Single-Particle Optical Imaging for Ultrasensitive Bioanalysis
Abstract
:1. Introduction
2. Ultrasensitive Bioanalysis by Different OSPI Techniques
2.1. Single-Particle Fluorescence Imaging

2.2. Single-Particle ECL Imaging

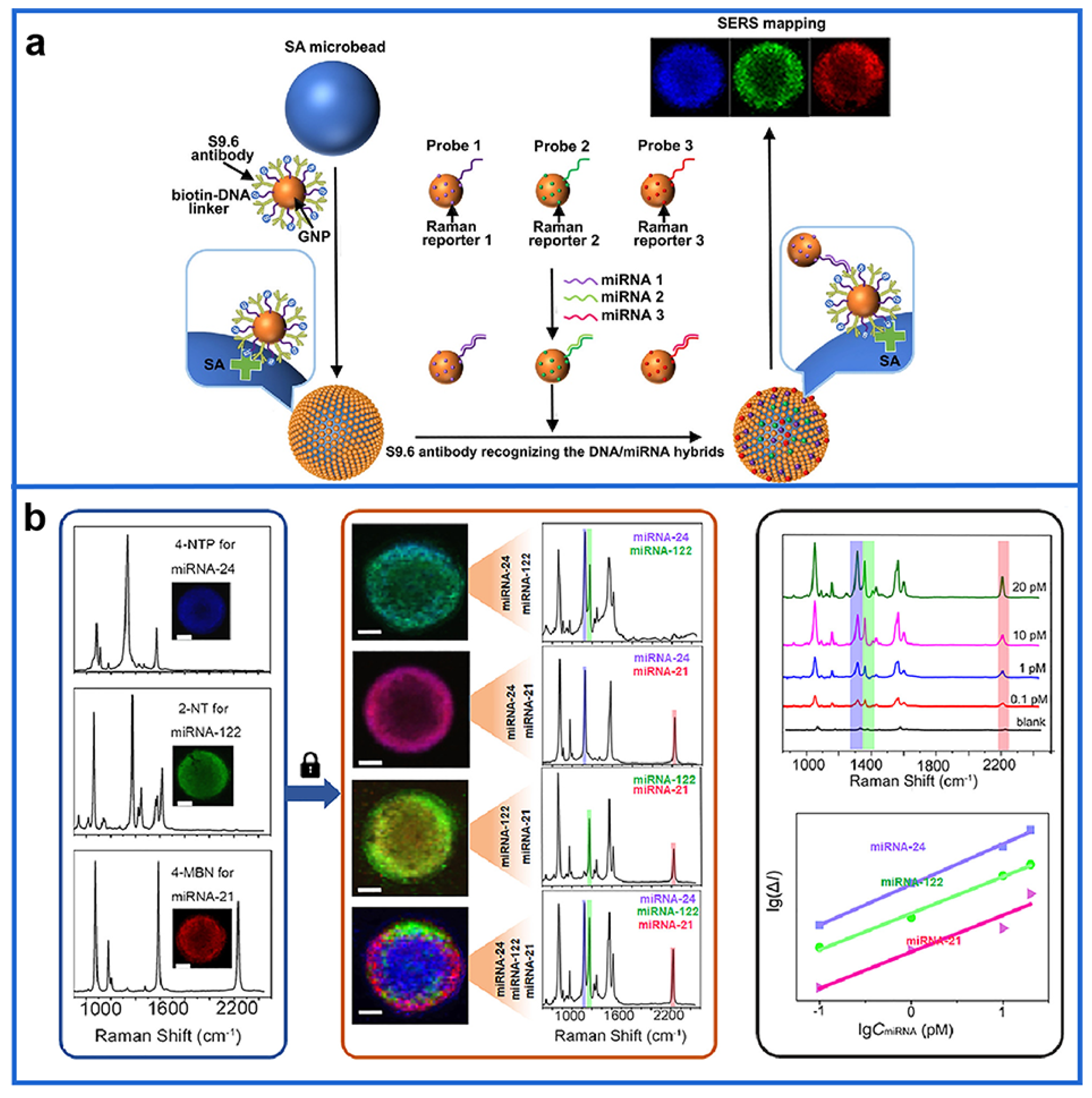
2.3. Single-Particle SERS Imaging
2.4. Single-Particle Dark-Field Scattering
3. Summery and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baud, D.; Qi, X.; Nielsen-Saines, K.; Musso, D.; Pomar, L.; Favre, G. Real estimates of mortality following COVID-19 infection. Lancet Infect. Dis. 2020, 20, 773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- GBD 2016 Dementia Collaborators. Global, regional, and national burden of Alzheimer’s disease and other dementias, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 88–106. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Ostrom, Q.T.; Kruchko, C.; Patil, N.; Tihan, T.; Cioffi, G.; Fuchs, H.E.; Waite, K.A.; Jemal, A.; Siegel, R.L.; et al. Brain and other central nervous system tumor statistics, 2021. CA Cancer J. Clin. 2021, 71, 381–406. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Zheng, R.; Zhang, S.; Zeng, H.; Wang, S.; Chen, R.; Wei, W.; He, J. Patterns and trends of cancer incidence in children and adolescents in China, 2011–2015: A population-based cancer registry study. Cancer Med. 2021, 10, 4575–4586. [Google Scholar] [CrossRef] [PubMed]
- GBD 2016 Brain and Other CNS Cancer Collaborators. Global, regional, and national burden of brain and other CNS cancer, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 376–393. [Google Scholar] [CrossRef] [Green Version]
- De Rubis, D.; Krishnan, S.R.; Bebawy, M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol. Sci. 2019, 40, 172–186. [Google Scholar] [CrossRef] [PubMed]
- Miralles, R.M.; Petit, J.; Gine, L.; Balaguero, L. Metastatic cancer spread at the laparoscopic puncture site. Report of a case in a patient with carcinoma of the ovary. Case Report. Eur. J. Gynaecol. Oncol. 1989, 10, 442–444. [Google Scholar] [PubMed]
- Parikh, A.R.; Leshchiner, I.; Elagina, L.; Goyal, L.; Levovitz, C.; Siravegna, G.; Livitz, D.; Rhrissorrakrai, K.; Martin, E.E.; Van Seventer, E.E.; et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 2019, 25, 1415–1421. [Google Scholar] [CrossRef] [PubMed]
- Kunjachan, S.; Ehling, J.; Storm, G.; Kiessling, F.; Lammers, T. Noninvasive imaging of nanomedicines and nanotheranostics: Principles, progress, and prospects. Chem. Rev. 2015, 115, 10907–10937. [Google Scholar] [CrossRef] [PubMed]
- Aberle, D.R.; Abtin, F.; Brown, K. Computed tomography screening for lung cancer: Has it finally arrived? Implications of the national lung screening trial. J. Clin. Oncol. 2013, 31, 1002–1008. [Google Scholar] [CrossRef] [Green Version]
- Chawla, S.C.; Federman, N.; Zhang, D.; Nagata, K.; Nuthakki, S.; McNitt-Gray, M.; Boechat, M.I. Estimated cumulative radiation dose from PET/CT in children with malignancies: A 5-year retrospective review. Pediatr. Radiol. 2010, 40, 681–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Månberg, A.; Skene, N.; Sanders, F.; Trusohamn, M.; Remnestål, J.; Szczepińska, A.; Aksoylu, I.S.; Lönnerberg, P.; Ebarasi, L.; Wouters, S.; et al. Altered perivascular fibroblast activity precedes ALS disease onset. Nat. Med. 2021, 27, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Alarcón-Sánchez, B.R.; Pérez-Carreón, J.I.; Villa-Treviño, S.; Arellanes-Robledo, J. Molecular alterations that precede the establishment of the hallmarks of cancer: An approach on the prevention of hepatocarcinogenesis. Biochem. Pharmacol. 2021, 194, 114818. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Qing, Y.; Li, M.; Sun, L.; Zhang, J.; Feng, L.; Li, J.; Chen, T.; Wang, J.; Wan, C. Salivary metabolomics reveals that metabolic alterations precede the onset of schizophrenia. J. Proteome. Res. 2021, 20, 5010–5023. [Google Scholar] [CrossRef]
- Acuña, A.I.; Esparza, M.; Kramm, C.; Beltrán, F.A.; Parra, A.V.; Cepeda, C.; Toro, C.A.; Vidal, R.L.; Hetz, C.; Concha, I.I.; et al. A failure in energy metabolism and antioxidant uptake precede symptoms of Huntington’s disease in mice. Nat. Commun. 2013, 4, 2917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snyder, M.W.; Kircher, M.; Hill, A.J.; Daza, R.M.; Shendure, J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 2016, 164, 57–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Avila, G.; Sommer, B.; Mendoza-Posada, D.A.; Ramos, C.; Garcia-Hernandez, A.A.; Falfan-Valencia, R. Matrix metalloproteinases participation in the metastatic process and their diagnostic and therapeutic applications in cancer. Crit. Rev. Oncol. Hematol. 2019, 137, 57–83. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Shi, K.; Yang, S.; Liu, J.; Zhou, Q.; Wang, G.; Song, J.; Li, Z.; Zhang, Z.; Yuan, W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol. Cancer. 2018, 17, 147. [Google Scholar] [CrossRef] [PubMed]
- Lehmann-Werman, R.; Neiman, D.; Zemmour, H.; Moss, J.; Magenheim, J.; Vaknin-Dembinsky, A.; Rubertsson, S.; Nellgård, B.; Blennow, K.; Zetterberg, H.; et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl. Acad. Sci. USA 2016, 113, E1826–E1834. [Google Scholar] [CrossRef] [Green Version]
- Danh, K.; Karp, D.G.; Singhal, M.; Tankasala, A.; Gebhart, D.; de Jesus Cortez, F.; Tandel, D.; Robinson, P.V.; Seftel, D.; Stone, M.; et al. Detection of neutralizing antibodies against multiple SARS-CoV-2 strains in dried blood spots using cell-free PCR. Nat. Commun. 2022, 13, 4212. [Google Scholar] [CrossRef] [PubMed]
- Engvall, E.; Perlmann, P. Enzyme-linked immunosorbent assay (ELISA) quantitative assay of immunoglobulin G. Immunochemistry 1971, 8, 871–874. [Google Scholar] [CrossRef] [PubMed]
- Klumpp-Thomas, C.; Kalish, H.; Drew, M.; Hunsberger, S.; Snead, K.; Fay, M.P.; Mehalko, J.; Shunmugavel, A.; Wall, V.; Frank, P.; et al. Standardization of ELISA protocols for serosurveys of the SARS-CoV-2 pandemic using clinical and at-home blood sampling. Nat. Commun. 2021, 12, 113. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.H.; Mix, S.; Xu, Z.; Taba, B.; Budvytiene, I.; Berliner, A.N.; Queralto, N.; Churi, Y.S.; Huang, R.S.; Eiden, M.; et al. Colorimetric sensor array allows fast detection and simultaneous identification of sepsis-causing bacteria in spiked blood culture. J. Clin. Microbiol. 2014, 52, 592–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Z.; Lv, T.; Zhu, K.; Li, Y.; Wang, L.; Gooding, J.J.; Liu, G.; Liu, B. Paper-based ratiometric fluorescence analytical devices towards point-of-care testing of human serum albumin. Angew. Chem. Int. Ed. 2020, 59, 3131–3136. [Google Scholar] [CrossRef]
- De, M.; Rana, S.; Akpinar, H.; Miranda, O.R.; Arvizo, R.R.; Bunz, U.H.; Rotello, V.M. Sensing of proteins in human serum using conjugates of nanoparticles and green fluorescent protein. Nat. Chem. 2009, 1, 461–465. [Google Scholar] [CrossRef]
- Yu, Q.; Xue, L.; Hiblot, J.; Griss, R.; Fabritz, S.; Roux, C.; Binz, P.A.; Haas, D.; Okun, J.G.; Johnsson, K. Semisynthetic sensor proteins enable metabolic assays at the point of care. Science 2018, 361, 1122–1126. [Google Scholar] [CrossRef]
- Zhang, H.; Cheng, X.; Richter, M.; Greene, M.I. A sensitive and high-throughput assay to detect low-abundance proteins in serum. Nat. Med. 2006, 12, 473–477. [Google Scholar] [CrossRef]
- Vujovic, S.; Henderson, S.; Presneau, N.; Odell, E.; Jacques, T.S.; Tirabosco, R.; Boshoff, C.; Flanagan, A.M. Brachyury, a crucial regulator of notochordal development, is a novel biomarker for chordomas. J. Pathol. 2006, 209, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Galasko, D. Biomarkers for Alzheimer’s disease-clinical needs and application. J. Alzheimers Dis. 2005, 8, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Barletta, J.M.; Edelman, D.C.; Constantine, N.T. Lowering the detection limits of HIV-1 viral load using real-time immuno-PCR for HIV-1 p24 antigen. Am. J. Clin. Pathol. 2004, 122, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Damon, D.E.; Davis, K.M.; Moreira, C.R.; Capone, P.; Cruttenden, R.; Badu-Tawiah, A.K. Direct biofluid analysis using hydrophobic paper spray mass spectrometry. Anal. Chem. 2016, 88, 1878–1884. [Google Scholar] [CrossRef]
- De la Rica, R.; Stevens, M.M. Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye. Nat. Nanotechnol. 2012, 7, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Jin, S.; Shi, W.; Oh, J.; Cao, H.; Jung, Y.M. Catalyzed deposition of signal reporter for highly sensitive surface-enhanced Raman spectroscopy immunoassay based on tyramine signal amplification strategy. Anal. Chem. 2018, 90, 13159–13162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.P.; Xun, L.; Xie, X.S. Single-molecule enzymatic dynamics. Science 1998, 282, 1877–1882. [Google Scholar] [CrossRef] [Green Version]
- Dickson, R.M.; Cubitt, A.B.; Tsien, R.Y.; Moerner, W.E. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature 1997, 388, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Lerner, E.; Cordes, T.; Ingargiola, A.; Alhadid, Y.; Chung, S.; Michalet, X.; Weiss, S. Toward dynamic structural biology: Two decades of single-molecule Förster resonance energy transfer. Science 2018, 359, eaan1133. [Google Scholar] [CrossRef] [Green Version]
- Zheng, M.; Tian, S.Z.; Capurso, D.; Kim, M.; Maurya, R.; Lee, B.; Piecuch, E.; Gong, L.; Zhu, J.J.; Li, Z.; et al. Multiplex chromatin interactions with single-molecule precision. Nature 2019, 566, 558–562. [Google Scholar] [CrossRef]
- Boersma, S.; Khuperkar, D.; Verhagen, B.M.; Sonneveld, S.; Grimm, J.B.; Lavis, L.D.; Tanenbaum, M.E. Multi-color single-molecule imaging uncovers extensive heterogeneity in mRNA decoding. Cell 2019, 178, 458–472. [Google Scholar] [CrossRef]
- Song, Y.; Sandford, E.; Tian, Y.; Yin, Q.; Kozminski, A.G.; Su, S.-H.; Cai, T.; Ye, Y.; Chung, M.T.; Lindstrom, R.; et al. Rapid single-molecule digital detection of protein biomarkers for continuous monitoring of systemic immune disorders. Blood 2021, 137, 1591–1602. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Huang, Y.; Hou, G.; Xiao, A.; Chen, P.; Liang, H.; Huang, Y.; Zhao, X.; Liang, L.; Feng, X.; et al. Single-molecule detection of biomarker and localized cellular photothermal therapy using an optical microfiber with nanointerface. Sci. Adv. 2019, 5, eaax4659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, W.C.; Horn, T.; Zayats, M.; Rizk, G.; Major, S.; Zhu, H.; Russell, J.; Xu, Z.; Rothman, R.E.; Celedon, A. Ultra-sensitive and rapid detection of nucleic acids and microorganisms in body fluids using single-molecule tethering. Nat. Commun. 2020, 11, 4774. [Google Scholar] [CrossRef] [PubMed]
- Norman, M.; Gilboa, T.; Walt, D.R. High-sensitivity single molecule array assays for pathological isoforms in Parkinson’s disease. Clin. Chem. 2022, 68, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Farka, Z.; Mickert, M.J.; Pastucha, M.; Mikušová, Z.; Skládal, P.; Gorris, H.H. Advances in optical single-molecule detection: An route to supersensitive bioaffinity assays. Angew. Chem. Int. Ed. 2020, 59, 10746–10773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.-L.; Huo, M.-Z.; Ying, Y.-L.; Long, Y.-T. Biological nanopore approach for single-molecule protein sequencing. Angew. Chem. Int. Ed. 2021, 60, 14738–14749. [Google Scholar] [CrossRef]
- Bernardi, R.C.; Durner, E.; Schoeler, C.; Malinowska, K.H.; Carvalho, B.G.; Bayer, E.A.; Luthey-Schulten, Z.; Gaub, H.E.; Nash, M.A. Mechanisms of nanonewton mechanostability in a protein complex revealed by molecular dynamics simulations and single-molecule force spectroscopy. J. Am. Chem. Soc. 2019, 141, 14752–14763. [Google Scholar] [CrossRef] [Green Version]
- Pei, Y.; Bian, T.; Liu, Y.; Liu, Y.; Xie, Y.; Song, J. Single-molecule resettable DNA computing via magnetic tweezers. Nano. Lett. 2022, 22, 3003–3010. [Google Scholar] [CrossRef]
- Walt, D.R. Optical methods for single molecule detection and analysis. Anal. Chem. 2013, 85, 1258–1263. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Ma, G.; Dong, W.; Wan, Z.; Wang, S.; Tao, N. Plasmonic scattering imaging of single proteins and binding kinetics. Nat. Methods 2020, 17, 1010–1017. [Google Scholar] [CrossRef]
- Jain, A.; Liu, R.; Ramani, B.; Arauz, E.; Ishitsuka, Y.; Ragunathan, K.; Park, J.; Chen, J.; Xiang, Y.K.; Ha, T. Probing cellular protein complexes using single-molecule pull-down. Nature 2011, 473, 484–488. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Q.; Lavis, L.D. Development of photostable fluorophores for molecular imaging. Curr. Opin. Chem. Biol. 2017, 39, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Grimm, J.B.; Lavis, L.D. Caveat fluorophores: An insider’s guide to small-molecule fluorescent labels. Nat. Methods 2022, 19, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.R.; Harootunian, A.T.; Buechler, Y.J.; Taylor, S.S.; Tsien, R.Y. Fluorescence ratio imaging of cyclic AMP in single cells. Nature 1991, 349, 694–697. [Google Scholar] [CrossRef]
- Wu, S.; Wang, D.; Liu, J.; Feng, Y.; Weng, J.; Li, Y.; Gao, X.; Liu, J.; Wang, W. The dynamic multisite interactions between two intrinsically disordered proteins. Angew. Chem. Int. Ed. 2017, 56, 7515–7519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willkomm, S.; Jakob, L.; Kramm, K.; Graus, V.; Neumeier, J.; Meister, G.; Grohmann, D. Single-molecule FRET uncovers hidden conformations and dynamics of human Argonaute 2. Nat. Commun. 2022, 13, 3825. [Google Scholar] [CrossRef]
- Pan, R.; Li, G.; Liu, S.; Zhang, X.; Liu, J.; Su, Z.; Wu, Y. Emerging nanolabels-based immunoassays: Principle and applications in food safety. TrAC, Trends Anal. Chem. 2021, 145, 116462. [Google Scholar] [CrossRef]
- Yang, W.; Wei, Z.; Nie, Y.; Tian, Y. Optical detection and imaging of nonfluorescent matter at the single-molecule/particle level. J. Phys. Chem. Lett. 2022, 13, 9618–9631. [Google Scholar] [CrossRef]
- Zhan, C.; Chen, X.-J.; Yi, J.; Li, J.-F.; Wu, D.-Y.; Tian, Z.-Q. From plasmon-enhanced molecular spectroscopy to plasmon-mediated chemical reactions. Nat. Rev. Chem. 2018, 2, 216–230. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, Y.; Peng, C.S.; Yang, T.; Joubert, L.-M.; Chu, S. Single upconversion nanoparticle imaging at sub-10 W cm-2 irradiance. Nat. Photonics 2018, 12, 548–553. [Google Scholar] [CrossRef]
- Richter, M.M. Electrochemiluminescence (ECL). Chem. Rev. 2004, 104, 3003–3036. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Chen, H.-Y.; Xu, J.-J. Electrogenerated chemiluminescence detection of single entities. Chem. Sci. 2021, 12, 5720–5736. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.W.; Sandoghdar, V. Interferometric scattering microscopy: Seeing single nanoparticles and molecules via Rayleigh scattering. Nano Lett. 2019, 19, 4827–4835. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Shen, G.; Wang, H.; Li, H.; Zhang, T.; Tao, N.; Ding, X.; Yu, H. Interferometric plasmonic imaging and detection of single exosomes. Proc. Natl. Acad. Sci. USA 2018, 115, 10275–10280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, D.; Zhao, X.; Liu, Y.-N.; Chen, H.-B.; Lv, W.-L.; Qian, C.; Liu, X.-W. Label-free probing of molecule binding kinetics using single-particle interferometric imaging. Anal. Chem. 2021, 93, 7965–7969. [Google Scholar] [CrossRef] [PubMed]
- Ohannesian, N.; Misban, I.; Lin, S.H.; Shin, W.-C. Plasmonic nano-aperture label-free imaging (PANORAMA). Nat. Commun. 2020, 11, 5805. [Google Scholar] [CrossRef]
- Chen, S.; Su, Y.-W.; Sun, J.; Chen, T.; Zheng, Y.; Sui, L.-J.; Yang, S.; Liu, C.; Wang, P.; Li, T.; et al. Label-free single-particle imaging approach for ultra-rapid detection of pathogenic bacterial in clinical samples. Proc. Natl. Acad. Sci. USA 2022, 119, e2206990119. [Google Scholar] [CrossRef]
- Di Antonio, M.; Ponjavic, A.; Radzevičius, A.; Ranasinghe, R.T.; Catalano, M.; Zhang, X.; Shen, J.; Needham, L.-M.; Lee, S.F.; Klenerman, D.; et al. Single-molecule visualization of DNA G-quadruplex formation in live cells. Nat. Chem. 2020, 12, 832–837. [Google Scholar] [CrossRef]
- Asher, W.B.; Geggier, P.; Holsey, M.D.; Gilmore, G.T.; Pati, A.K.; Meszaros, J.; Terry, D.S.; Mathiasen, S.; Kaliszewski, M.J.; McCauley, M.D.; et al. Single-molecule FRET imaging of GPCR dimers in living cells. Nat. Methods 2021, 18, 397–405. [Google Scholar] [CrossRef]
- Ebrahimi, S.; Akhlaghi, Y.; Kompany-Zareh, M.; Rinnan, Å. Nucleic acid based fluorescent nanothermometers. ACS Nano 2014, 8, 10372–10382. [Google Scholar] [CrossRef]
- Fang, X.; Zheng, Y.; Duan, Y.; Liu, Y.; Zhong, W. Recent advances in design of fluorescence-based assays for high throughput screening. Anal. Chem. 2019, 91, 482–504. [Google Scholar] [CrossRef]
- Zare, I.; Yaraki, M.T.; Speranza, G.; Najafabadi, A.H.; Shourangiz-Haghighi, A.; Nik, A.B.; Manshian, B.B.; Saraiva, C.; Soenen, S.J.; Kogan, M.J.; et al. Gold nanostructures: Synthesis, properties, and neurological applications. Chem. Soc. Rev. 2022, 51, 2601–2680. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, M.; Zheng, G.; Dai, Z.; Ma, Y. Recent progress in sensing application of metal nanoarchitecture-enhanced fluorescence. Nanoscale Adv. 2021, 3, 2448–2465. [Google Scholar] [CrossRef] [PubMed]
- Aubert, T.; Soenen, S.J.; Wassmuth, D.; Cirillo, M.; Deun, R.V.; Braeckmans, K.; Hens, Z. Bright and stable CdSe/CdS@SiO2 nanoparticles suitable for long-term cell labeling. ACS Appl. Mater. Interfaces 2014, 6, 11714–11723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.-Y.; Chen, G.; Gu, Y.-P.; Cui, R.; Zhang, Z.-L.; Yu, Z.-L.; Tang, B.; Zhao, Y.-F.; Pang, D.-W. Ultrasmall magnetically engineered Ag2Se quantum dots for instant efficient labeling and whole-body high-resolution multimodal real-time tracking of cell-derived microvesicles. J. Am. Chem. Soc. 2016, 138, 1893–1903. [Google Scholar] [CrossRef] [PubMed]
- Chu, B.; Wang, H.; Song, B.; Peng, F.; Su, Y.; He, Y. Fluorescent and photostable silicon nanoparticles sensors for real-time and long-term intracellular pH measurement in live cells. Anal. Chem. 2016, 88, 9235–9242. [Google Scholar] [CrossRef]
- Iyer, G.; Michalet, X.; Chang, Y.-P.; Pinaud, F.F.; Matyas, S.E.; Payne, G.; Weiss, S. High affinity scFv-hapten pair as a tool for quantum dot labeling and tracking of single proteins in live cells. Nano Lett. 2008, 8, 4618–4623. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Wang, H.; Zhu, Y.; Zhang, R.; Cogger, V.C.; Liu, X.; Xu, Z.P.; Grice, J.; Roberts, M.S. Short- and long-term tracking of anionic ultrasmall nanoparticles in kideny. ACS Nano 2016, 10, 387–395. [Google Scholar] [CrossRef]
- Wolfbeis, O.S. An overview of nanoparticles commonly used in fluorescent bioimaging. Chem. Soc. Rev. 2015, 44, 4743–4768. [Google Scholar] [CrossRef] [Green Version]
- Hariri, A.A.; Newman, S.S.; Tan, S.; Mamerow, D.; Adams, A.M.; Maganzini, N.; Zhong, B.L.; Eisenstein, M.; Dunn, A.R.; Soh, H.T. Improved immunoassay sensitivity and specificity using single-molecule colocalization. Nat. Commun. 2022, 13, 5359. [Google Scholar] [CrossRef]
- Fedyuk, V.; Erez, N.; Furth, N.; Beresh, O.; Andreishcheva, E.; Shinde, A.; Jones, D.; Zakai, B.B.; Mavor, Y.; Peretz, T.; et al. Multiplexed, single-molecule, epigenetic analysis of plasma-isolated nucleosomes for cancer diagnostics. Nat. Biotechnol. 2022, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.-P.; Wang, S.-C.; Su, Y.-P.; Tseng, S.-H.; He, L.; Wu, A.A.; Roden, R.B.S.; Xiao, J.; Hung, C.-F. Protein detection in blood with single-molecule imaging. Sci. Adv. 2021, 7, eabg6522. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Huang, X.; Liu, J.; Liu, B. Simultaneous and ultrasensitive detection of multiple microRNAs by single-molecule fluorescence imaging. Chem. Sci. 2020, 11, 3812–3819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Liu, Y.; Liu, Y.; Tian, T.; Yang, B.; Huang, X.; Liu, J.; Liu, B. Construction of dual-color probes with target-triggered signal amplification for in situ single-molecule imaging of microRNA. ACS Nano 2020, 14, 8116–8125. [Google Scholar] [CrossRef]
- Fan, Z.; Wang, Y.-J.; Huang, X.; Li, B.; Wang, S.; Liu, Y.; Liu, Y.-J.; Liu, B. Spatially resolved single-molecule profiling of microRNAs in migrating cells driven by microconfinement. Chem. Sci. 2022, 13, 11197–11204. [Google Scholar] [CrossRef]
- Liu, Y.; Li, B.; Wang, Y.-J.; Fan, Z.; Du, Y.; Li, B.; Liu, Y.-J.; Liu, B. In situ single-molecule imaging of microRNAs in switchable migrating cells under biomimetic confinement. Anal. Chem. 2022, 94, 4030–4038. [Google Scholar] [CrossRef]
- Chan, W.C.W.; Nie, S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science 1998, 281, 2016–2018. [Google Scholar] [CrossRef] [Green Version]
- Sukhanova, A.; Devy, L.; Venteo, L.; Kaplan, H.; Artemyev, M.; Oleinikov, V.; Klinov, D.; Pluot, M.; Cohen, J.H.; Nabiev, I. Biocompatible fluorescent nanocrystals for immunolabeling of membrane proteins and cells. Anal. Biochem. 2004, 324, 60–67. [Google Scholar] [CrossRef]
- Resch-Genger, U.; Grabolle, M.; Cavaliere-Jaricot, S.; Nitschke, R.; Nann, T. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods 2008, 5, 763–775. [Google Scholar] [CrossRef]
- Leatherdale, C.A.; Woo, W.-K.; Mikulec, F.V.; Bawendi, M.G. On the absorption cross section of CdSe nanocrystal quantum dots. J. Phys. Chem. B 2002, 106, 7619–7622. [Google Scholar] [CrossRef]
- Mazumder, S.; Dey, R.; Mitra, M.K.; Mukherjee, S.; Das, G.C. Review: Biofunctionalized quantum dots in biology and medicine. J. Nanomate. 2009, 2009, 815734. [Google Scholar] [CrossRef] [Green Version]
- Medinta, I.L.; Clapp, A.R.; Mattoussi, H.; Goldman, E.R.; Fisher, B.; Mauro, J.M. Self-assembled nanoscale biosensors based on quantum dots FRET donors. Nat. Mater. 2003, 2, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Qiu, J.-G.; Ma, F.; Zhang, C.-Y. Advances in single-molecule fluorescent nanosensors. Wires. Nanomed. Nanobi. 2021, 12, e1716. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Li, C.-C.; Zhang, C.-Y. Nucleic acid amplification-integrated single-molecule fluorescence imaging for in vitro and in vivo biosensing. Chem. Commun. 2021, 57, 13415–13428. [Google Scholar] [CrossRef]
- Ma, F.; Zhang, Q.; Zhang, C.-Y. Catalytic self-assembly of quantum-dot-based microRNA nanosensor directed by toehold-mediated strand displacement cascade. Nano Lett. 2019, 19, 6370–6376. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Q.-N.; Zhou, K.; Xu, Q.; Zhang, C.-Y. Identification of specific N6-methyladenosine RNA demethylase FTO inhibitors by single-quantum-dot-based FRET nanosensors. Anal. Chem. 2020, 92, 13936–13944. [Google Scholar] [CrossRef]
- Li, C.-C.; Hu, J.; Luo, X.; Hu, J.; Zhang, C.-Y. Development of a single quantum dot-mediated FRET nanosensor for sensitive detection of single-nucleotide polymorphism in cancer. Anal. Chem. 2021, 93, 14568–14576. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Q.; Cui, L.; Liu, W.; Qiu, J.-G.; Zhang, C.-Y. Zirconium ion-mediated assembly of a single quantum dot-based nanosensor for kinase assay. Chem. Commun. 2021, 57, 6376–6379. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, K.; Yao, Y.; Liu, Y.; Ji, J.; Huang, X.; Liu, J.; Liu, B. Single-molecule fluorescence imaging for untrasensitive DNA methyltransferase activity measurement and inhibitor screening. Anal. Chem. 2019, 91, 9500–9507. [Google Scholar] [CrossRef]
- Xiong, Y.; Huang, Q.; Canady, T.D.; Barya, P.; Liu, S.; Arogundade, Q.H.; Race, C.M.; Che, C.; Wang, X.; Zhou, L.; et al. Photonic crystal enhanced fluorescence emission and blinking suppression for single quantum dot digital resolution biosensing. Nat. Commun. 2022, 13, 4647. [Google Scholar] [CrossRef]
- Algar, W.R.; Massey, M.; Rees, K.; Higgins, R.; Krause, K.D.; Darwish, G.H.; Peveler, W.J.; Xiao, Z.; Tsai, H.-Y.; Gupta, R.; et al. Photoluminescent nanoparticle for chemical and biological analysis and imaging. Chem. Rev. 2021, 121, 9243–9358. [Google Scholar] [CrossRef]
- Zhang, Z.; Shikha, S.; Liu, J.; Zhang, J.; Mei, Q.; Zhang, Y. Upconversion nanoprobes: Recent advances in sensing applications. Anal. Chem. 2019, 91, 548–568. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wei, L.; Pan, L.; Yi, Z.; Wang, X.; Ye, Z.; Xiao, L.; Li, H.-W.; Wang, J. Homogeneous immunosorbent assay based on single-particle enumeration using upconversion nanoparticles for sensitive detection of cancer biomarkers. Anal. Chem. 2018, 90, 4807–4814. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Han, Y.; Wang, S.; Ye, Z.; Wei, L.; Xiao, L. Single-particle LRET aptasensor for the sensitive detection of aflatoxin B1 with upconversion nanoparticles. Anal. Chem. 2019, 91, 11856–11863. [Google Scholar] [CrossRef] [PubMed]
- Mickert, M.J.; Farka, Z.; Kostiv, U.; Hlaváček, A.; Horák, D.; Skládal, P.; Gorris, H.H. Measurement of sub-femtomolar concentrations of prostate-specific antigen through single-molecule counting with an upconversion-linked immunosorbent assay. Anal. Chem. 2019, 91, 9435–9441. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, R.; Li, B.; Fan, Y.; Wu, Y.; Zhu, X.; Zhang, F. Precise in vivo inflammation imaging using in situ responsive cross-linking of glutathione-modified ultra-small NIR-II lanthanide nanoparticles. Angew. Chem. Int. Ed. 2019, 58, 2050–2054. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhao, M.; Lin, J.; Huang, P.; Chen, X. Management of fluorescent organic/inorganic nanohybrids for biomedical applications in the NIR-II region. Chem. Soc. Rev. 2022, 51, 7692–7714. [Google Scholar] [CrossRef] [PubMed]
- Piwoński, H.; Wang, Y.; Li, W.; Michinobu, T.; Habuchi, S. Millimeter-deep detection of single shortwave-infrared-emitting polymer dots through turbid media. Nano Lett. 2020, 20, 8803–8810. [Google Scholar] [CrossRef]
- Voci, S.; Goudeau, B.; Valenti, G.; Lesch, A.; Jovic, M.; Rapino, S.; Paolucci, F.; Arbault, S.; Sojic, N. Surface-confined electrochemiluminescence microscopy of cell membranes. J. Am. Chem. Soc. 2018, 140, 14753–14760. [Google Scholar] [CrossRef]
- Wang, Y.; Jin, R.; Sojic, N.; Jiang, D.; Chen, H.-Y. Intracellular wireless analysis of single cells by bipolar electrochemiluminescence confined in a nanopipette. Angew. Chem. Int. Ed. 2020, 59, 10416–10420. [Google Scholar] [CrossRef]
- Chen, M.-M.; Xu, C.-H.; Zhao, W.; Chen, H.-Y.; Xu, J.-J. Super-resolution electrogenerated chemiluminescence microscopy for single-nanocatalyst imaging. J. Am. Chem. Soc. 2021, 143, 18511–18518. [Google Scholar] [CrossRef]
- Zhu, M.-J.; Pan, J.-B.; Wu, Z.-Q.; Gao, X.-Y.; Zhao, W.; Xia, X.-H.; Xu, J.-J.; Chen, H.-Y. Electrogenerated chemiluminescence imaging of electrocatalysis at a single Au-Pt janus nanoparticle. Angew. Chem. Int. Ed. 2018, 57, 4010–4014. [Google Scholar] [CrossRef] [PubMed]
- Zanut, A.; Fiorani, A.; Canola, S.; Saito, T.; Ziebart, N.; Rapino, S.; Rebeccani, S.; Barbon, A.; Irie, T.; Josel, H.-P.; et al. Insights into the mechanism of coreactant electrochemiluminescence facilitating enhanced bioanalytical performance. Nat. Commun. 2020, 11, 2668. [Google Scholar] [CrossRef] [PubMed]
- Sentic, M.; Milutinovic, M.; Kanoufi, F.; Manojlovic, D.; Arbault, S.; Sojic, N. Mapping electrogenerated chemiluminescence reactivity in space: Mechanistic insight into model systems used in immunoassays. Chem. Sci. 2014, 5, 2568–2572. [Google Scholar] [CrossRef]
- Dong, J.; Lu, Y.; Xu, Y.; Chen, F.; Yang, J.; Chen, Y.; Feng, J. Direct imaging of single-molecule electrochemical reactions in solution. Nature 2021, 596, 244–249. [Google Scholar] [CrossRef]
- Ma, C.; Wei, H.-F.; Wang, M.-X.; Wu, S.; Chang, Y.-C.; Zhang, J.; Jiang, L.-P.; Zhu, W.; Chen, Z.; Lin, Y. Hydrogen evolution reaction monitored by electro-chemiluminescence blinking at single-nanoparticle level. Nano Lett. 2020, 20, 5008–5016. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, H.; Li, B.; Liu, J.; Jiang, D.; Liu, B.; Sojic, N. Single biomolecule imaging by electrochemiluminescence. J. Am. Chem. Soc. 2021, 143, 17910–17914. [Google Scholar] [CrossRef]
- Li, B.; Huang, X.; Lu, Y.; Fan, Z.; Li, B.; Jiang, D.; Sojic, N.; Liu, B. High electrochemiluminescence from Ru(bpy)32+ embedded metal-organic fromeworks to visualize single molecule movement at the cellular membrane. Adv. Sci. 2022, 9, 2204715. [Google Scholar] [CrossRef]
- Xu, L.; Li, Y.; Wu, S.; Liu, X.; Su, B. Imaging latent fingerprints by electrochemiluminescence. Angew. Chem. Int. Ed. 2012, 51, 8068–8072. [Google Scholar] [CrossRef]
- Wang, N.; Gao, H.; Li, Y.; Li, G.; Chen, W.; Jin, Z.; Lei, J.; Wei, Q.; Ju, H. Dual intramolecular electron transfer for in situ coreactant-embedded electro-chemiluminescence microimaging of membrane protein. Angew. Chem. Int. Ed. 2021, 60, 197–201. [Google Scholar] [CrossRef]
- Qin, X.; Zhang, X.; Wang, M.; Dong, Y.; Liu, J.; Zhu, Z.; Li, M.; Yang, D.; Shao, Y. Fabrication of tris(bipyridine)ruthenium(II)-functionalized metal-organic framework thin films by electrochemically assisted self-assembly technique for electrochemiluminescent immunoassay. Anal. Chem. 2018, 90, 11622–11628. [Google Scholar] [CrossRef]
- Ding, H.; Guo, W.; Su, B. Imaging cell-matrix adhesions and collective migration of living cells by electrochemiluminescence microscopy. Angew. Chem. Int. Ed. 2020, 59, 449–456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Colin, C.; Descamps, J.; Arbault, S.; Sojic, N. Shadow electro-chemiluminescence microscopy of single mitochondria. Angew. Chem. Int. Ed. 2021, 60, 18742–18749. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jin, R.; Jiang, D.; Chen, H.-Y. Electrochemiluminescence-based capacitance microscopy for label-free imaging of antigens on the cellular plasma membrane. J. Am. Chem. Soc. 2019, 141, 10294–10299. [Google Scholar] [CrossRef]
- Zong, C.; Xu, M.; Xu, L.-J.; Wei, T.; Ma, X.; Zheng, X.-S.; Hu, R.; Ren, B. Surface-enhanced Raman spectroscopy for bioanalysis: Reliability and challenges. Chem. Rev. 2018, 118, 4946–4980. [Google Scholar] [CrossRef]
- Blackie, E.J.; Le Ru, E.C.; Etchegoin, P.G. Single-molecule surface-enhanced Raman spectroscopy of nonresonant molecules. J. Am. Chem. Soc. 2009, 131, 14466–14472. [Google Scholar] [CrossRef]
- Lindquist, N.C.; de Albuquerque, C.D.L.; Sobral-Filho, R.G.; Paci, I.; Brolo, A.G. High-speed imaging of surface-enhanced Raman scattering fluctuations from individual nanoparticles. Nat. Nanotechnol. 2019, 14, 981–987. [Google Scholar] [CrossRef]
- de Albuquerque, C.D.L.; Hokanson, K.M.; Thorud, S.R.; Sobral-Filho, R.G.; Lindquist, N.C.; Brolo, A.G. Dynamic imaging of multiple SERS hotspots on single nanoparticles. ACS Photonics 2020, 7, 434–443. [Google Scholar] [CrossRef]
- Li, J.; Wuethrich, A.; Sina, A.A.I.; Cheng, H.-H.; Wang, Y.; Behren, A.; Mainwaring, P.N.; Trau, M. A digital single-molecule nanopillar SERS platform for predicting and monitoring immune toxicities in immunotherapy. Nat. Commun. 2021, 12, 1087. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sina, A.A.I.; Antaw, F.; Fielding, D.; Möller, A.; Lobb, R.; Wuethrich, A.; Trau, M. Digital decoding of single extracellular vesicle phenotype differentiates early malignant and benign lung lesions. Adv. Sci. 2022, 9, 2204207. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Hu, C.; Jia, D.; Fan, W.; Ren, W.; Liu, C. Amplification-free and mix-and-read analysis of multiplexed microRNAs on a single plasmonic microbead. Nano Lett. 2021, 21, 6718–6724. [Google Scholar] [CrossRef]
- Zhang, Y.; de Aberasturi, D.J.; Henriksen-Lacey, M.; Langer, J.; Liz-Marzán, L.M. Live-cell surface-enhanced Raman spectroscopy imaging of intracellular pH: From two dimensions to three dimensions. ACS Sens. 2020, 5, 3194–3206. [Google Scholar] [CrossRef]
- Zheng, X.S.; Zong, C.; Wang, X.; Ren, B. Cell-penetrating peptide conjugated SERS nanosensor for in situ intracellular pH imaging of single living cells during cell cycle. Anal. Chem. 2019, 91, 8383–8389. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Bando, K.; Mochizuki, K.; Taguchi, A.; Fujita, K.; Kawata, S. Quantitative evaluation of surface-enhanced Raman scattering nanoparticles for intracellular pH sensing at a single particle level. Anal. Chem. 2019, 91, 3254–3262. [Google Scholar] [CrossRef] [PubMed]
- Laing, S.; Jamieson, L.E.; Faulds, K.; Graham, D. Surface-enhanced Raman spectroscopy for in vivo biosensing. Nat. Rev. Chem. 2017, 1, 0060. [Google Scholar] [CrossRef] [Green Version]
- Gao, P.F.; Lei, G.; Huang, C.Z. Dark-field microscopy: Recent advances in accurate analysis and emerging applications. Anal. Chem. 2021, 93, 4707–4726. [Google Scholar] [CrossRef]
- Xiao, L.; Wei, L.; He, Y.; Yeung, E.S. Single molecule biosensing using color coded plasmon resonant metal nanoparticles. Anal. Chem. 2010, 82, 6308–6314. [Google Scholar] [CrossRef] [PubMed]
- Schultz, S.; Smith, D.R.; Mock, J.J.; Schultz, D.A. Single-target molecule detection with nonbleaching multicolor optical immunolabels. Proc. Natl. Acad. Sci. USA 2000, 97, 996–1001. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Wang, X.; Feng, J.; Huang, C.; Fan, Z. Individual plasmonic nanoprobes for biosensing and bioimaging: Recent advances and perspectives. Small 2021, 17, 2004287. [Google Scholar] [CrossRef]
- Tian, T.; Yao, Y.; Yang, B.; Zhang, K.; Liu, B. Ultrasensitive amplification-free detection of protein kinase based on catalyzed assembly and enumeration of gold nanoparticles. Chem. Commun. 2019, 55, 2505–2508. [Google Scholar] [CrossRef]
- Tian, T.; Zhao, J.; Wang, Y.; Li, B.; Qiao, L.; Zhang, K.; Liu, B. Transpeptidation-mediated single-particle imaging assay for sensitive and specific detection of sortase with dark-field optical microscopy. Biosens. Bioelectron. 2021, 178, 113003. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Jeong, S.; Gonzalez, G.; Marks, H.; Ho, A.; Roussakis, E.; Krauledat, P.B.; Hansen, P.; Evans, C.L. Assessment of glial fibrillary acidic protein binding to the surface of leukocytes with dark-field imaging and computational analysis. Adv. Funct. Mater. 2021, 31, 2009229. [Google Scholar] [CrossRef]
- Markhali, B.P.; Sriram, M.; Bennett, D.T.; Khiabani, P.S.; Hoque, S.; Tilley, R.D.; Bakthavahsalam, P.; Gooding, J.J. Single particle detection of protein molecules using dark-field microscopy to avoid signals from nonspecific adsorption. Biosens. Bioelectron. 2020, 169, 112612. [Google Scholar] [CrossRef]
- Wang, G.; Guo, Y.; Liu, Y.; Zhou, W.; Wang, G. Algorithm-assisted detection and imaging of microRNAs in living cancer cells via the disassembly of plasmonic core-satellite probes coupled with strand displacement amplification. ACS Sens. 2021, 6, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, K.; Wei, W.; Liu, Y.; Liu, S. Multifunctional plasmonic core-satellites nanoprobe for cancer diagnosis ans therapy based on a cascade reaction induced by microRNA. Anal. Chem. 2021, 93, 9521–9530. [Google Scholar] [CrossRef]
- Liu, J.J.; Yan, H.H.; Zhang, Q.; Gao, P.F.; Li, C.M.; Liang, G.L.; Huang, C.Z.; Wang, J. High-resolution vertical polarization excited dark-field microscopic imaging of anisotropic gold nanorods for the sensitive detection and spatial imaging of intracellular microRNA-21. Anal. Chem. 2020, 92, 13118–13125. [Google Scholar] [CrossRef]
- Ma, J.; Gao, M.X.; Zuo, H.; Li, Y.F.; Gao, P.F.; Huang, C.Z. Distance-dependence study of plasmon resonance energy transfer with DNA spacers. Anal. Chem. 2020, 92, 14278–14283. [Google Scholar] [CrossRef]
- Visser, E.W.A.; Yan, J.; van IJzendoorn, L.J.; Prins, M.W.J. Continuous biomarker monitoring by particle mobility sensing with single molecule resolution. Nat. Commun. 2018, 9, 2541. [Google Scholar] [CrossRef] [Green Version]
- Liang, K.; Liu, F.; Fan, J.; Sun, D.; Liu, C.; Lyonm, C.J.; Bernard, D.W.; Li, Y.; Yokoi, K.; Katz, M.H.; et al. Nanoplasmonic quantification of tumor-derived extracellular vesicles in plasma microsamples for diagnosis and treatment monitoring. Nat. Biomed. Eng. 2017, 1, 0021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, W.; LaCourse, S.M.; Song, B.; Singh, D.K.; Khanna, M.; Olivo, J.; Stern, J.; Escudero, J.N.; Vergara, C.; Zhang, F.; et al. Diagnosis of paediatric tuberculosis by optically detecting two virulence factors on extracellular vesicles in blood samples. Nat. Biomed. Eng. 2022, 6, 979–991. [Google Scholar] [CrossRef]
- Huang, M.; Yang, J.; Wang, T.; Song, J.; Xia, J.; Wu, L.; Wang, W.; Wu, Q.; Zhu, Z.; Song, Y.; et al. Homogeneous, low-volume, efficient, and sensitive quantitation of circulating exosomal PD-L1 for cancer diagnosis and immunotherapy response prediction. Angew. Chem. Int. Ed. 2020, 59, 4800–4805. [Google Scholar] [CrossRef]
- Gidi, Y.; Bayram, S.; Ablenas, C.J.; Blum, A.S.; Cosa, G. Efficient one-step PEG-silane passivation of glass surfaces for single-molecule fluorescence studies. ACS Appl. Mater. Interfaces 2018, 10, 39505–39511. [Google Scholar] [CrossRef]
- Sriram, M.; Markhali, B.P.; Nicovich, P.R.; Bennett, D.T.; Reece, P.J.; Hibbert, D.B.; Tilley, R.D.; Gaus, K.; Vivekchand, S.R.C.; Gooding, J.J. A rapid readout for many single plasmonic nanoparticles using dark-field microscopy and digital color analysis. Biosens. Bioelectron. 2018, 117, 530–536. [Google Scholar] [CrossRef]
- Park, K.-D.; May, M.A.; Leng, H.; Wang, J.; Kropp, J.A.; Gougousi, T. Tip-enhanced strong coupling spectroscopy, imaging, and control of a single quantum emitter. Sci. Adv. 2019, 5, eaav5931. [Google Scholar] [CrossRef] [Green Version]
- Niekamp, S.; Stuurman, N.; Vale, R.D. A 6-nm ultra-photostable DNA FluoroCube for fluorescence imaging. Nat. Methods 2020, 17, 437–441. [Google Scholar] [CrossRef]
- Huang, Q.; Chen, X.; Meng, Q.-F.; Yue, L.; Jiang, W.; Zhao, X.-Z.; Rao, L.; Chen, X.; Chen, S. Microfluidics-assisted fluorescence mapping of DNA phosphorothioation. Anal. Chem. 2022, 94, 10479–10486. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, C.; Zou, X.; Tian, X.; Hu, J.; Zhang, C. Simultaneous enzyme-free detection of multiple long noncoding RNAs in cancer cells at single-molecule/particle level. Nano Lett. 2021, 21, 4193–4210. [Google Scholar] [CrossRef]
- Norman, M.; Ter-Ovanesyan, D.; Trieu, W.; Lazarovits, R.; Kowal, E.J.K.; Lee, J.H.; Chen-Plotkin, A.S.; Regev, A.; Church, G.M.; Walt, D.R. L1CAM is not associated with extracellular vesicles in human cerebrospinal fluid or plasma. Nat. Methods 2021, 18, 631–634. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, S.; Chaurasia, A.; Ma, D.; Mlodzianoski, M.J.; Culurciello, E.; Huang, F. Analyzing complex single-molecule emission patterns with deep learning. Nat. Methods 2018, 15, 913–916. [Google Scholar] [CrossRef]
- Speiser, A.; Müller, L.-R.; Hoess, P.; Matti, U.; Obara, C.J.; Legant, W.R.; Kreshuk, A.; Macke, J.H.; Ries, J.; Turaga, S.C. Deep learning enables fast and dense single-molecule localization with high accuracy. Nat. Methods 2021, 18, 1082–1090. [Google Scholar] [CrossRef]
- Song, M.K.; Chen, S.X.; Hu, P.P.; Huang, C.Z.; Zhou, J. Automated plasmonic resonance scattering imaging analysis via deep learning. Anal. Chem. 2021, 93, 2619–2626. [Google Scholar] [CrossRef]

| Methods | Probes | Targets | Linear Range | Detection Limit | Ref. |
|---|---|---|---|---|---|
| Fluorescence | Cy3, Cy5 | miRNA-21, miRNA-122 | 10 fM–1 nM, 10 fM–1 nM | 5 fM, 5 fM | [83] |
| Fluorescence | QDs, Cy5 | miRNA-21 | 1 fM–1 pM | 1 fM | [95] |
| Fluorescence | QDs | DNA MTase | 0.001–1 U/mL | 0.0005 U/mL | [99] |
| Fluorescence | QDs | miRNA-375 | / | 10 aM | [100] |
| Fluorescence | UCNPs | PSA 1 | 0–500 pM | 1 pM | [103] |
| Fluorescence | UCNPs | Aflatoxin B1 | 3.31–125 ng/mL | 0.17 ng/mL | [104] |
| Fluorescence | UCNPs | PSA | 0.1–1000 pg/mL | 23 fg/mL | [105] |
| ECL | RuDSNS/ AuNPs | CK19 2 | 0.01–10 ng/mL | 0.12 pg/mL | [117] |
| SERS | Labeled AuNPs | miRNA-21 | 0.1–100 pM | 0.1 pM | [131] |
| DFM | AuNPs | SrtA 3 | 0.05–50 nM | 7.9 pM | [141] |
| DFM | AuNPs | IL-6 4 | / | 7 pg/mL | [143] |
| DFM | AuNPs | miRNA-21 | 10–200 pM | 2 pM | [144] |
| DFM | AuNRs, AuNPs | miRNA-21 | 0.02–1 nM | 2 pM | [146] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Li, B.; Liu, B.; Zhang, K. Single-Particle Optical Imaging for Ultrasensitive Bioanalysis. Biosensors 2022, 12, 1105. https://doi.org/10.3390/bios12121105
Liu Y, Li B, Liu B, Zhang K. Single-Particle Optical Imaging for Ultrasensitive Bioanalysis. Biosensors. 2022; 12(12):1105. https://doi.org/10.3390/bios12121105
Chicago/Turabian StyleLiu, Yujie, Binxiao Li, Baohong Liu, and Kun Zhang. 2022. "Single-Particle Optical Imaging for Ultrasensitive Bioanalysis" Biosensors 12, no. 12: 1105. https://doi.org/10.3390/bios12121105
APA StyleLiu, Y., Li, B., Liu, B., & Zhang, K. (2022). Single-Particle Optical Imaging for Ultrasensitive Bioanalysis. Biosensors, 12(12), 1105. https://doi.org/10.3390/bios12121105




