A 3D-Printed Microfluidic Device for qPCR Detection of Macrolide-Resistant Mutations of Mycoplasma pneumoniae
Abstract
:1. Introduction
2. Materials and Methods
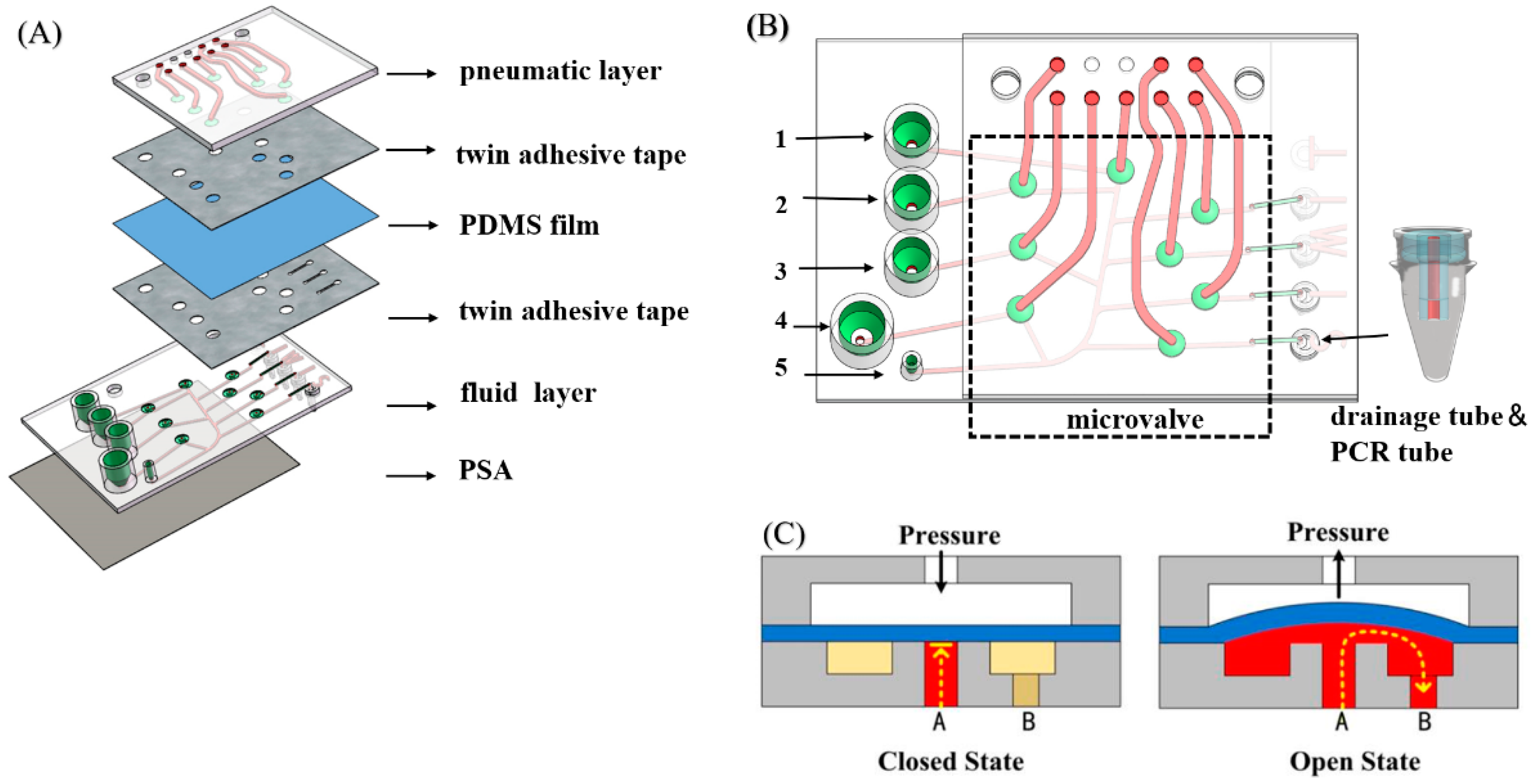
2.1. Device Design and Fabrication
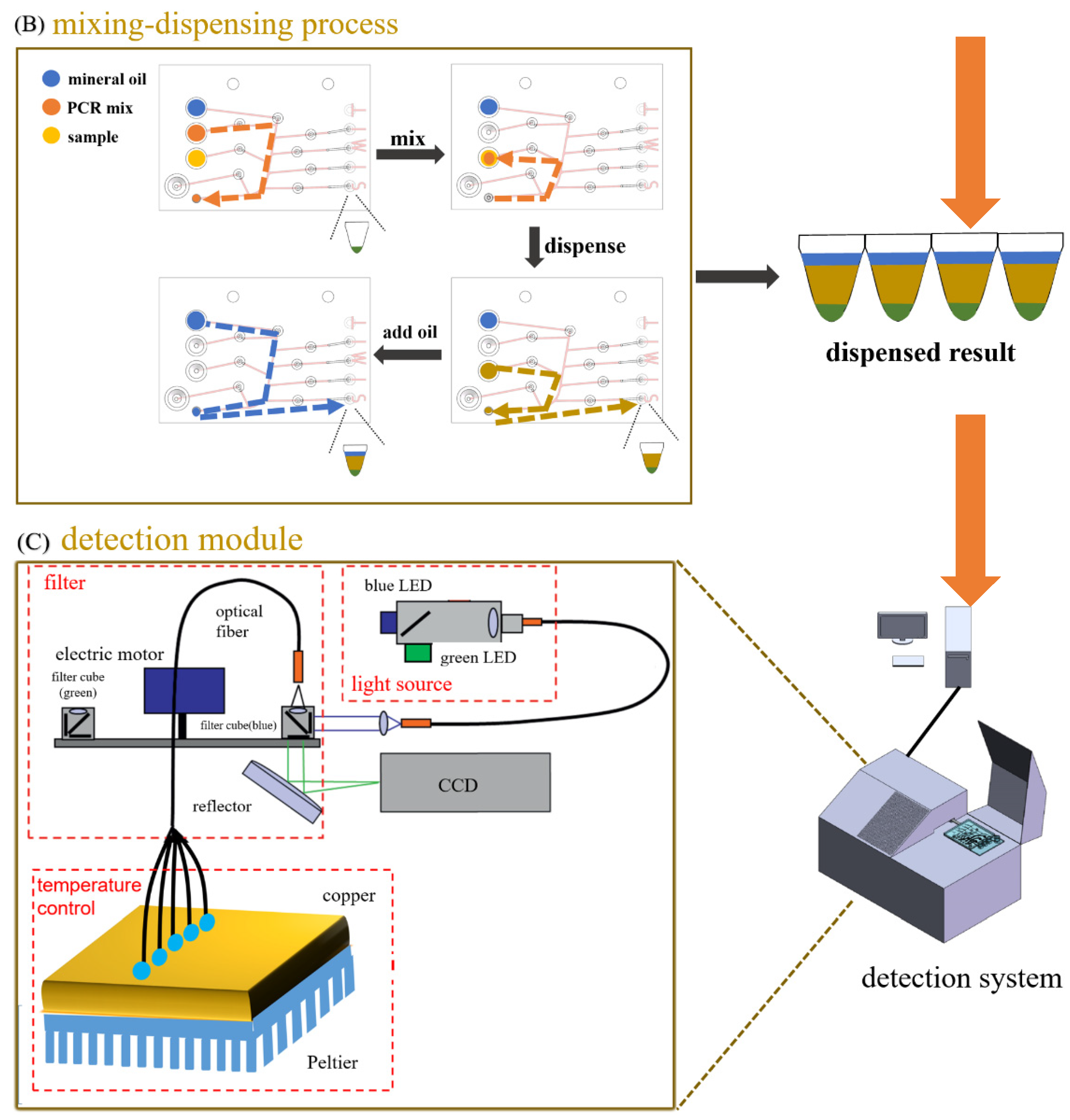
2.2. System Setup and Workflow
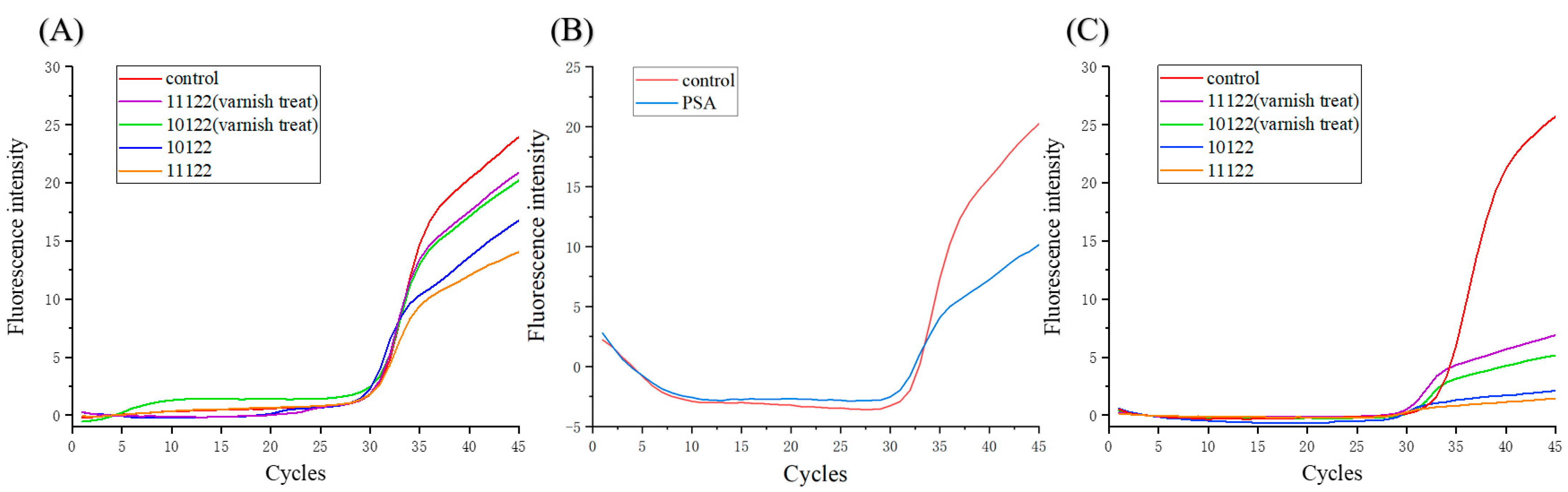
2.3. Analyzing the Effects of Materials and Coatings on PCR
2.4. qPCR Detection of Macrolide-Resistant Mutations of Mycoplasma pneumoniae
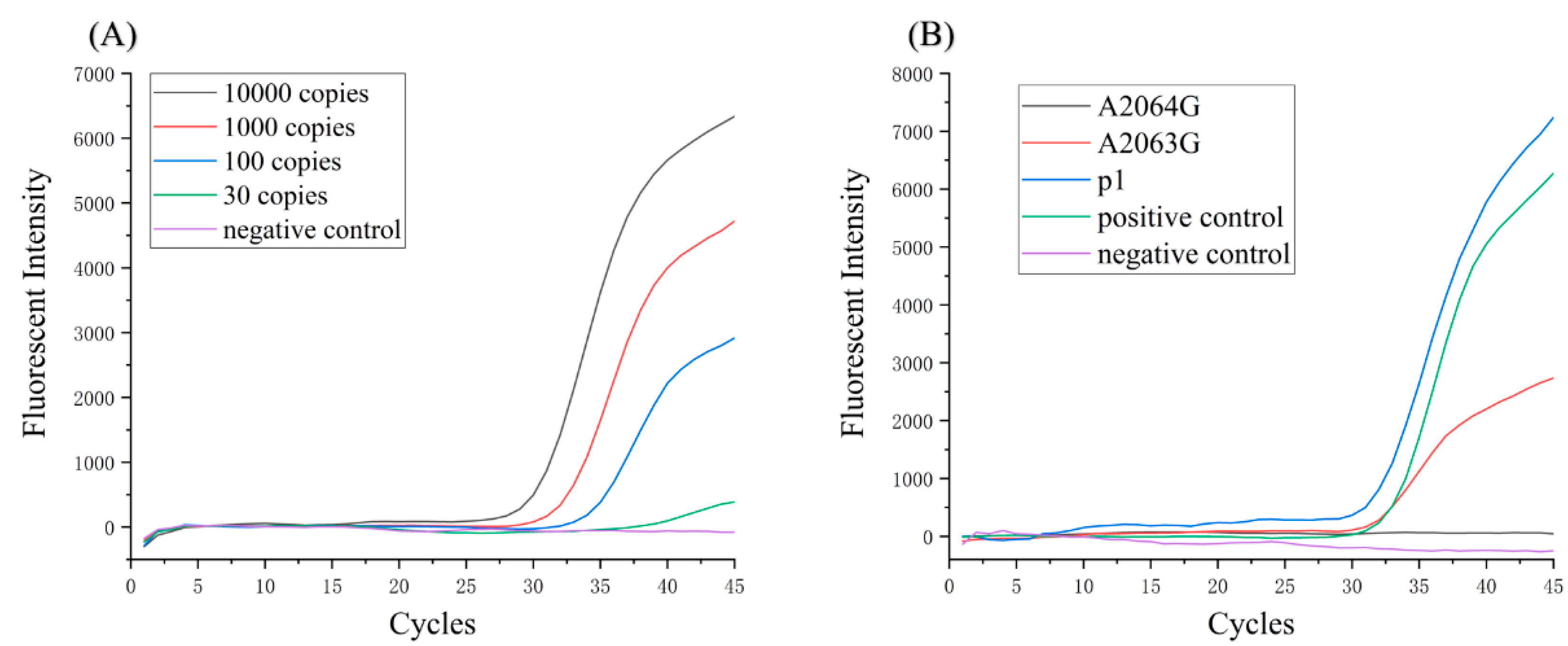
2.5. Sensitivity of Detection System
3. Results
3.1. Characterization of Bonding Strength and Microvalve Performance
3.2. Effects of Materials and Coatings on PCR
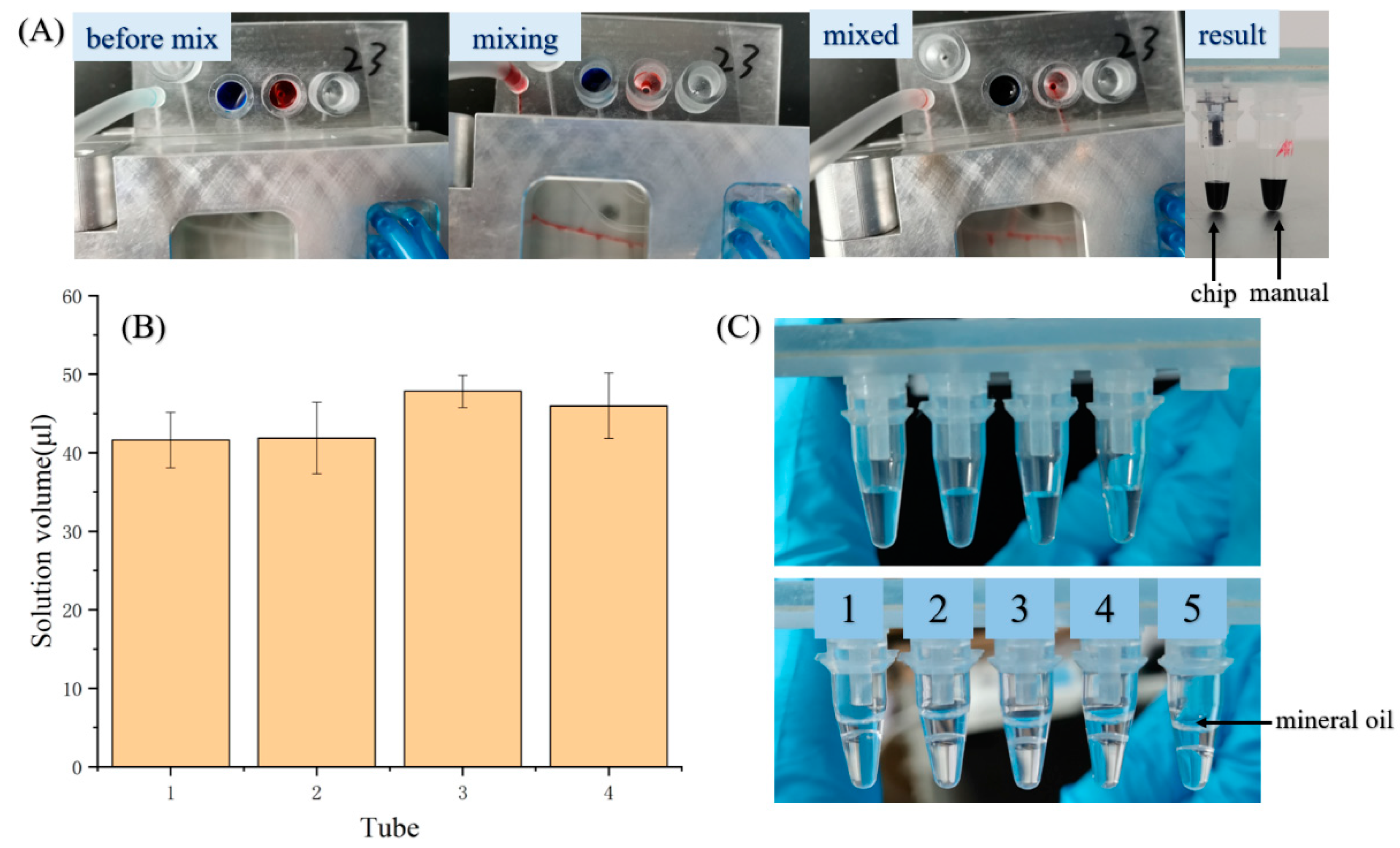
3.3. Sample Preparation
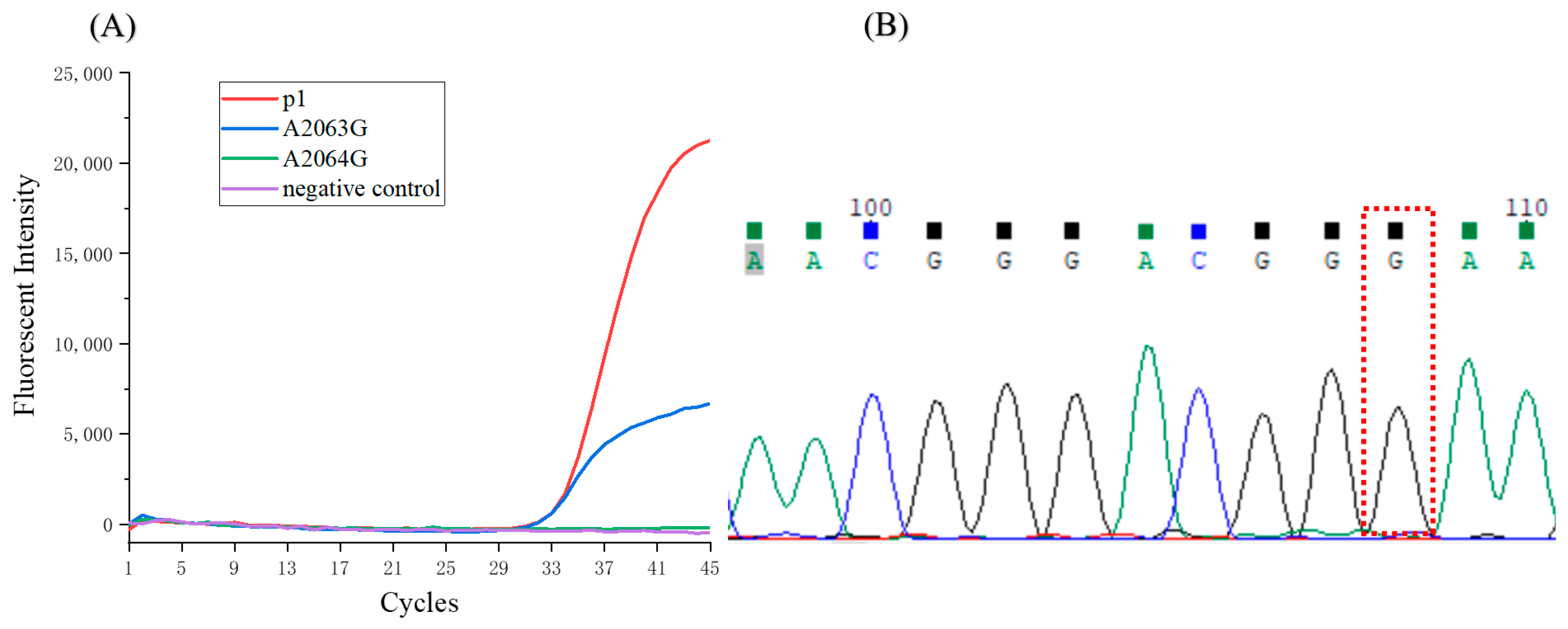
3.4. Macrolide-Resistant Mutations Detection
3.5. Detection of the Clinical Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Waites, K.B.; Talkington, D.F. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 2004, 17, 697–728. [Google Scholar] [CrossRef] [Green Version]
- Jain, S.; Self, W.H.; Wunderink, R.G.; Fakhran, S.; Balk, R.; Bramley, A.M.; Reed, C.; Grijalva, C.G.; Anderson, E.J.; Courtney, D.M. Community-acquired pneumonia requiring hospitalization among US adults. N. Engl. J. Med. 2015, 373, 415–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, S.; Williams, D.J.; Arnold, S.R.; Ampofo, K.; Bramley, A.M.; Reed, C.; Stockmann, C.; Anderson, E.J.; Grijalva, C.G.; Self, W.H. Community-acquired pneumonia requiring hospitalization among US children. N. Engl. J. Med. 2015, 372, 835–845. [Google Scholar] [CrossRef] [Green Version]
- Pereyre, S.; Goret, J.; Bébéar, C. Mycoplasma pneumoniae: Current knowledge on macrolide resistance and treatment. Front. Microbiol. 2016, 7, 974. [Google Scholar] [CrossRef] [Green Version]
- Zhao, F.; Liu, G.; Wu, J.; Cao, B.; Tao, X.; He, L.; Meng, F.; Zhu, L.; Lv, M.; Yin, Y. Surveillance of macrolide-resistant Mycoplasma pneumoniae in Beijing, China, from 2008 to 2012. Antimicrob. Agents Chemother. 2013, 57, 1521–1523. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Ye, X.; Zhang, H.; Xu, X.; Li, W.; Zhu, D.; Wang, M. Antimicrobial susceptibility of Mycoplasma pneumoniae isolates and molecular analysis of macrolide-resistant strains from Shanghai, China. Antimicrob. Agents Chemother. 2009, 53, 2160–2162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bébéar, C.; Pereyre, S. Mechanisms of drug resistance in Mycoplasma pneumoniae. Curr. Drug Targets-Infect. Disord. 2005, 5, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Xin, D.; Mi, Z.; Han, X.; Qin, L.; Li, J.; Wei, T.; Chen, X.; Ma, S.; Hou, A.; Li, G. Molecular mechanisms of macrolide resistance in clinical isolates of Mycoplasma pneumoniae from China. Antimicrob. Agents Chemother. 2009, 53, 2158–2159. [Google Scholar] [CrossRef] [Green Version]
- Daxboeck, F.; Krause, R.; Wenisch, C. Laboratory diagnosis of Mycoplasma pneumoniae infection. Clin. Microbiol. Infect. 2003, 9, 263–273. [Google Scholar] [CrossRef] [Green Version]
- Shin, D.J.; Andini, N.; Hsieh, K.; Yang, S.; Wang, T.H. Emerging Analytical Techniques for Rapid Pathogen Identification and Susceptibility Testing. Annu. Rev. Anal. Chem. 2019, 12, 41–67. [Google Scholar] [CrossRef] [PubMed]
- Ohst, C.; Saschenbrecker, S.; Stiba, K.; Steinhagen, K.; Probst, C.; Radzimski, C.; Lattwein, E.; Komorowski, L.; Stocker, W.; Schlumberger, W. Reliable Serological Testing for the Diagnosis of Emerging Infectious Diseases. Adv. Exp. Med. Biol. 2018, 1062, 19–43. [Google Scholar]
- Razmi, N.; Hasanzadeh, M.; Willander, M.; Nur, O. Recent Progress on the Electrochemical Biosensing of Escherichia coli O157:H7: Material and Methods Overview. Biosensors 2020, 10, 54. [Google Scholar] [CrossRef]
- Ali, A.A.; Altemimi, A.B.; Alhelfi, N.; Ibrahim, S.A. Application of Biosensors for Detection of Pathogenic Food Bacteria: A Review. Biosensors 2020, 10, 58. [Google Scholar] [CrossRef]
- Linger, Y.; Knickerbocker, C.; Sipes, D.; Golova, J.; Franke, M.; Calderon, R.; Lecca, L.; Thakore, N.; Holmberg, R.; Qu, P.; et al. Genotyping Multidrug-Resistant Mycobacterium tuberculosis from Primary Sputum and Decontaminated Sediment with an Integrated Microfluidic Amplification Microarray Test. J. Clin. Microbiol. 2018, 56, e01652-17. [Google Scholar] [CrossRef] [Green Version]
- Gopfert, L.; Elsner, M.; Seidel, M. Isothermal haRPA detection of blaCTX-M in bacterial isolates from water samples and comparison with qPCR. Anal. Methods 2021, 13, 552–557. [Google Scholar] [CrossRef]
- Kuhnemund, M.; Hernandez-Neuta, I.; Sharif, M.I.; Cornaglia, M.; Gijs, M.A.; Nilsson, M. Sensitive and inexpensive digital DNA analysis by microfluidic enrichment of rolling circle amplified single-molecules. Nucleic Acids Res. 2017, 45, e59. [Google Scholar] [CrossRef] [Green Version]
- Moon, J.; Kwon, H.J.; Yong, D.; Lee, I.C.; Kim, H.; Kang, H.; Lim, E.K.; Lee, K.S.; Jung, J.; Park, H.G.; et al. Colorimetric Detection of SARS-CoV-2 and Drug-Resistant pH1N1 Using CRISPR/dCas9. ACS Sens. 2020, 5, 4017–4026. [Google Scholar] [CrossRef]
- Dreyer, V.; Utpatel, C.; Kohl, T.A.; Barilar, I.; Groschel, M.I.; Feuerriegel, S.; Niemann, S. Detection of low-frequency resistance-mediating SNPs in next-generation sequencing data of Mycobacterium tuberculosis complex strains with binoSNP. Sci. Rep. 2020, 10, 7874. [Google Scholar] [CrossRef] [PubMed]
- Baltekin, O.; Boucharin, A.; Tano, E.; Andersson, D.I.; Elf, J. Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging. Proc. Natl. Acad. Sci. USA 2017, 114, 9170–9175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, D.K.; Ali, M.M.; Zhang, K.; Huang, S.S.; Peterson, E.; Digman, M.A.; Gratton, E.; Zhao, W. Rapid detection of single bacteria in unprocessed blood using Integrated Comprehensive Droplet Digital Detection. Nat. Commun. 2014, 5, 5427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Li, H.; Xie, H.; Xu, D. A novel method combining aptamer-Ag10NPs based microfluidic biochip with bright field imaging for detection of KPC-2-expressing bacteria. Anal. Chim. Acta 2020, 1132, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.V.; Nguyen, V.D.; Nguyen, H.Q.; Chau, T.H.; Lee, E.Y.; Seo, T.S. Nucleic acid diagnostics on the total integrated lab-on-a-disc for point-of-care testing. Biosens. Bioelectron. 2019, 141, 111466. [Google Scholar] [CrossRef]
- Ma, Y.D.; Li, K.H.; Chen, Y.H.; Lee, Y.M.; Chou, S.T.; Lai, Y.Y.; Huang, P.C.; Ma, H.P.; Lee, G.B. A sample-to-answer, portable platform for rapid detection of pathogens with a smartphone interface. Lab Chip 2019, 19, 3804–3814. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ma, Z.; Qin, J.; Shen, Z.; Liu, Q.; Chen, X.; Wang, H.; An, Z.; Liu, W.; Li, M. A versatile loop-mediated isothermal amplification microchip platform for Streptococcus pneumoniae and Mycoplasma pneumoniae testing at the point of care. Biosens Bioelectron. 2019, 126, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.Y.; Wang, C.H.; Che, Y.J.; Kao, C.Y.; Wu, J.J.; Lee, G.B. An integrated microfluidic system for diagnosis of the resistance of Helicobacter pylori to quinolone-based antibiotics. Biosens. Bioelectron. 2016, 78, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Naderi, A.; Bhattacharjee, N.; Folch, A. Digital Manufacturing for Microfluidics. Annu. Rev. Biomed. Eng. 2019, 21, 325–364. [Google Scholar] [CrossRef]
- Nielsen, A.V.; Beauchamp, M.J.; Nordin, G.P.; Woolley, A.T. 3D Printed Microfluidics. Annu. Rev. Anal. Chem. 2020, 13, 45–65. [Google Scholar] [CrossRef] [PubMed]
- Unger, M.A.; Chou, H.P.; Thorsen, T.; Scherer, A.; Quake, S.R. Monolithic Microfabricated Valves and Pumps by Multilayer Soft Lithography. Science 2000, 288, 113–116. [Google Scholar] [CrossRef] [Green Version]
- Eddings, M.A.; Johnson, M.A.; Gale, B.K. Determining the optimal PDMS–PDMS bonding technique for microfluidic devices. J. Micromech. Microeng. 2008, 18, 067001. [Google Scholar] [CrossRef]
- Lee, Y.S.; Bhattacharjee, N.; Folch, A. 3D-printed Quake-style microvalves and micropumps. Lab Chip 2018, 18, 1207–1214. [Google Scholar] [CrossRef]
- Tzivelekis, C.; Sgardelis, P.; Waldron, K.; Whalley, R.; Huo, D.; Dalgarno, K. Fabrication routes via projection stereolithography for 3D-printing of microfluidic geometries for nucleic acid amplification. PLoS ONE 2020, 15, e0240237. [Google Scholar] [CrossRef] [PubMed]


| Primer | Probe | Other | Sample Treated by the Device | |
|---|---|---|---|---|
| Tube 1 (conserved sequence) | P1 gene | P1 gene | / | Plasmid/clinical sample |
| Tube 2 (A2063G mutation) | 23S rRNA | A2063G | / | plasmid/clinical sample |
| Tube 3 (A2064G mutation) | 23S rRNA | A2064G | / | plasmid/clinical sample |
| Tube 4 (positive control) | 23S rRNA | A2063G | A2063G plasmid | plasmid/clinical sample |
| Tube 5 (negative control) | 23S rRNA | A2063G | water | / |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, A.; Wu, Z.; Huang, Y.; Zhou, H.; Wu, L.; Jia, C.; Chen, Q.; Zhao, J. A 3D-Printed Microfluidic Device for qPCR Detection of Macrolide-Resistant Mutations of Mycoplasma pneumoniae . Biosensors 2021, 11, 427. https://doi.org/10.3390/bios11110427
Wang A, Wu Z, Huang Y, Zhou H, Wu L, Jia C, Chen Q, Zhao J. A 3D-Printed Microfluidic Device for qPCR Detection of Macrolide-Resistant Mutations of Mycoplasma pneumoniae . Biosensors. 2021; 11(11):427. https://doi.org/10.3390/bios11110427
Chicago/Turabian StyleWang, Anyan, Zhenhua Wu, Yuhang Huang, Hongbo Zhou, Lei Wu, Chunping Jia, Qiang Chen, and Jianlong Zhao. 2021. "A 3D-Printed Microfluidic Device for qPCR Detection of Macrolide-Resistant Mutations of Mycoplasma pneumoniae " Biosensors 11, no. 11: 427. https://doi.org/10.3390/bios11110427
APA StyleWang, A., Wu, Z., Huang, Y., Zhou, H., Wu, L., Jia, C., Chen, Q., & Zhao, J. (2021). A 3D-Printed Microfluidic Device for qPCR Detection of Macrolide-Resistant Mutations of Mycoplasma pneumoniae . Biosensors, 11(11), 427. https://doi.org/10.3390/bios11110427






