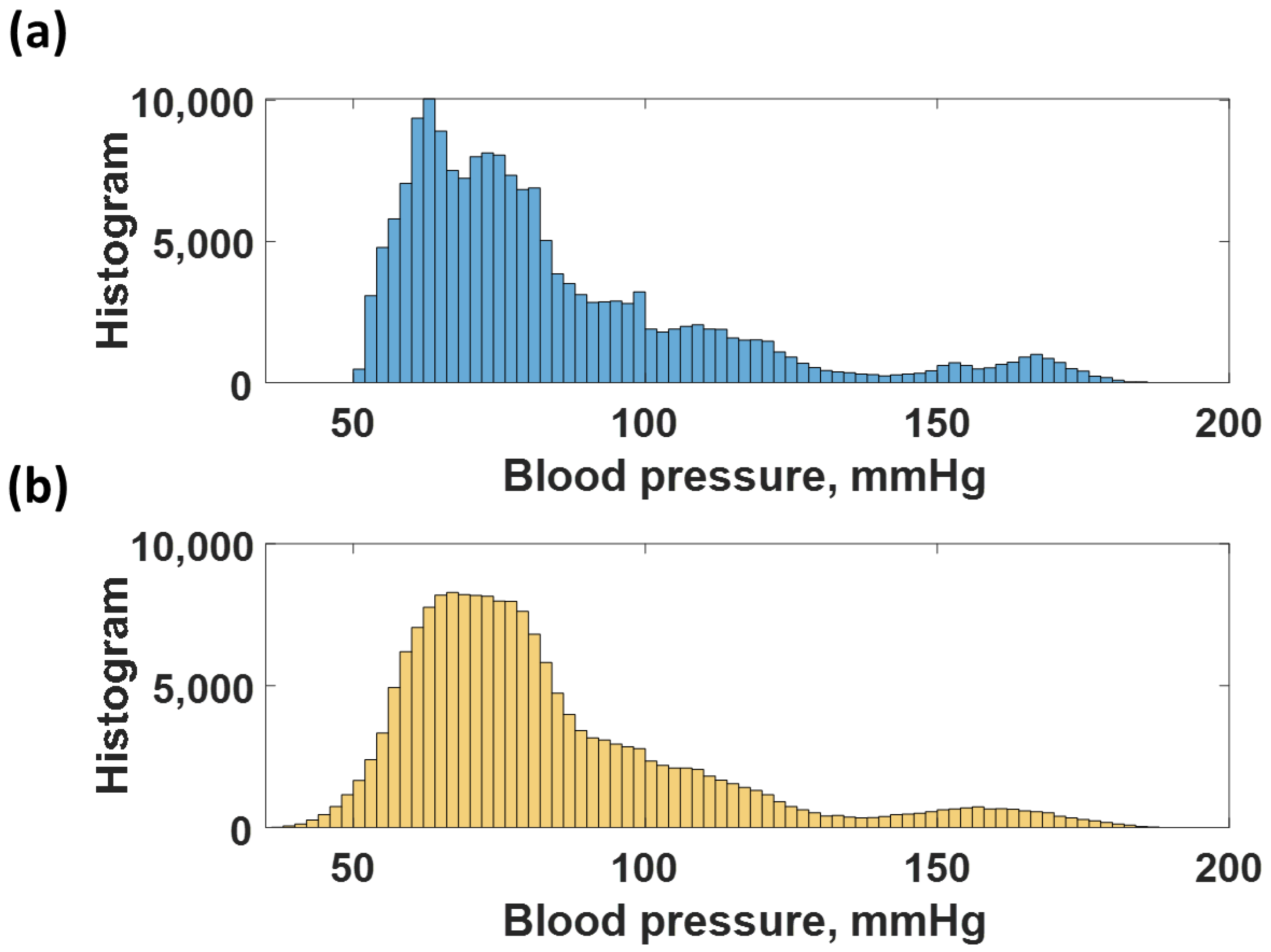
2.2. Training Dataset and Test Dataset
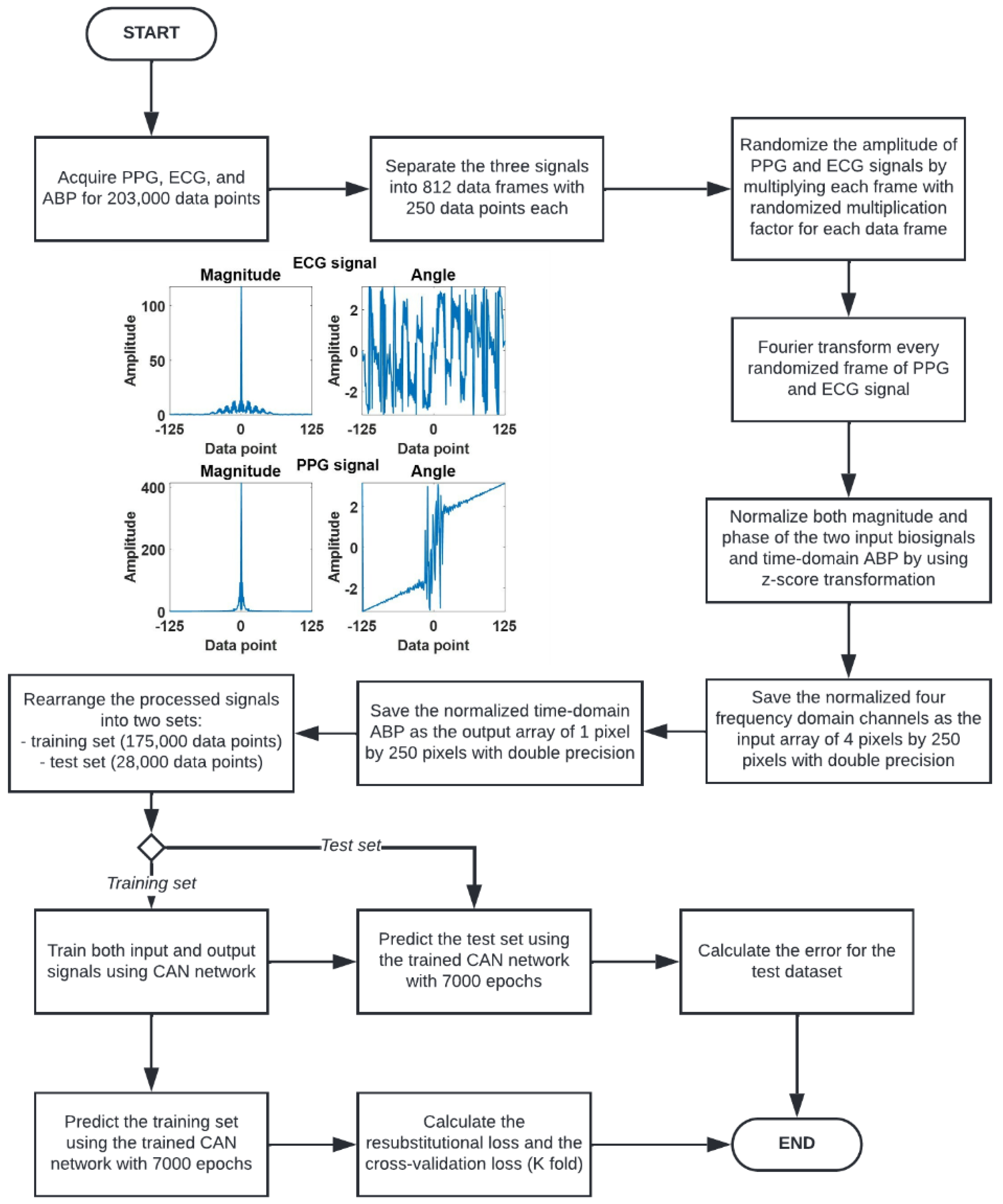
2.3. Signal Preconditioning
- 1.
- Acquire the PPG, ECG, and ABP signals for 203,000 data points.
- 2.
- Separate the three signals to 203,000 data points into 812 data frames at 250 data points for each data frame. Each frame of the 250 data points represented a 2 s data window.
- 3.
- Randomize the amplitude of PPG and ECG signals by multiplying each 2 s window with two randomized multiplication factors for each signal using a uniform random number generator ranging from 0 to 1.
- 4.
- Fourier transform every randomized 2 s PPG and ECG signal window. The Fourier transform gives out signals in terms of amplitude and phase, leading to 4 frequency domain channels: the amplitude of ECG, the phase of ECG, the amplitude of PPG, and the phase of PPG, respectively.
- 5.
- The four frequency domain channels are then z-score transformed to ensure that the frequency data is appropriate for neural network training. These four channels are then saved as an array of 4 pixels by 250 pixels with double precision.
- 6.
- The label of the input arrays is prepared by z-transforming the corresponding ABP signal in the time domain. Note that the input to the CAN was the four channels of frequency-domain data, and the label is the corresponding ABP signal in the time domain.
2.4. Context Aggregation Neural Network (CAN) Training
- 1.
- Resubstitution RMSE is computed by evaluating the root mean square error between the training labels and outputs predicted with the training data using the network trained using the labels and the training dataset [33,34,35]. In other words, how accurately the network can predict the labels of the training dataset.
- 2.
- Cross-validation RMSE or K-fold cross-validation; the network performance is evaluated using validation K-fold of 5 by separating the training and validation dataset into five sub-datasets. Note that the members of each sub-datasets were chosen at random and then trained five separate networks using each sub-dataset. The cross-validation RMSE was then computed as the average RMSE error of the five networks.
3. Results
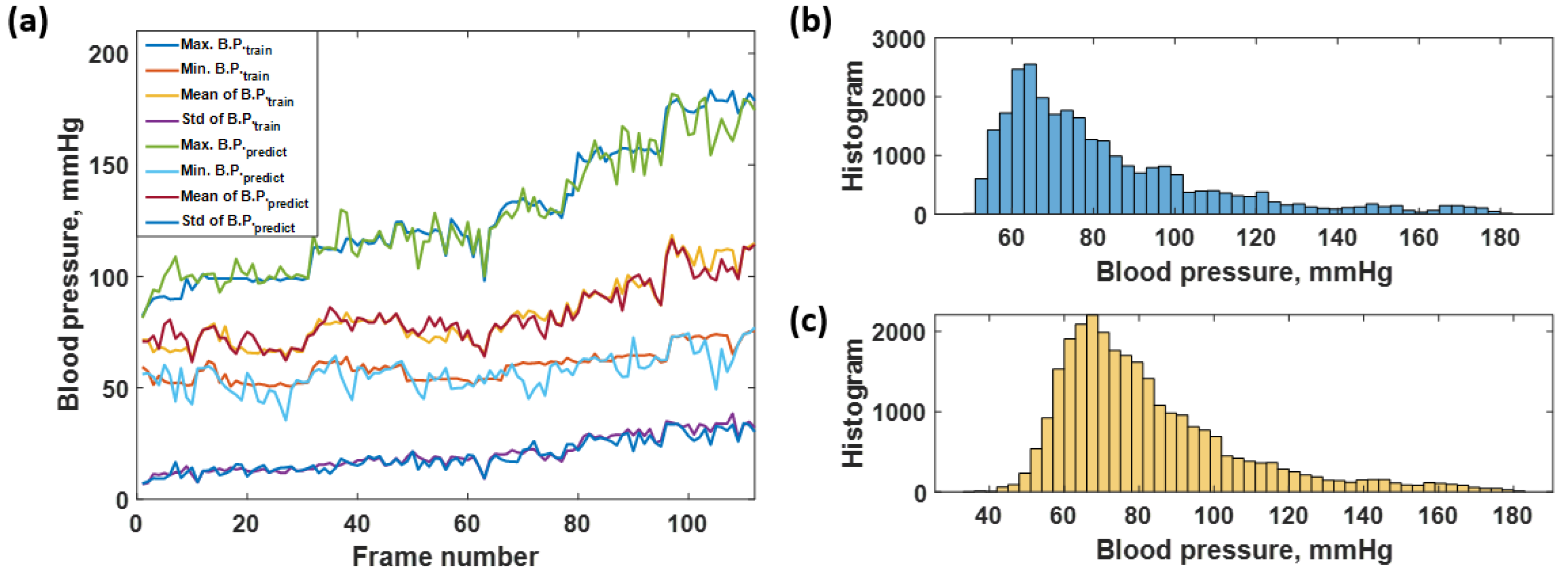
3.1. Network Training
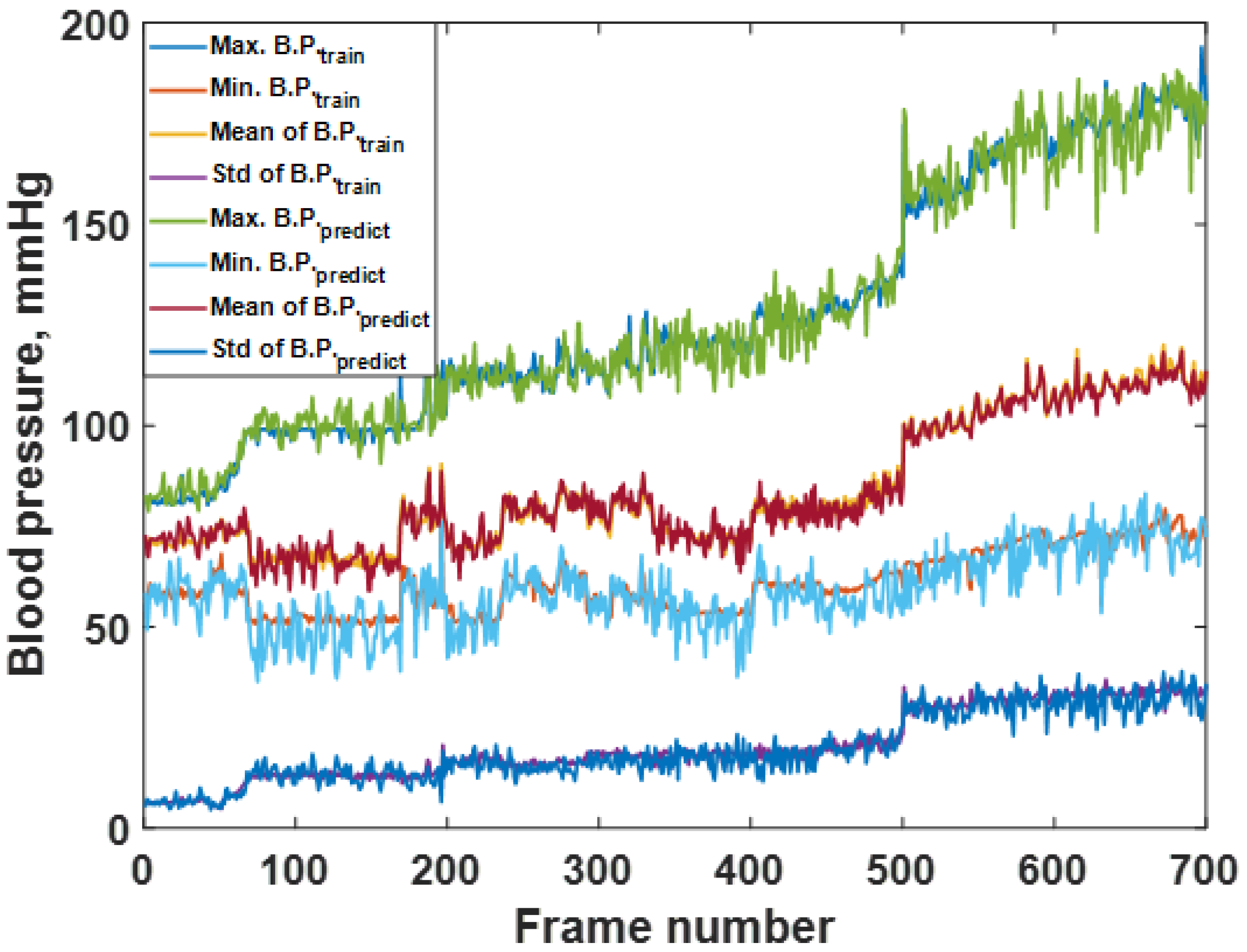
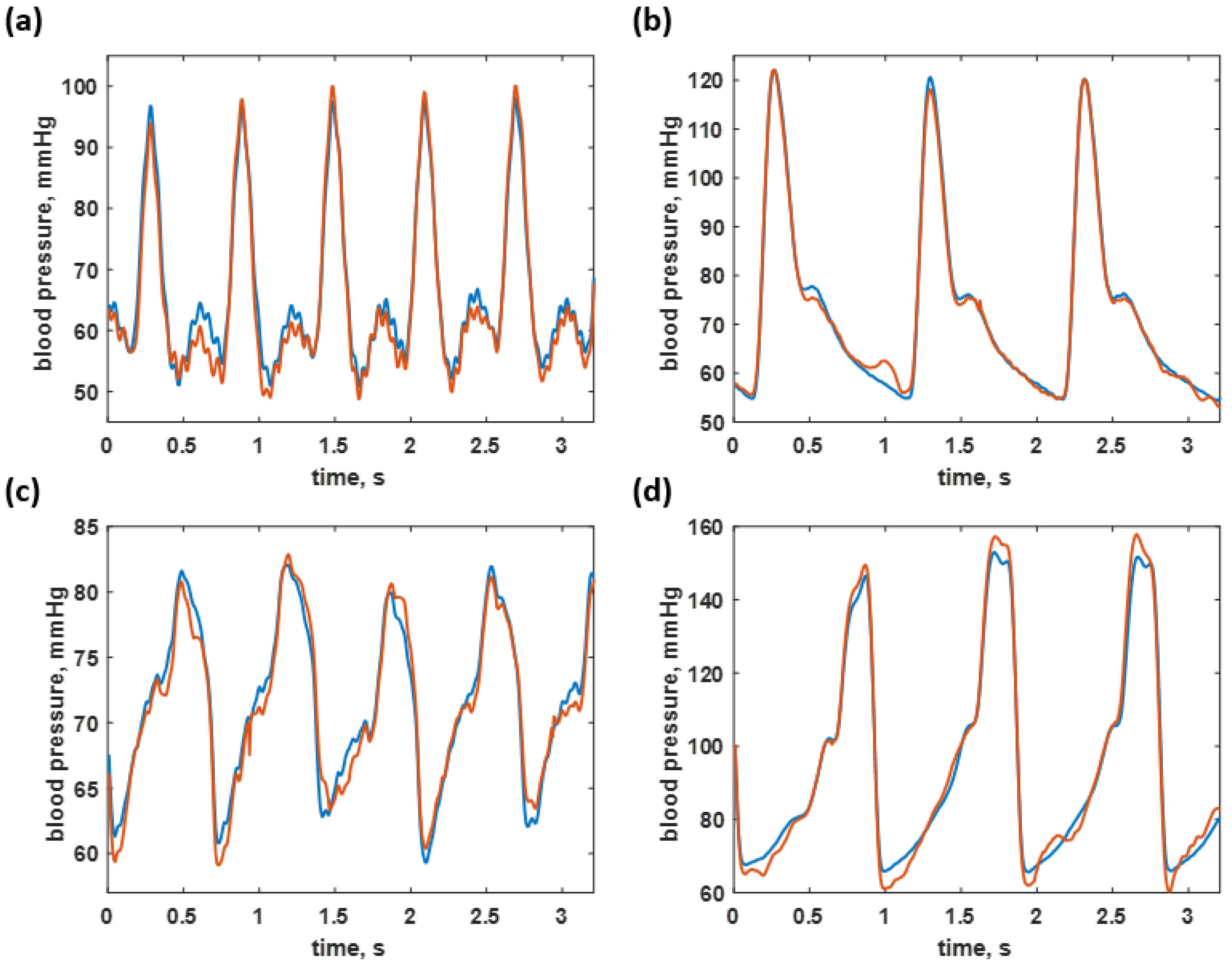
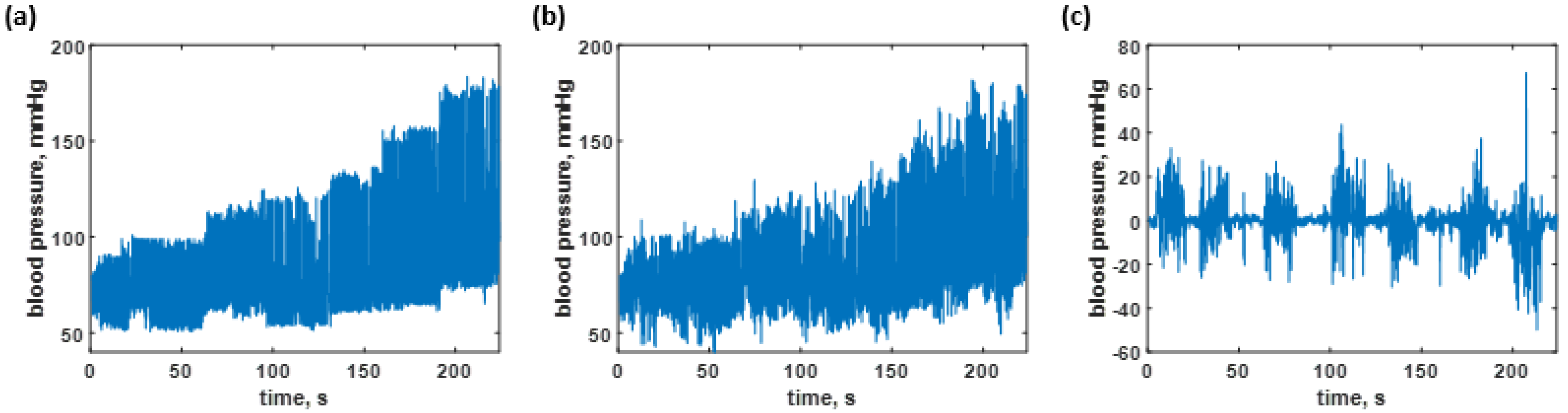
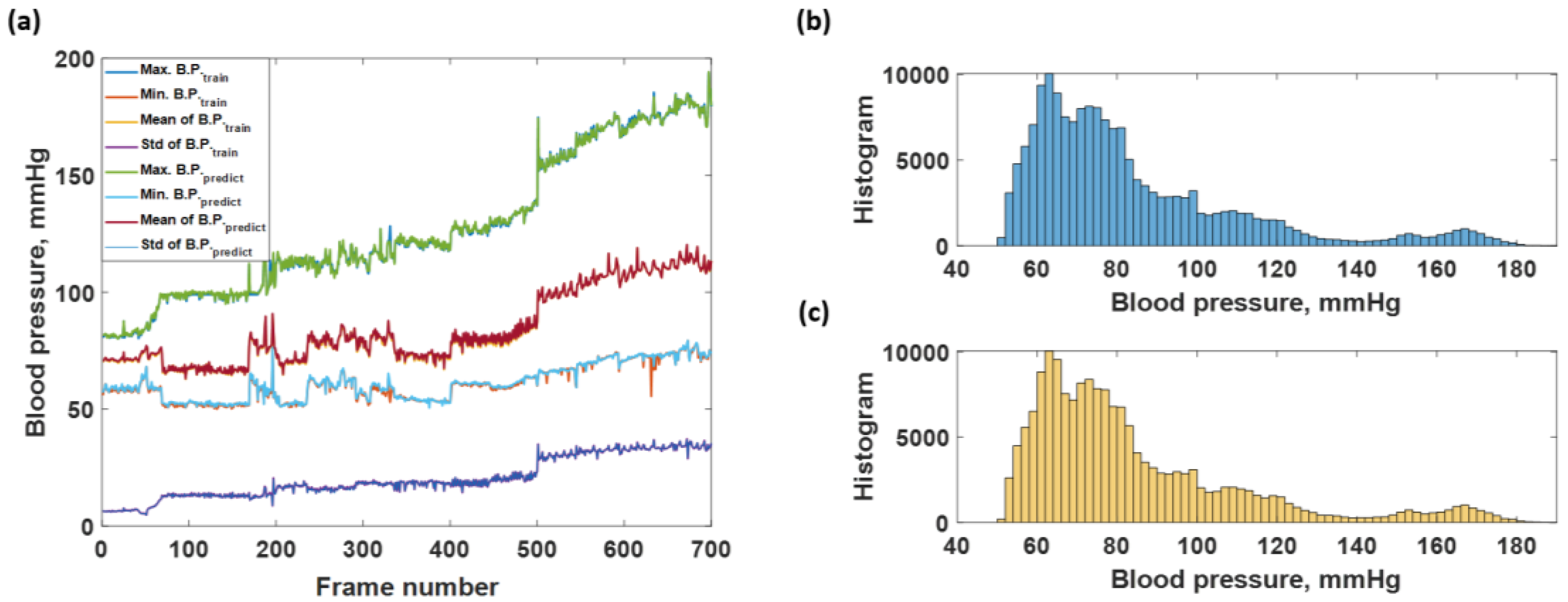
3.2. Blood Pressure Prediction of the Trained Sequences
3.3. Blood Pressure Prediction of the Test Dataset
3.4. Performance Comparison to Networks Trained without the Proposed Preprocessing Method
3.5. The CAN Network Prediction Compared to Other Methods
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perloff, D.; Grim, C.; Flack, J.; Frohlich, E.D.; Hill, M.; McDonald, M.; Morgenstern, B.Z. Human blood pressure determination by sphygmomanometry. Circulation 1993, 88, 2460–2470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rademacher, E.R.; Jacobs, D.R., Jr.; Moran, A.; Steinberger, J.; Prineas, R.J.; Sinaiko, A. Relation of blood pressure and body mass index during childhood to cardiovascular risk factor levels in young adults. J. Hypertens. 2009, 27, 1766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedner, J.; Ejnell, H.; Sellgren, J.; Hedner, T. Development of hypertension? J. Hypertens. 1988, 6, S529–S531. [Google Scholar] [CrossRef] [PubMed]
- Balanda, K.; Fahy, L.; Barron, S.; McLaughlin, A. Making Chronic Conditions Count: Hypertension, Stroke, Coronary Heart Disease, Diabetes; Institute of Public Health: Dublin, Ireland, 2010; pp. 15–79. [Google Scholar]
- Walther, D.; Curjuric, I.; Dratva, J.; Schaffner, E.; Quinto, C.; Schmidt-Trucksäss, A.; Eze, I.C.; Burdet, L.; Pons, M.; Gerbase, M.W. Hypertension, diabetes and lifestyle in the long-term—Results from a swiss population-based cohort. Prev. Med. 2017, 97, 56–61. [Google Scholar] [CrossRef] [Green Version]
- Chobanian, A.V.; Bakris, G.L.; Black, H.R.; Cushman, W.C.; Green, L.A.; Izzo, J.L., Jr.; Jones, D.W.; Materson, B.J.; Oparil, S.; Wright, J.T., Jr. Seventh report of the joint national committee on prevention, detection, evaluation, and treatment of high blood pressure. Hypertension 2003, 42, 1206–1252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muntner, P.; Shimbo, D.; Carey, R.M.; Charleston, J.B.; Gaillard, T.; Misra, S.; Myers, M.G.; Ogedegbe, G.; Schwartz, J.E.; Townsend, R.R. Measurement of blood pressure in humans: A scientific statement from the American Heart Association. Hypertension 2019, 73, e35–e66. [Google Scholar] [CrossRef]
- Speechly, C.; Bignell, N.; Turner, M. Sphygmomanometer calibration: Why, how and how often? Aust. Fam. Physician 2007, 36, 834–838. [Google Scholar]
- Parati, G.; Faini, A.; Castiglioni, P. Accuracy of blood pressure measurement: Sphygmomanometer calibration and beyond. J. Hypertens. 2006, 24, 1915–1918. [Google Scholar] [CrossRef]
- Tomitani, N.; Kanegae, H.; Suzuki, Y.; Kuwabara, M.; Kario, K. Stress-induced blood pressure elevation self-measured by a wearable watch-type device. Am. J. Hypertens. 2021, 34, 377–382. [Google Scholar] [CrossRef]
- Kario, K.; Shimbo, D.; Tomitani, N.; Kanegae, H.; Schwartz, J.E.; Williams, B. The first study comparing a wearable watch-type blood pressure monitor with a conventional ambulatory blood pressure monitor on in-office and out-of-office settings. J. Clin. Hypertens. 2020, 22, 135–141. [Google Scholar] [CrossRef] [Green Version]
- Kuwabara, M.; Harada, K.; Hishiki, Y.; Kario, K. Validation of two watch-type wearable blood pressure monitors according to the ANSI/AAMI/ISO81060-2: 2013 guidelines: Omron HEM-6410T-ZM and HEM-6410T-ZL. J. Clin. Hypertens. 2019, 21, 853–858. [Google Scholar] [CrossRef] [Green Version]
- Bote, J.M.; Recas, J.; Hermida, R. Evaluation of blood pressure estimation models based on pulse arrival time. Comput. Electr. Eng. 2020, 84, 106616. [Google Scholar] [CrossRef]
- Sun, S.; Bezemer, R.; Long, X.; Muehlsteff, J.; Aarts, R. Systolic blood pressure estimation using PPG and ECG during physical exercise. Physiol. Meas. 2016, 37, 2154. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Kobayashi, T.; Ichikawa, S.; Takeuchi, Y.; Togawa, T. Continuous estimation of systolic blood pressure using the pulse arrival time and intermittent calibration. Med. Biol. Eng. Comput. 2000, 38, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Solà, J.; Delgado-Gonzalo, R. The Handbook of Cuffless Blood Pressure Monitoring; Springer: Berlin/Heidelberg, Germany, 2019; pp. 978–983. Available online: https://link.springer.com/book/10.1007/978-3-030-24701-0 (accessed on 10 December 2021).
- Khalid, S.G.; Liu, H.; Zia, T.; Zhang, J.; Chen, F.; Zheng, D. Cuffless blood pressure estimation using single channel photoplethysmography: A two-step method. IEEE Access 2020, 8, 58146–58154. [Google Scholar] [CrossRef]
- Ding, X.-R.; Zhang, Y.-T.; Liu, J.; Dai, W.-X.; Tsang, H.K. Continuous cuffless blood pressure estimation using pulse transit time and photoplethysmogram intensity ratio. IEEE Trans. Biomed. Eng. 2015, 63, 964–972. [Google Scholar] [CrossRef]
- Lin, W.-H.; Wang, H.; Samuel, O.W.; Li, G. Using a new PPG indicator to increase the accuracy of PTT-based continuous cuffless blood pressure estimation. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju, Korea, 11–15 July 2017; pp. 738–741. [Google Scholar]
- Wong, M.Y.-M.; Poon, C.C.-Y.; Zhang, Y.-T. An evaluation of the cuffless blood pressure estimation based on pulse transit time technique: A half year study on normotensive subjects. Cardiovasc. Eng. 2009, 9, 32–38. [Google Scholar] [CrossRef]
- Tang, Z.; Sekine, M.; Tamura, T.; Yoshida, M.; Chen, W. A chair for cuffless real-time estimation of systolic blood pressure based on pulse transit time. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 5118–5121. [Google Scholar]
- Shriram, R.; Wakankar, A.; Daimiwal, N.; Ramdasi, D. Continuous cuffless blood pressure monitoring based on PTT. In Proceedings of the 2010 International Conference on Bioinformatics and Biomedical Technology, Chengdu, China, 16–18 April 2010; pp. 51–55. [Google Scholar]
- Tang, Z.; Tamura, T.; Sekine, M.; Huang, M.; Chen, W.; Yoshida, M.; Sakatani, K.; Kobayashi, H.; Kanaya, S. A chair–based unobtrusive cuffless blood pressure monitoring system based on pulse arrival time. IEEE J. Biomed. Health Inform. 2016, 21, 1194–1205. [Google Scholar] [CrossRef]
- Yan, W.-R.; Peng, R.-C.; Zhang, Y.-T.; Ho, D. Cuffless continuous blood pressure estimation from pulse morphology of photoplethysmograms. IEEE Access 2019, 7, 141970–141977. [Google Scholar] [CrossRef]
- Dey, J.; Gaurav, A.; Tiwari, V.N. Instabp: Cuff-less blood pressure monitoring on smartphone using single ppg sensor. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 5002–5005. [Google Scholar]
- Wang, W.; Mohseni, P.; Kilgore, K.; Najafizadeh, L. PulseLab: An Integrated and Expandable Toolbox for Pulse Wave Velocity-based Blood Pressure Estimation. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Guadalajara, Mexico, 1–5 November 2021; pp. 5654–5657. [Google Scholar]
- Kotzur, L.; Markewitz, P.; Robinius, M.; Stolten, D. Impact of different time series aggregation methods on optimal energy system design. Renew. Energy 2018, 117, 474–487. [Google Scholar] [CrossRef] [Green Version]
- Thadson, K.; Visitsattapongse, S.; Pechprasarn, S. Deep learning-based single-shot phase retrieval algorithm for surface plasmon resonance microscope based refractive index sensing application. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lateef, F.; Ruichek, Y. Survey on semantic segmentation using deep learning techniques. Neurocomputing 2019, 338, 321–348. [Google Scholar] [CrossRef]
- Kachuee, M.; Kiani, M.M.; Mohammadzade, H.; Shabany, M. Cuff-less high-accuracy calibration-free blood pressure estimation using pulse transit time. In Proceedings of the 2015 IEEE International Symposium on Circuits and Systems (ISCAS), Lisbon, Portugal, 24–27 May 2015; pp. 1006–1009. [Google Scholar]
- Gu, S.; Li, Y.; Gool, L.V.; Timofte, R. Self-guided network for fast image denoising. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Korea, 27–28 October 2019; pp. 2511–2520. [Google Scholar]
- Albawi, S.; Mohammed, T.A.; Al-Zawi, S. Understanding of a convolutional neural network. In Proceedings of the 2017 International Conference on Engineering and Technology (ICET), Antalya, Turkey, 21–24 August 2017; pp. 1–6. [Google Scholar]
- Allwein, E.L.; Schapire, R.E.; Singer, Y. Reducing multiclass to binary: A unifying approach for margin classifiers. J. Mach. Learn. Res. 2000, 1, 113–141. [Google Scholar]
- Escalera, S.; Pujol, O.; Radeva, P. On the decoding process in ternary error-correcting output codes. IEEE Trans. Pattern Anal. Mach. Intell. 2008, 32, 120–134. [Google Scholar] [CrossRef] [PubMed]
- Escalera, S.; Pujol, O.; Radeva, P. Separability of ternary codes for sparse designs of error-correcting output codes. Pattern Recognit. Lett. 2009, 30, 285–297. [Google Scholar] [CrossRef]
- Shahbabu, B.; Dasgupta, A.; Sarkar, K.; Sahoo, S.K. Which is more accurate in measuring the blood pressure? A digital or an aneroid sphygmomanometer. J. Clin. Diagn. Res. 2016, 10, LC11. [Google Scholar] [CrossRef]
- Thadson, K.; Sasivimolkul, S.; Suvarnaphaet, P.; Visitsattapongse, S.; Pechprasarn, S. Measurement precision enhancement of surface plasmon resonance based angular scanning detection using deep learning. Sci. Rep. 2022, 12, 1–14. [Google Scholar] [CrossRef]
- Hamdia, K.M.; Marino, M.; Zhuang, X.; Wriggers, P.; Rabczuk, T. Sensitivity analysis for the mechanics of tendons and ligaments: Investigation on the effects of collagen structural properties via a multiscale modeling approach. Int. J. Numer. Methods Biomed. Eng. 2019, 35, e3209. [Google Scholar] [CrossRef]
- Li, H.; Deng, J.; Feng, P.; Pu, C.; Arachchige, D.D.; Cheng, Q. Short-Term Nacelle Orientation Forecasting Using Bilinear Transformation and ICEEMDAN Framework. Front. Energy Res. 2021, 9, 697. [Google Scholar] [CrossRef]
- Li, H.; Deng, J.; Yuan, S.; Feng, P.; Arachchige, D.D. Monitoring and Identifying Wind Turbine Generator Bearing Faults Using Deep Belief Network and EWMA Control Charts. Front. Energy Res. 2021, 9, 770. [Google Scholar] [CrossRef]





| Systolic Blood Pressure Errors (Mean ± Standard Deviation) | Diastolic Blood Pressure Errors (Mean ± Standard Deviation) | Operating Range Systolic Blood Pressure | Operating Range Diastolic Blood Pressure | |
|---|---|---|---|---|
| PTT-based Methods | ||||
| Two-step algorithm developed by machine learning [17] | 0.07 ± 7.1 mmHg | −0.08 ± 6.0 mmHg | Reduced accuracy for hypotension | Reduced accuracy for hypotension |
| B.P. estimation based on PTT and photoplethysmography intensity ratio (PIR) [18] | −0.37 ± 5.21 mmHg | −0.08 ± 4.06 mmHg | Reduced accuracy for hypertension | Reduced accuracy for hypertension |
| B.P. estimation based on PTT and intensity ratio of the first derivative wave of PPG (1st-dPIR) [19] | 2.88 ± 7.75 mmHg | 2.80 ± 4.38 mmHg | ||
| Proceeding PTT-based method on the repeatability test [20] | 0.0 ± 5.3 mmHg | 0.0 ± 2.9 mmHg | 80–150 mmHg | 60–120 mmHg |
| Proceeding PTT-based method using regression coefficients [20] | 1.4 ± 10.2 mmHg | 2.1 ± 7.3 mmHg | 80–150 mmHg | 60–120 mmHg |
| PAT-based Methods | ||||
| Estimating beat-by-beat blood pressure using Chen’s method [21] | −0.5 ± 5.3 mmHg | 4.1 ± 3.4 mmHg | ||
| Standard pulse arrival time based method calculations [22] | 0 ± 3 mmHg | 0 ± 3 mmHg | ||
| Using a linear correlation of systolic blood pressure and a non-linear correlation of diastolic blood pressure and PAT [23] | 0.2 ± 5.8 mmHg | 0.4 ± 5.7 mmHg | ||
| Model-driven method: Logarithmic [26] | −0.512 ± 8.793 mmHg | −0.148 ± 3.622 mmHg | ||
| Model-driven method: Inverse [26] | −0.008 ± 8.203 mmHg | −0.078 ± 3.448 mmHg | ||
| Model-driven method: Inverse Square [26] | −0.358 ± 8.084 mmHg | −0.066 ± 3.574 mmHg | ||
| Other Methods | ||||
| Estimating blood pressure based on pulse morphology of PPG [24] | 0.043 ± 5.001 mmHg | 0.011 ± 3.689 mmHg | ||
| Blood pressure prediction based on demographic and physiological partitioning [25] | Mean absolute error = 6.9 mmHg | Mean absolute error = 5 mmHg | 80–220 mmHg | 45–120 mmHg |
| Layer | Activations | Learnable Variable | Descriptions |
|---|---|---|---|
| Image input | 4 × 250 × 1 | – | 4 × 500 × 1 images |
| Convolutional | 4 × 250 × 32 | Weights 2 × 2 × 1 × 32, Bias 1 × 1 × 32 | 1 padding |
| Batch normalization | Offset 1 × 1 × 32, Scale 1 × 1 × 32 | – | |
| Adaptive normalization | – | – | |
| Leaky ReLU | – | Scale 0.2 | |
| Convolutional | Weights 3 × 3 × 32 × 32, Bias 1 × 1 × 32 | 2 padding, 1 Stride, 2 dilation | |
| Batch normalization | Offset 1 × 1 × 32, Scale 1 × 1 × 32 | – | |
| Adaptive normalization | – | – | |
| Leaky ReLU | – | Scale 0.2 | |
| Convolutional | Weights 3 × 3 × 32 × 32, Bias 1 × 1 × 32 | 4 padding, 1 Stride, 4 dilation | |
| Batch normalization | Offset 1 × 1 × 32, Scale 1 × 1 × 32 | – | |
| Adaptive normalization | – | – | |
| Leaky ReLU | – | Scale 0.2 | |
| Convolutional | Weights 3 × 3 × 32 × 32, Bias 1 × 1 × 32 | 8 padding, 1 Stride, 8 dilation | |
| Batch normalization | Offset 1 × 1 × 32, Scale 1 × 1 × 32 | – | |
| Adaptive normalization | – | – | |
| Leaky ReLU | – | Scale 0.2 | |
| Convolutional | Weights 3 × 3 × 32 × 32, Bias 1 × 1 × 32 | 16 padding, 1 Stride, 16 dilation | |
| Batch normalization | Offset 1 × 1 × 32, Scale 1 × 1 × 32 | – | |
| Adaptive normalization | 4 × 250 × 32 | – | – |
| Leaky ReLU | – | Scale 0.2 | |
| Convolutional | Weights 3 × 3 × 32 × 32, Bias 1 × 1 × 32 | 32 padding, 1 Stride, 32 dilation | |
| Batch normalization | Offset 1 × 1 × 32, Scale 1 × 1 × 32 | – | |
| Adaptive normalization | – | – | |
| Leaky ReLU | – | Scale 0.2 | |
| Convolutional | Weights 3 × 3 × 32 × 32, Bias 1 × 1 × 32 | 64 padding, 1 Stride, 64 dilation | |
| Batch normalization | Offset 1 × 1 × 32, Scale 1 × 1 × 32 | – | |
| Adaptive normalization | – | – | |
| Leaky ReLU | – | Scale 0.2 | |
| Convolutional | Weights 3 × 3 × 32 × 32, Bias 1 × 1 × 32 | 128 padding, 1 Stride, 128 dilation | |
| Batch normalization | Offset 1 × 1 × 32, Scale 1 × 1 × 32 | – | |
| Adaptive normalization | – | – | |
| Leaky ReLU | – | Scale 0.2 | |
| Convolutional | Weights 3 × 3 × 32 × 32, Bias 1 × 1 × 32 | 1 padding, 1 Stride | |
| Batch normalization | Offset 1 × 1 × 32, Scale 1 × 1 × 32 | – | |
| Adaptive normalization | – | – | |
| Leaky ReLU | 4 × 250 × 32 | – | Scale 0.2 |
| Convolutional | 4 × 250 × 3 | Weights 3 × 3 × 32 × 32, Bias 1 × 1 × 32 | 0 padding, 1 Stride |
| Regression output | 1 × 1 × 250 | – | Mean square error |
| Hyperparameter | Parameter Value |
|---|---|
| Initial Learn Rate | |
| Gradient Decay Factor | 0.9000 |
| Squared Gradient Decay Factor | 0.9990 |
| Epsilon () | |
| Learn Rate Schedule | piecewise |
| Learn Rate Drop Factor | 0.0100 |
| Learn Rate Drop Period | 125,000 |
| L2 Regularization | |
| Gradient Threshold Method | L2 norm |
| Gradient Threshold | 1 |
| Maximum Epochs | 7000 |
| Mini Batch Size | 1 |
| Input and Label Shuffle | every epoch |
| Layer | Activations | Learnable Variable | Descriptions |
|---|---|---|---|
| Sequence input | 2 | – | Sequence input with 2 dimensions |
| LSTM | 400 | InputWeights 1600 × 2, RecurrentWeights 1600 × 400, Bias 1600 × 1 | LSTM with 400 hidden units |
| Fully Connected | 1 | Weights 1 × 400, Bias 1 × 1 | 1 fully connected layer |
| Regression Output | 1 | – | Mean-squared-error with response |
| Hyperparameter | Parameter Value |
|---|---|
| Initial Learn Rate | |
| Gradient Decay Factor | 0.9000 |
| Squared Gradient Decay Factor | 0.9990 |
| Epsilon () | |
| Learn Rate Schedule | piecewise |
| Learn Rate Drop Factor | 0.0100 |
| Learn Rate Drop Period | 125,000 |
| L2 Regularization | |
| Gradient Threshold Method | L2 norm |
| Gradient Threshold | 1 |
| Maximum Epochs | 7000 |
| Mini Batch Size | 2 |
| Input and Label Shuffle | once |
| RMSEs for the Resubstitution Performance | ||
| RMSE | CAN with the Preprocessing | LSTM without the Preprocessing |
| The average maximum B.P. | 4.9590 mmHg | 0.6804 mmHg |
| The average minimum B.P | 5.0880 mmHg | 0.8556 mmHg |
| The average mean B.P. | 1.9776 mmHg | 0.3992 mmHg |
| The standard deviation | 2.4064 mmHg | 0.1348 mmHg |
| RMSEs for the Test Dataset Responses | ||
| RMSE | CAN with the Preprocessing | LSTM without the Preprocessing |
| The average maximum B.P. | 7.1455 mmHg | 9.5528 mmHg |
| The average minimum B.P | 6.0862 mmHg | 7.3774 mmHg |
| The average mean B.P. | 4.2381 mmHg | 7.2500 mmHg |
| The standard deviation | 2.3218 mmHg | 2.5795 mmHg |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Treebupachatsakul, T.; Boosamalee, A.; Shinnakerdchoke, S.; Pechprasarn, S.; Thongpance, N. Cuff-Less Blood Pressure Prediction from ECG and PPG Signals Using Fourier Transformation and Amplitude Randomization Preprocessing for Context Aggregation Network Training. Biosensors 2022, 12, 159. https://doi.org/10.3390/bios12030159
Treebupachatsakul T, Boosamalee A, Shinnakerdchoke S, Pechprasarn S, Thongpance N. Cuff-Less Blood Pressure Prediction from ECG and PPG Signals Using Fourier Transformation and Amplitude Randomization Preprocessing for Context Aggregation Network Training. Biosensors. 2022; 12(3):159. https://doi.org/10.3390/bios12030159
Chicago/Turabian StyleTreebupachatsakul, Treesukon, Apivitch Boosamalee, Siratchakrit Shinnakerdchoke, Suejit Pechprasarn, and Nuntachai Thongpance. 2022. "Cuff-Less Blood Pressure Prediction from ECG and PPG Signals Using Fourier Transformation and Amplitude Randomization Preprocessing for Context Aggregation Network Training" Biosensors 12, no. 3: 159. https://doi.org/10.3390/bios12030159
APA StyleTreebupachatsakul, T., Boosamalee, A., Shinnakerdchoke, S., Pechprasarn, S., & Thongpance, N. (2022). Cuff-Less Blood Pressure Prediction from ECG and PPG Signals Using Fourier Transformation and Amplitude Randomization Preprocessing for Context Aggregation Network Training. Biosensors, 12(3), 159. https://doi.org/10.3390/bios12030159





