Quantum Simulation of the Silicene and Germanene for Sensing and Sequencing of DNA/RNA Nucleobases
Abstract
1. Introduction
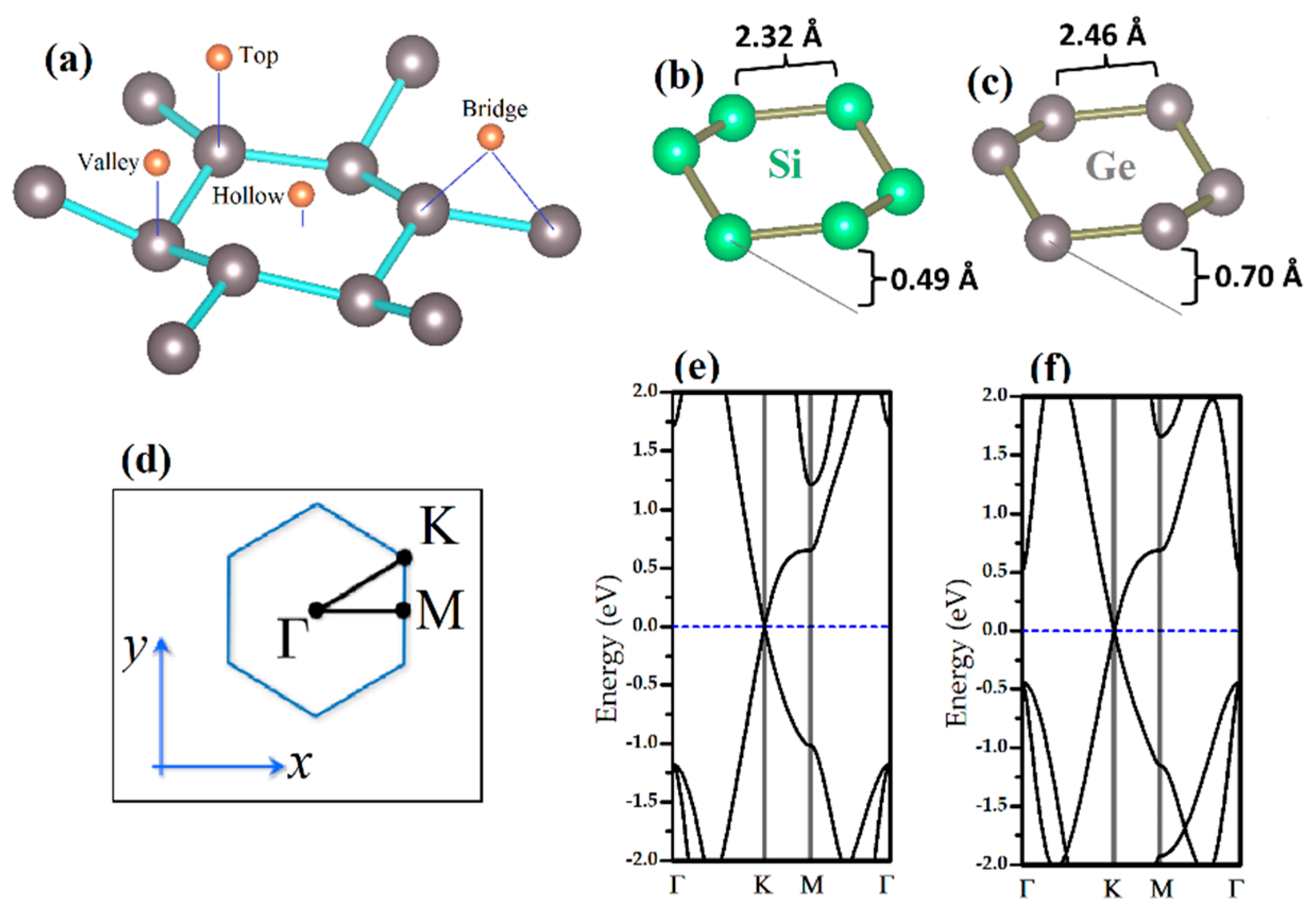
2. Materials and Methods
3. Results and Discussion
3.1. Binding Mechanism of Bare Silicene and Germanene Monolayers with Nucleobases
3.2. Binding Mechanism of Li/Au-Doped Silicene and Germanene Monolayers with Nucleobases
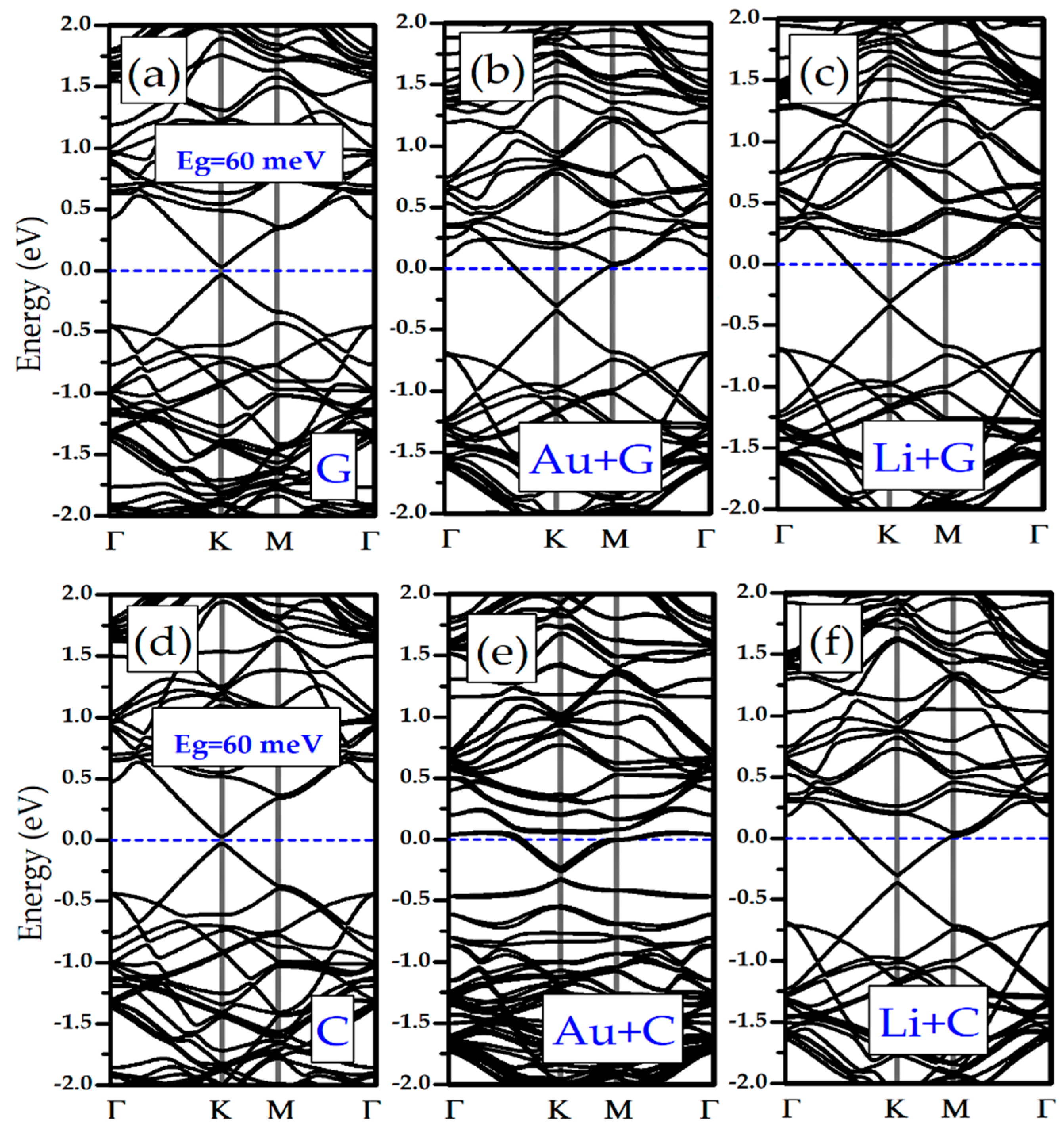
3.3. Electronic Properties of Li/Au-Doped Silicene and Germanene Monolayers with Nucleobases
3.4. Electronic Transport Properties
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Geim, A.K.; Novoselov, K.S. The rise of graphene. Nat. Mater. 2007, 6, 183–191. [Google Scholar] [CrossRef]
- Vogt, P.; De Padova, P.; Quaresima, C.; Avila, J.; Frantzeskakis, E.; Asensio, M.C.; Resta, A.; Ealet, B.; Le Lay, G. Silicene: Compelling experimental evidence for graphene like two-dimensional silicon. Phys. Rev. Lett. 2012, 108, 155501. [Google Scholar]
- Derivaz, M.; Dentel, D.; Stephan, R.; Hanf, M.C.; Mehdaoui, A.; Sonnet, P.; Pirri, C. Continuous germanene layer on Al (111). Nano Lett. 2015, 15, 2510–2516. [Google Scholar] [CrossRef]
- Cassabois, G.; Valvin, P.; Gil, B. Hexagonal boron nitride is an indirect bandgap semiconductor. Nat. Photonics 2016, 10, 262–266. [Google Scholar] [CrossRef]
- Li, Y.; Wang, H.; Xie, L.; Liang, Y.; Hong, G.; Dai, H. MoS2 nanoparticles grown on graphene: An advanced catalyst for the hydrogen evolution reaction. J. Am. Chem. Soc. 2011, 133, 7296–7299. [Google Scholar] [CrossRef]
- Zhu, F.F.; Chen, W.J.; Xu, Y.; Gao, C.L.; Guan, D.D.; Liu, C.H.; Qian, D.; Zhang, C.; Jia, J.F. Epitaxial growth of two-dimensional stanene. Nat. Mater. 2015, 14, 1020–1025. [Google Scholar] [CrossRef]
- Kalantar-zadeh, K.; Ou, J.Z. Biosensors based on two-dimensional MoS2. ACS Sens. 2016, 1, 5–16. [Google Scholar] [CrossRef]
- Huang, K.; Li, Z.; Lin, J.; Han, G.; Huang, P. Two-dimensional transition metal carbides and nitrides (MXenes) for biomedical applications. Chem. Soc. Rev. 2018, 47, 5109–5124. [Google Scholar] [CrossRef]
- Zhao, H.; Ding, R.; Zhao, X.; Li, Y.; Qu, L.; Pei, H.; Yildirimer, L.; Wu, Z.; Zhang, W. Graphene-based nanomaterials for drug and/or gene delivery, bioimaging, and tissue engineering. Drug Discov. Today 2017, 22, 1302–1317. [Google Scholar] [CrossRef]
- Suvarnaphaet, P.; Pechprasarn, S. Graphene-based materials for biosensors: A review. Sensors 2017, 17, 2161. [Google Scholar]
- Heerema, S.J.; Dekker, C. Graphene nanodevices for DNA sequencing. Nat. Nanotechnol. 2016, 11, 127–136. [Google Scholar] [CrossRef]
- Lee, J.H.; Choi, Y.K.; Kim, H.J.; Scheicher, R.H.; Cho, J.H. Physisorption of DNA nucleobases on h-BN and graphene: VdW-corrected DFT calculations. J. Phys. Chem. C 2013, 117, 13435–13441. [Google Scholar] [CrossRef]
- Gowtham, S.; Scheicher, R.H.; Ahuja, R.; Pandey, R.; Karna, S.P. Physisorption of nucleobases on graphene: Density-functional calculations. Phys. Rev. B 2007, 76, 033401. [Google Scholar] [CrossRef]
- Amorim, R.G.; Scheicher, R.H. Silicene as a new potential DNA sequencing device. Nanotechnology 2015, 26, 154002. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, D. Interaction of Nucleic Acid Bases (NABs) with Graphene (GR) and Boron Nitride Graphene (BNG). J. Mol. Struct. 2020, 1222, 128889. [Google Scholar] [CrossRef]
- Gürel, H.H.; Salmankurt, B. Binding mechanisms of DNA/RNA nucleobases adsorbed on graphene under charging: First-principles van der Waals study. Mater. Res. Express 2017, 4, 065401. [Google Scholar] [CrossRef]
- Gorkan, T.; Kadioglu, Y.; Aktürk, E.; Ciraci, S. Interactions of selected organic molecules with a blue phosphorene monolayer: Self-assembly, solvent effect, enhanced binding and fixation through coadsorbed gold clusters. Phys. Chem. Chem. Phys. 2020, 22, 26552–26561. [Google Scholar] [CrossRef]
- Vovusha, H.; Sanyal, B. Adsorption of nucleobases on 2D transition-metal dichalcogenides and graphene sheet: A first principles density functional theory study. RSC Adv. 2015, 5, 67427–67434. [Google Scholar] [CrossRef]
- Corteés-Arriagada, D. Phosphorene as a template material for physisorption of DNA/RNA nucleobases and resembling of base pairs: A cluster DFT study and comparisons with graphene. J. Phys. Chem. C 2018, 122, 4870–4880. [Google Scholar] [CrossRef]
- Hussain, T.; Vovusha, H.; Kaewmaraya, T.; Amornkitbamrung, V.; Ahuja, R. Adsorption characteristics of DNA nucleobases, aromatic amino acids and heterocyclic molecules on silicene and germanene monolayers. Sens. Actuators B Chem. 2018, 255, 2713–2720. [Google Scholar] [CrossRef]
- Le, D.; Kara, A.; Schröder, E.; Hyldgaard, P.; Rahman, T.S. Physisorption of nucleobases on graphene: A comparative van der Waals study. J. Phys. Condens. Matter 2012, 24, 424210. [Google Scholar] [CrossRef]
- Farimani, A.B.; Min, K.; Aluru, N.R. DNA base detection using a single-layer MoS2. ACS Nano 2014, 8, 7914–7922. [Google Scholar] [CrossRef]
- Srimathi, U.; Nagarajan, V.; Chandiramouli, R. Detection of nucleobases using 2D germanane nanosheet: A first-principles study. Comput. Theor. Chem. 2018, 1130, 68–76. [Google Scholar] [CrossRef]
- Ramraj, A.; Hillier, I.H.; Vincent, M.A.; Burton, N.A. Assessment of approximate quantum chemical methods for calculating the interaction energy of nucleic acid bases with graphene and carbon nanotubes. Chem. Phys. Lett. 2010, 484, 295–298. [Google Scholar] [CrossRef]
- Sadeghi, M.; Jahanshahi, M.; Ghorbanzadeh, M.; Najafpour, G. Adsorption of DNA/RNA nucleobases onto single-layer MoS2 and Li-Doped MoS2: A dispersion-corrected DFT study. Appl. Surf. Sci. 2018, 434, 176–187. [Google Scholar] [CrossRef]
- Panigrahi, S.; Bhattacharya, A.; Banerjee, S.; Bhattacharyya, D. Interaction of nucleobases with wrinkled graphene surface: Dispersion corrected DFT and AFM studies. J. Phys. Chem. C 2012, 116, 4374–4379. [Google Scholar] [CrossRef]
- Ayatollahi, A.; Roknabadi, M.R.; Behdani, M.; Shahtahmassebi, N.; Sanyal, B. Density functional investigations on the adsorption characteristics of nucleobases on germanene nanoribbons. Phys. E Low-Dimens. Syst. Nanostruct. 2020, 117, 113772. [Google Scholar] [CrossRef]
- Bhavadharani, R.K.; Nagarajan, V.; Chandiramouli, R. Density functional study on the binding properties of nucleobases to stanane nanosheet. Appl. Surf. Sci. 2018, 462, 831–839. [Google Scholar] [CrossRef]
- Sharma, M.; Kumar, A.; Ahluwalia, P.K. Optical fingerprints and electron transport properties of DNA bases adsorbed on monolayer MoS2. RSC Adv. 2016, 6, 60223–60230. [Google Scholar] [CrossRef]
- Feng, B.; Ding, Z.; Meng, S.; Yao, Y.; He, X.; Cheng, P.; Chen, L.; Wu, K. Evidence of silicene in honeycomb structures of silicon on Ag (111). Nano Lett. 2012, 12, 3507–3511. [Google Scholar] [CrossRef]
- Chen, L.; Liu, C.C.; Feng, B.; He, X.; Cheng, P.; Ding, Z.; Meng, S.; Yao, Y.; Wu, K. Evidence for Dirac fermions in a honeycomb lattice based on silicon. Phys. Rev. Lett. 2012, 109, 056804. [Google Scholar] [CrossRef]
- Chen, L.; Li, H.; Feng, B.; Ding, Z.; Qiu, J.; Cheng, P.; Wu, K.; Meng, S. Spontaneous symmetry breaking and dynamic phase transition in monolayer silicene. Phys. Rev. Lett. 2013, 110, 085504. [Google Scholar] [CrossRef]
- Meng, L.; Wang, Y.; Zhang, L.; Du, S.; Wu, R.; Li, L.; Zhang, L.; Zhou, H.; Hofer, W.A.; Gao, H.J. Buckled silicene formation on Ir (111). Nano Lett. 2013, 13, 685–690. [Google Scholar] [CrossRef]
- Fleurence, A.; Friedlein, R.; Ozaki, T.; Kawai, H.; Wang, Y.; Yamada-Takamura, Y. Experimental evidence for epitaxial silicene on diboride thin films. Phys. Rev. Lett. 2012, 108, 245501. [Google Scholar] [CrossRef]
- Li, L.; Lu, S.Z.; Pan, J.; Qin, Z.; Wang, Y.Q.; Wang, Y.; Wang, Y.; Cao, G.; Du, S.; Gao, H.J. Buckled germanene formation on Pt (111). Adv. Mater. 2014, 26, 4820–4824. [Google Scholar] [CrossRef]
- Soler, J.M.; Artacho, E.; Gale, J.D.; García, A.; Junquera, J.; Ordejón, P.; Sánchez-Portal, D. The SIESTA method for ab initio order-N materials simulation. J. Phys. Condens. Matter 2002, 14, 2745. [Google Scholar] [CrossRef]
- Brandbyge, M.; Mozos, J.L.; Ordejón, P.; Taylor, J.; Stokbro, K. Density-functional method for nonequilibrium electron transport. Phys. Rev. B 2002, 65, 165401. [Google Scholar] [CrossRef]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized gradient approximation made simple. Phys. Rev. Lett. 1996, 77, 3865. [Google Scholar] [CrossRef]
- Hamann, D.R.; Schlüter, M.; Chiang, C. Norm-conserving pseudopotentials. Phys. Rev. Lett. 1979, 43, 1494. [Google Scholar] [CrossRef]
- Monkhorst, H.J.; Pack, J.D. Special points for Brillouin-zone integrations. Phys. Rev. B 1976, 13, 5188. [Google Scholar] [CrossRef]
- Grimme, S. Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J. Comput. Chem. 2006, 27, 1787–1799. [Google Scholar] [CrossRef]
- Momma, K.; Izumi, F. VESTA 3 for three-dimensional visualization of crystal, volumetric and morphology data. J. Appl. Crystallogr. 2011, 44, 1272–1276. [Google Scholar] [CrossRef]
- Rocha, A.R.; García-Suárez, V.M.; Bailey, S.; Lambert, C.; Ferrer, J.; Sanvito, S. Spin and molecular electronics in atomically generated orbital landscapes. Phys. Rev. B 2006, 73, 085414. [Google Scholar] [CrossRef]
- Sahin, H.; Peeters, F.M. Adsorption of alkali, alkaline-earth, and 3d transition metal atoms on silicene. Phys. Rev. B 2013, 87, 085423. [Google Scholar] [CrossRef]
- Ye, M.; Quhe, R.; Zheng, J.; Ni, Z.; Wang, Y.; Yuan, Y.; Tse, G.; Shi, J.; Gao, Z.; Lu, J. Tunable band gap in germanene by surface adsorption. Phys. E Low-Dimens. Syst. Nanostruct. 2014, 59, 60–65. [Google Scholar] [CrossRef]
- Kaloni, T.P.; Schwingenschlögl, U. Effects of heavy metal adsorption on silicene. Phys. Status Solidi (RRL)—Rapid Res. Lett. 2014, 8, 685–687. [Google Scholar] [CrossRef]
- Li, C.; Yang, S.; Li, S.S.; Xia, J.B.; Li, J. Au-decorated silicene: Design of a high-activity catalyst toward CO oxidation. J. Phys. Chem. C 2013, 117, 483–488. [Google Scholar] [CrossRef]
- Sosa, A.N.; de Santiago, F.; Miranda, Á.; Trejo, A.; Salazar, F.; Pérez, L.A.; Cruz-Irisson, M. Alkali and transition metal atom-functionalized germanene for hydrogen storage: A DFT investigation. Int. J. Hydrogen Energy 2020. [Google Scholar] [CrossRef]
- Xu, Q.; Si, X.; She, W.; Yang, G.; Fan, X.; Zheng, W. First-Principle Calculation of Optimizing Performance of Germanene-Based Supercapacitors by Vacancies and Metal Atoms. J. Phys. Chem. C 2020, 124, 12346–12358. [Google Scholar] [CrossRef]
- Song, J.; Li, M.; Liang, H.; Lou, H. Adsorption patterns of aromatic amino acids on monolayer MoS2 and Au-modified MoS2 surfaces: A first-principles study. Comput. Theor. Chem. 2017, 1118, 115–122. [Google Scholar] [CrossRef]
- Kadioglu, Y.; Gorkan, T.; Uzengi Akturk, O.; Akturk, E.; Ciraci, S. Enhanced interactions of amino acids and nucleic acid bases with bare black phosphorene monolayer mediated by coadsorbed species. J. Phys. Chem. C 2019, 123, 23691–23704. [Google Scholar] [CrossRef]
- Min, S.K.; Kim, W.Y.; Cho, Y.; Kim, K.S. Fast DNA sequencing with a graphene-based nanochannel device. Nat. Nanotechnol. 2011, 6, 162–165. [Google Scholar]

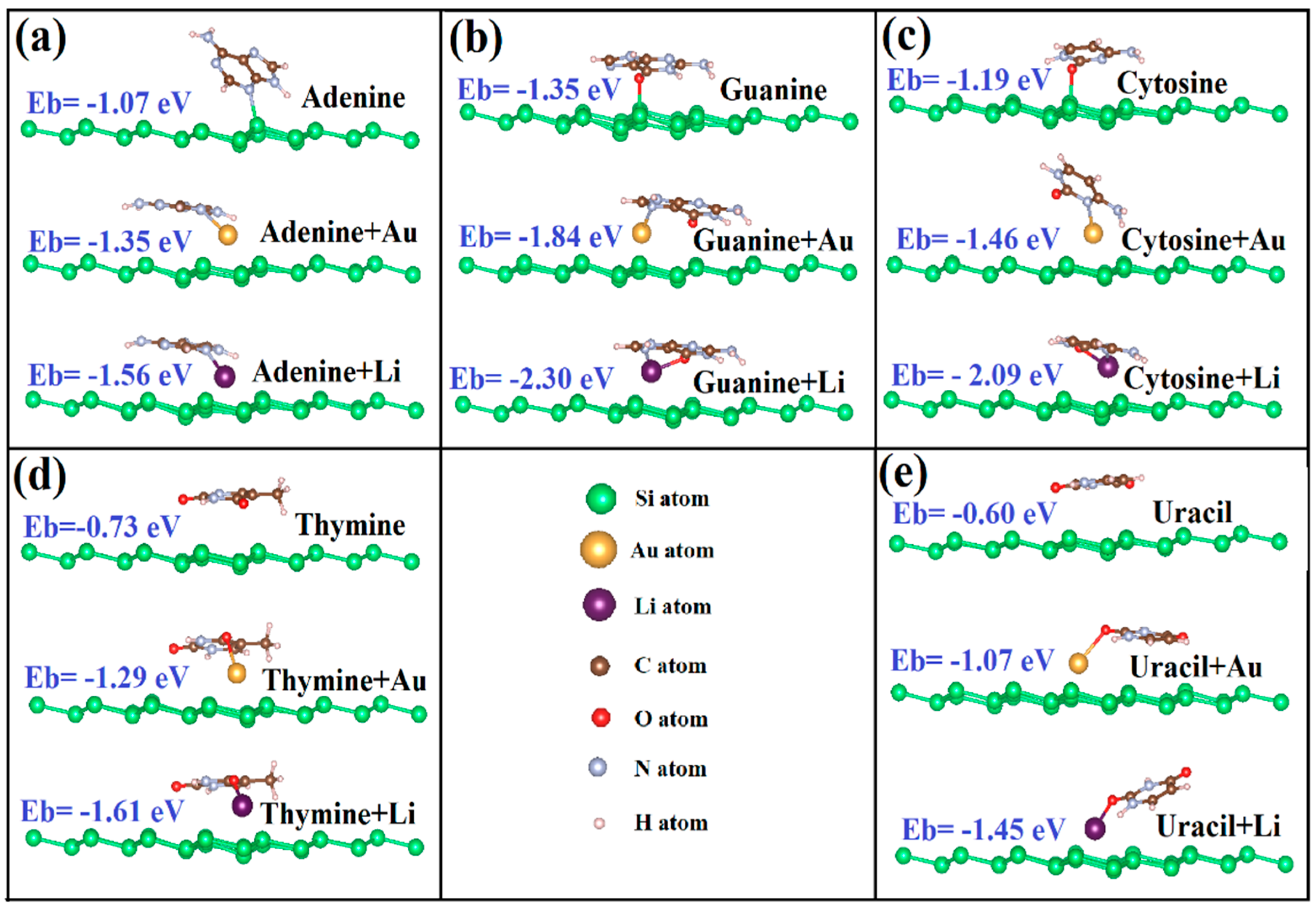
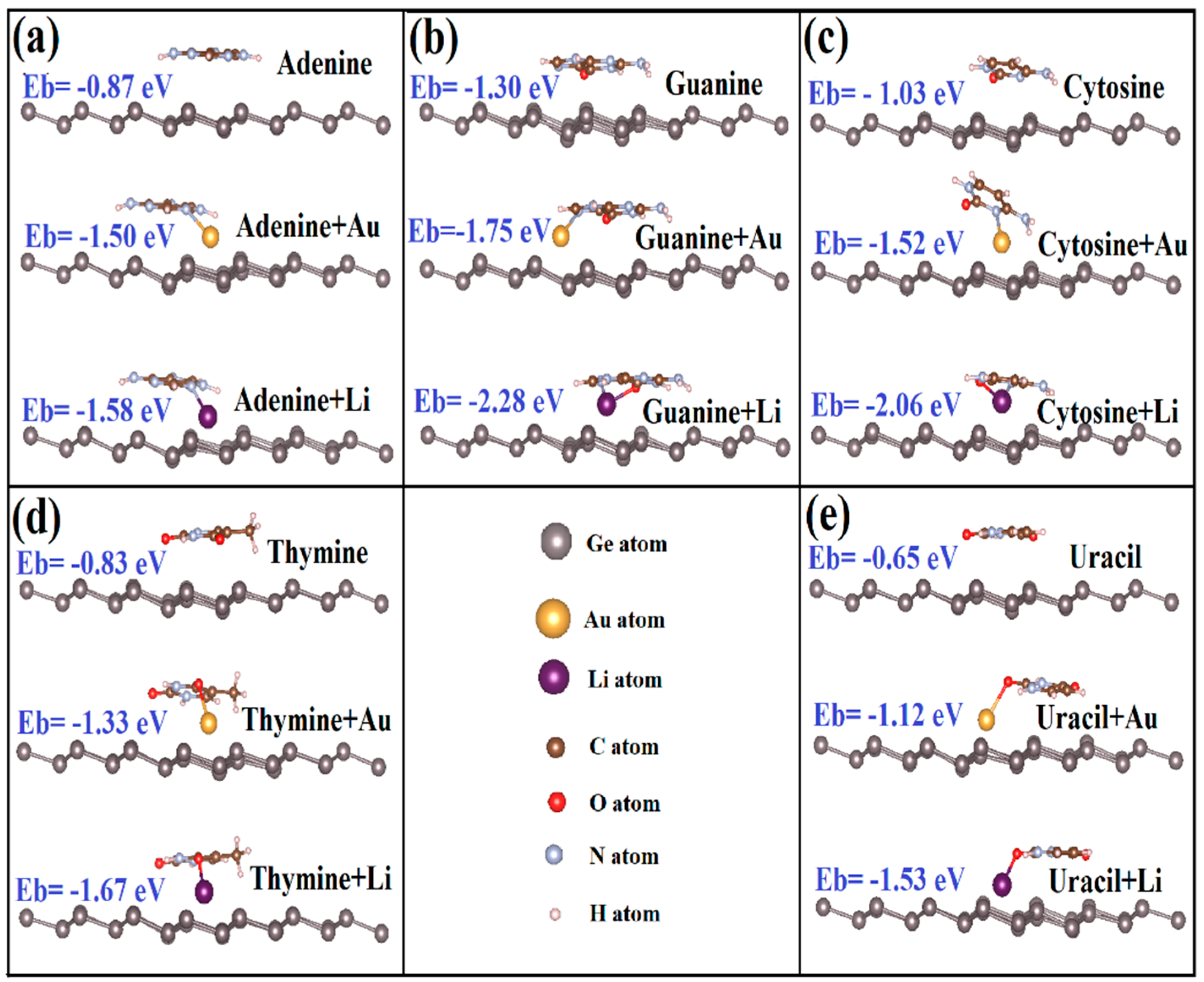
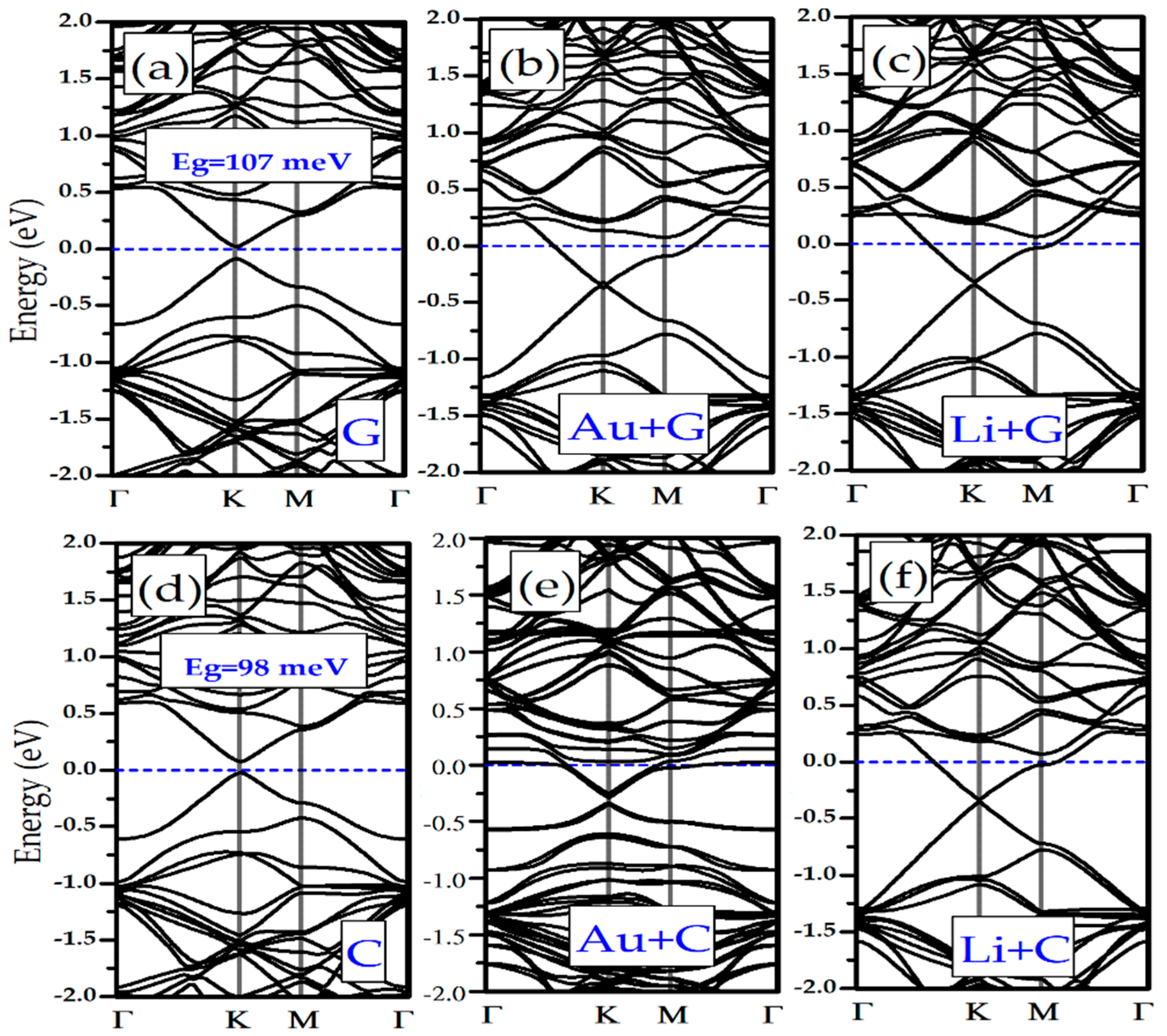



| Layer | G | A | T | C | U |
|---|---|---|---|---|---|
| Si | −1.35 | −1.07 | −0.73 | −1.19 | −0.58 |
| Si [14] | −0.77 | −0.60 | −0.60 | −0.88 | |
| Si [20] | −1.16 | −0.54 | −0.70 | −1.13 | −0.43 |
| Si + Au | −1.84 | −1.35 | −1.29 | −1.46 | −1.07 |
| Si + Li | −2.30 | −1.56 | −1.61 | −2.09 | −1.45 |
| Ge | −1.30 | −0.87 | −0.83 | −1.03 | −0.65 |
| Ge [20] | −0.95 | −0.70 | −0.57 | −0.56 | −0.51 |
| Ge + Au | −1.75 | −1.50 | −1.33 | −1.52 | −1.12 |
| Ge + Li | −2.28 | −1.58 | −1.67 | −2.06 | −1.53 |
| System | G | A | T | C | U |
|---|---|---|---|---|---|
| Si | (Si-O) | (Si-N) | (Si-O) | (Si-O) | (Si-O) |
| 1.95 | 2.02 | 2.77 | 1.92 | 3.28 | |
| Si [14] | (Si-O) | (Si-H) | (Si-H) | (Si-O) | |
| 1.99 | 3.07 | 3.07 | 2.04 | ||
| Si [21] | (Si-O) | (Si-H) | (Si-O) | (Si-O) | (Si-H) |
| 1.82 | 3.22 | 1.85 | 1.81 | 3.13 | |
| Si + Au | (Au-N) | (Au-N) | (Au-O) | (Au-N) | (Au-O) |
| 2.43 | 2.4 | 2.38 | 2.39 | 2.42 | |
| Si + Li | (Li-O) | (Li-N) | (Li-O) | (Li-O) | (Li-O) |
| 2.09 | 2.04 | 1.86 | 2.02 | 1.88 |
| System | G | A | T | C | U |
|---|---|---|---|---|---|
| Ge | (Ge-O) | (Ge-H) | (Ge-O) | (Ge-N) | (Ge-O) |
| 2.41 | 3.17 | 2.96 | 2.61 | 3.09 | |
| Ge [21] | (Ge-O) | (Ge-H) | (Ge-H) | (Ge-O) | (Ge-O) |
| 2.26 | 3.05 | 3.02 | 3.09 | 3.12 | |
| Ge + Au | (Au-N) | (Au-N) | (Au-O) | (Au-N) | (Au-O) |
| 2.43 | 2.41 | 2.42 | 2.4 | 2.43 | |
| Ge + Li | (Li-O) | (Li-N) | (Li-O) | (Li-O) | (Li-O) |
| 2.12 | 2.06 | 1.9 | 2.03 | 1.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gürel, H.H.; Salmankurt, B. Quantum Simulation of the Silicene and Germanene for Sensing and Sequencing of DNA/RNA Nucleobases. Biosensors 2021, 11, 59. https://doi.org/10.3390/bios11030059
Gürel HH, Salmankurt B. Quantum Simulation of the Silicene and Germanene for Sensing and Sequencing of DNA/RNA Nucleobases. Biosensors. 2021; 11(3):59. https://doi.org/10.3390/bios11030059
Chicago/Turabian StyleGürel, Hikmet Hakan, and Bahadır Salmankurt. 2021. "Quantum Simulation of the Silicene and Germanene for Sensing and Sequencing of DNA/RNA Nucleobases" Biosensors 11, no. 3: 59. https://doi.org/10.3390/bios11030059
APA StyleGürel, H. H., & Salmankurt, B. (2021). Quantum Simulation of the Silicene and Germanene for Sensing and Sequencing of DNA/RNA Nucleobases. Biosensors, 11(3), 59. https://doi.org/10.3390/bios11030059





