On-Site Detection of Carcinoembryonic Antigen in Human Serum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Instruments
2.2. Synthesis of GNP@PDA
2.3. Preparation of GNP–mAb and GNP@PDA–mAb Conjugates
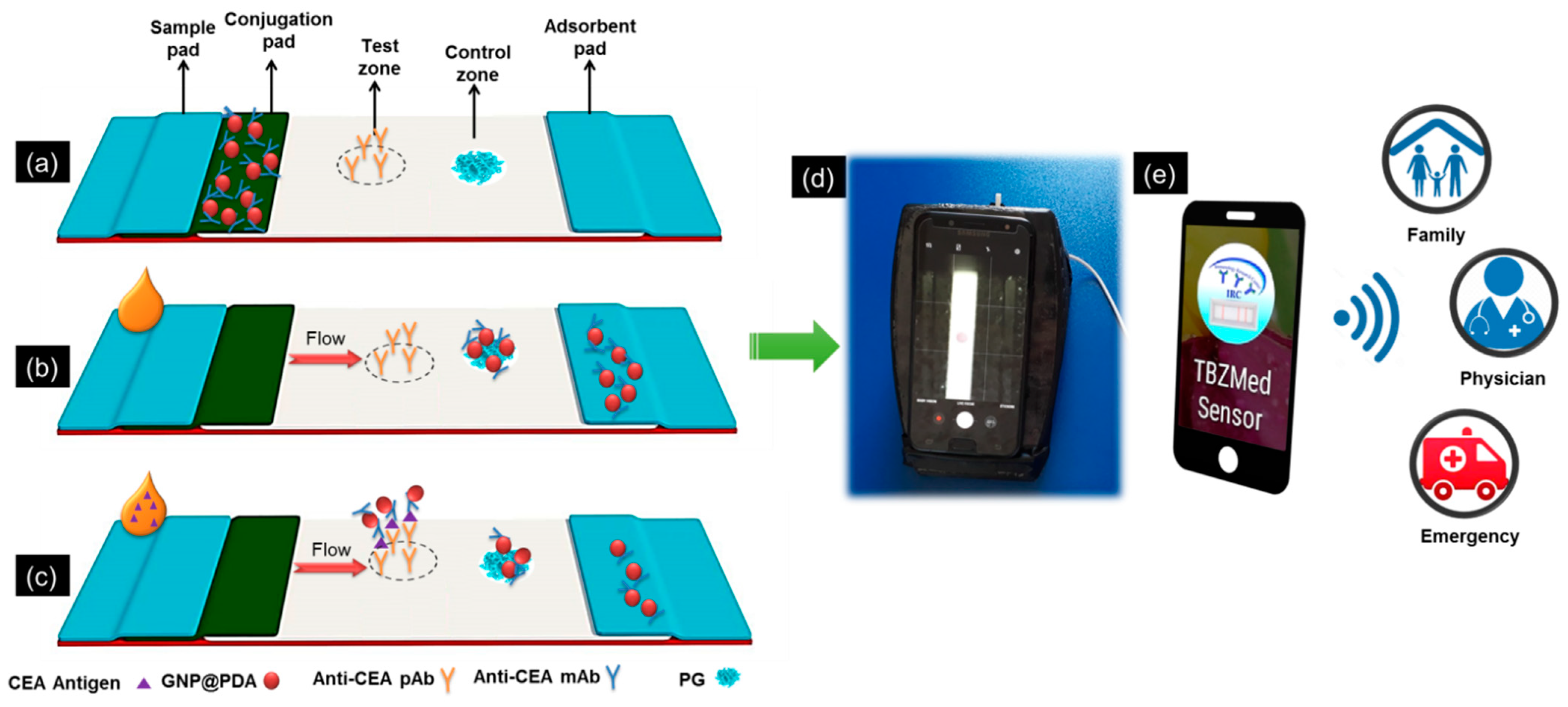
2.4. Preparation of the Lateral Flow Test Strips
2.5. The Fabrication and Setup of the Smartphone-Based Colorimetric Imaging Device
2.6. The App Development
 ) for possible real-time connectivity (Figure S3f).
) for possible real-time connectivity (Figure S3f).2.7. Lateral Flow Immunoassay Procedure
2.8. Clinical Samples Analysis
3. Results and Discussion
3.1. Characterization of GNP–mAb and GNP@PDA–mAb Conjugates
3.2. Optimization of Effective Factors on the Performance of the Developed Smartphone-Based LFIA Kit
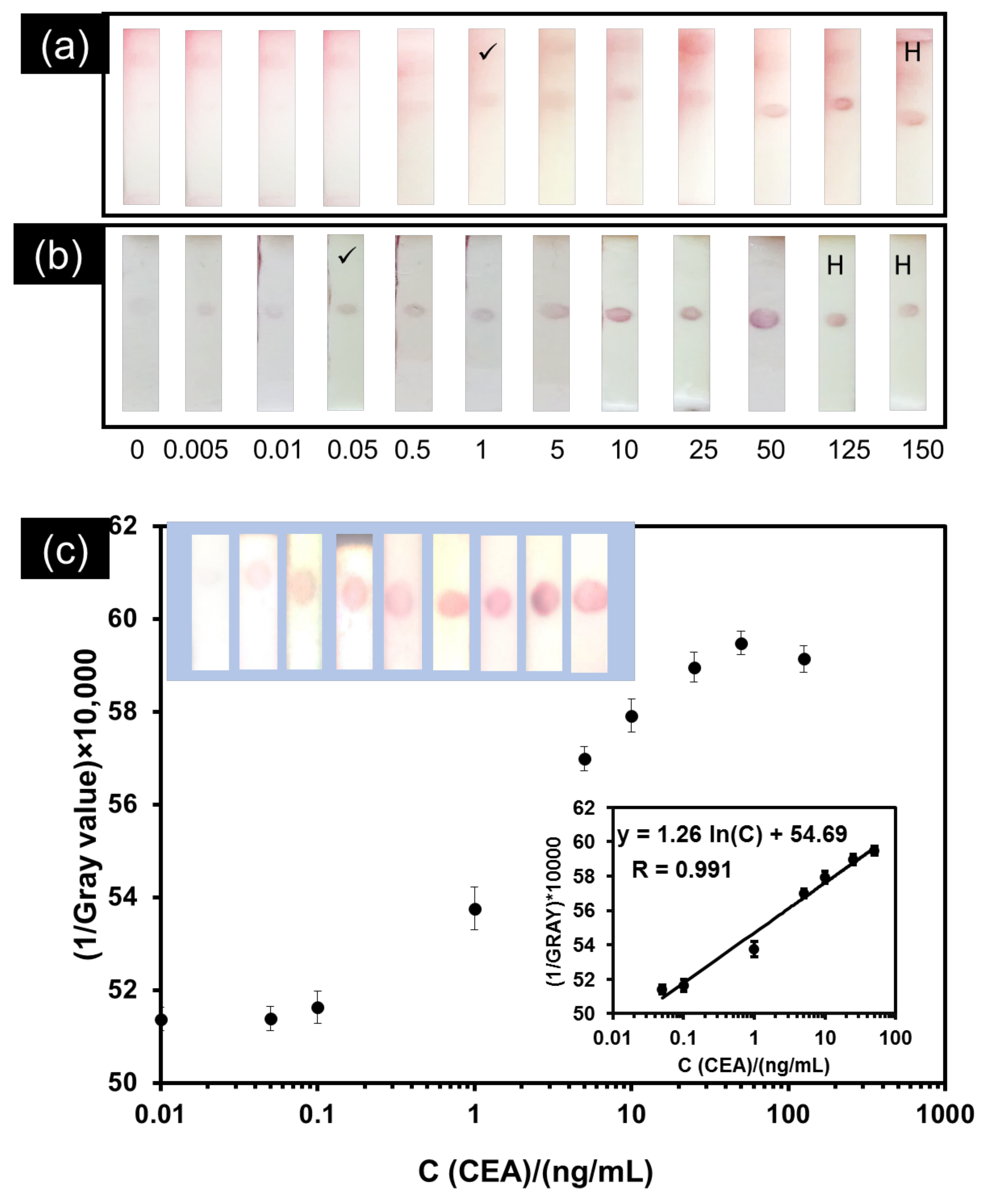
3.3. Analytical Performance of the Developed Smartphone-Based LFIA Kit
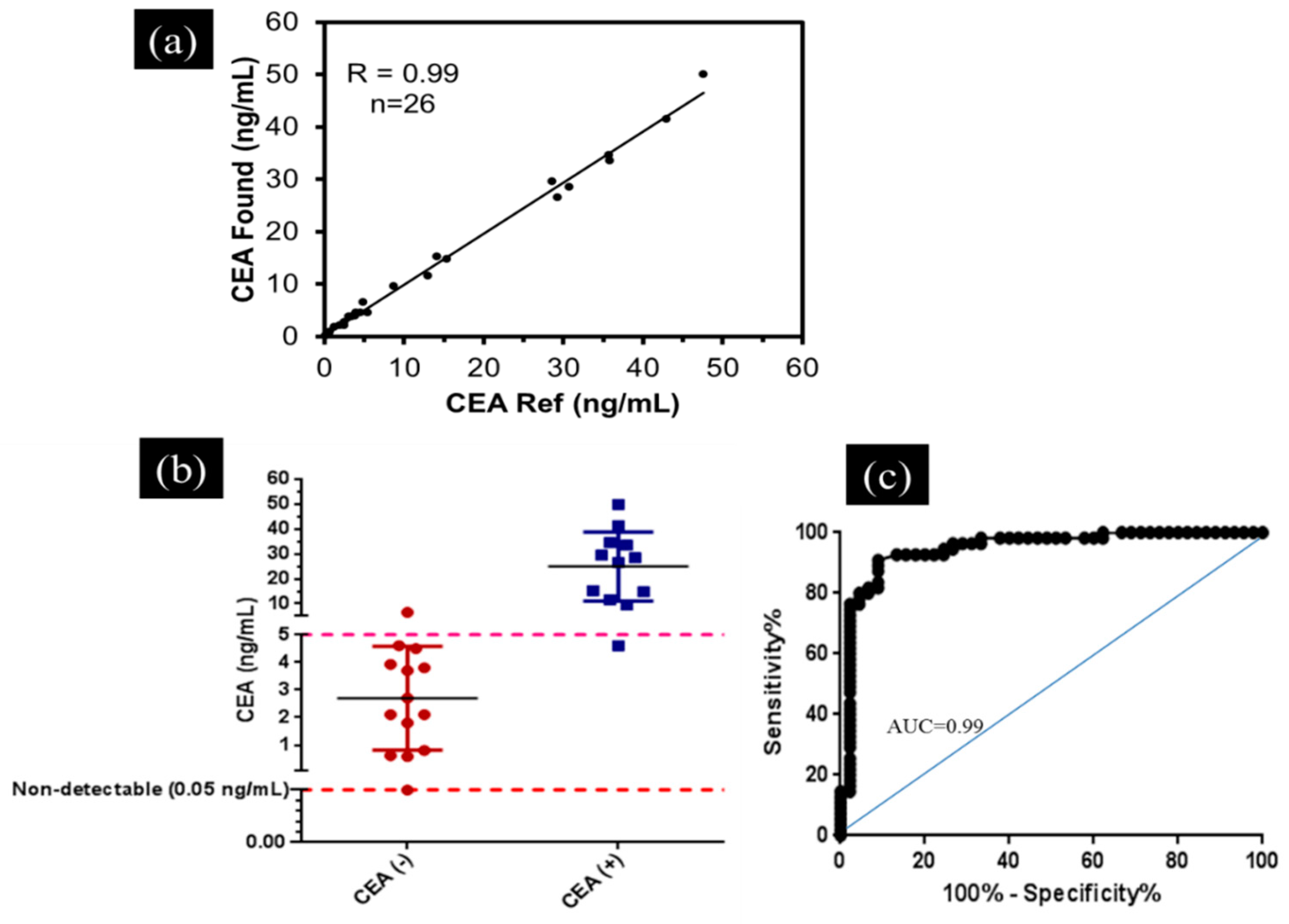
3.4. Application of the Developed Smartphone-Based LFIA Kit for the Detection of CEA in Clinical Serum Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Wender, R.C.; Brawley, O.W.; Fedewa, S.A.; Gansler, T.; Smith, R.A. A blueprint for cancer screening and early detection: Advancing screening’s contribution to cancer control. CA Cancer J. Clin. 2019, 69, 50–79. [Google Scholar] [CrossRef]
- Duffy, M.J. Carcinoembryonic antigen as a marker for colorectal cancer: Is it clinically useful? Clin. Chem. 2001, 47, 624–630. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Xu, S.; Fjaertoft, G.; Pauksen, K.; Håkansson, L.; Venge, P. An enzyme-linked immunosorbent assay for human carcinoembryonic antigen-related cell adhesion molecule 8, a biological marker of granulocyte activities in vivo. J. Immunol. Methods 2004, 293, 207–214. [Google Scholar] [CrossRef]
- Kuroki, M.; Yamaguchi, A.; Koga, Y.; Matsuoka, Y. Antigenic reactivities of purified preparations of carcinoembryonic antigen (CEA) and related normal antigens using four different radioimmunoassay systems for CEA. J. Immunol. Methods 1983, 60, 221–233. [Google Scholar] [CrossRef]
- Qu, S.; Liu, J.; Luo, J.; Huang, Y.; Shi, W.; Wang, B.; Cai, X. A rapid and highly sensitive portable chemiluminescent immunosensor of carcinoembryonic antigen based on immunomagnetic separation in human serum. Anal. Chim. Acta 2013, 766, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Falzarano, R.; Viggiani, V.; Michienzi, S.; Longo, F.; Tudini, S.; Frati, L.; Anastasi, E. Evaluation of a CLEIA automated assay system for the detection of a panel of tumor markers. Tumor Biol. 2013, 34, 3093–3100. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, T.; de la Guardia, M.; Baradaran, B. Lateral Flow Assays towards Point-of-Care Cancer Detection: A Review of Current Progress and Future Trends. TrAC Trends Anal. Chem. 2020, 125, 115842. [Google Scholar] [CrossRef]
- Golmohammadi, H.; Hamzei, Z.; Hosseinifard, M.; Ahmadi, S.H. Smart Fully Integrated Lab: A Smartphone-Based Compact Miniaturized Analytical/Diagnostic Device. Adv. Mater. Technol. 2020, 5, 2000742. [Google Scholar] [CrossRef]
- Mabey, D.; Peeling, R.W.; Ustianowski, A.; Perkins, M.D. Diagnostics for the developing world. Nat. Rev. Microbiol. 2004, 2, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Land, K.J.; Boeras, D.I.; Chen, X.-S.; Ramsay, A.R.; Peeling, R.W. REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol. 2019, 4, 46–54. [Google Scholar] [CrossRef]
- Mahmoudi, T.; de la Guardia, M.; Shirdel, B.; Mokhtarzadeh, A.; Baradarn, B. Recent Advancements in Structural Improvements of Lateral Flow Assays towards Point-of-Care Testing. TrAC Trends Anal. Chem. 2019, 116, 13–30. [Google Scholar] [CrossRef]
- Younis, M.R.; Wang, C.; Younis, M.A.; Xia, X.H. Smartphone-Based Biosensors. In Nanobiosensors: From Design to Applications; Wiley: Hoboken, NJ, USA, 2020; pp. 357–387. [Google Scholar] [CrossRef]
- Chen, Z.; Liang, R.; Guo, X.; Liang, J.; Deng, Q.; Li, M.; An, T.; Liu, T.; Wu, Y. Simultaneous quantitation of cytokeratin-19 fragment and carcinoembryonic antigen in human serum via quantum dot-doped nanoparticles. Biosens. Bioelectron. 2017, 91, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Wang, K.; Xiao, K.; Hou, Y.; Lu, W.; Xu, H.; Wo, Y.; Feng, S.; Cui, D. Carcinoembryonic antigen detection with “Handing”-controlled fluorescence spectroscopy using a color matrix for point-of-care applications. Biosens. Bioelectron. 2017, 90, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, H.; Wu, Z.; Dong, H.; Zhou, L.; Yang, D.; Ge, Y.; Jia, C.; Liu, H.; Jin, Q. Highly sensitive and selective lateral flow immunoassay based on magnetic nanoparticles for quantitative detection of carcinoembryonic antigen. Talanta 2016, 161, 205–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, W.; Wang, K.; Xiao, K.; Qin, W.; Hou, Y.; Xu, H.; Yan, X.; Chen, Y.; Cui, D.; He, J. Dual immunomagnetic nanobeads-based lateral flow test strip for simultaneous quantitative detection of carcinoembryonic antigen and neuron specific enolase. Sci. Rep. 2017, 7, 42414. [Google Scholar] [CrossRef]
- Zhou, Y.; Ding, L.; Wu, Y.; Huang, X.; Lai, W.; Xiong, Y. Emerging Strategies to Develop Sensitive AuNP-based ICTS Nanosensors. TrAC Trends Anal. Chem. 2019, 112, 147–160. [Google Scholar] [CrossRef]
- Khashayar, P.; Amoabediny, G.; Larijani, B.; Hosseini, M.; Vanfleteren, J. Fabrication and verification of conjugated aunp-antibody nanoprobe for sensitivity improvement in electrochemical biosensors. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Zhang, G.; Fang, B.; Xiong, Q.; Duan, H.; Lai, W. Lateral Flow Immunoassay Based on Polydopamine-Coated Gold Nanoparticles for the Sensitive Detection of Zearalenone in Maize. ACS Appl. Mater. Interfaces 2019, 11, 31283–31290. [Google Scholar] [CrossRef]
- Mahmoudi, T.; Shirdel, B.; Mansoori, B.; Baradaran, B. Dual sensitivity enhancement in gold nanoparticle-based lateral flow immunoassay for visual detection of carcinoembryonic antigen. Anal. Sci. Adv. 2020, 1, 161–172. [Google Scholar] [CrossRef]
- Byzova, N.A.; Safenkova, I.V.; Slutskaya, E.S.; Zherdev, A.V.; Dzantiev, B.B. Less is more: A comparison of antibody–gold nanoparticle conjugates of different ratios. Bioconjug. Chem. 2017, 28, 2737–2746. [Google Scholar] [CrossRef] [PubMed]
- Schiettecatte, J.; Anckaert, E.; Smitz, J. Interferences in immunoassays. Adv. Immunoass Technol. 2012, 3, 45–62. [Google Scholar]
- Mahmoudi, T.; Tazehkand, A.P.; Pourhassan-Moghaddam, M.; Alizadeh-Ghodsi, M.; Ding, L.; Baradaran, B.; Bazaz, S.R.; Jin, D.; Warkiani, M.E. PCR-free paper-based nanobiosensing platform for visual detection of telomerase activity via gold enhancement. Microchem. J. 2020, 154, 104594. [Google Scholar] [CrossRef]
- Jung, Y.; Heo, Y.; Lee, J.J.; Deering, A.; Bae, E. Smartphone-based lateral flow imaging system for detection of food-borne bacteria E. coli O157: H7. J. Microbiol. Methods 2020, 168, 105800. [Google Scholar] [CrossRef] [PubMed]
- De Puig, H.; Bosch, I.; Gehrke, L.; Hamad-Schifferli, K. Challenges of the nano–bio interface in lateral flow and dipstick immunoassays. Trends Biotechnol. 2017, 35, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- Qie, Z.; Yan, W.; Gao, Z.; Meng, W.; Xiao, R.; Wang, S. An anti-BSA antibody-based immunochromatographic assay for chloramphenicol and aflatoxin M 1 by using carboxy-modified CdSe/ZnS core–shell nanoparticles as label. Microchim. Acta 2020, 187, 10. [Google Scholar] [CrossRef]
- Parolo, C.; de la Escosura-Muñiz, A.; Merkoçi, A. Enhanced lateral flow immunoassay using gold nanoparticles loaded with enzymes. Biosens. Bioelectron. 2013, 40, 412–416. [Google Scholar] [CrossRef] [Green Version]
- Litvak, A.; Cercek, A.; Segal, N.; Reidy-Lagunes, D.; Stadler, Z.K.; Yaeger, R.D.; Kemeny, N.E.; Weiser, M.R.; Pessin, M.S.; Saltz, L. False-positive elevations of carcinoembryonic antigen in patients with a history of resected colorectal cancer. J. Natl. Compr. Cancer Netw. 2014, 12, 907–913. [Google Scholar] [CrossRef]
- Zou, K.H.; O’Malley, A.J.; Mauri, L. Receiver-operating characteristic analysis for evaluating diagnostic tests and predictive models. Circulation 2007, 115, 654–657. [Google Scholar] [CrossRef] [Green Version]
- Saisin, L.; Amarit, R.; Somboonkaew, A.; Gajanandana, O.; Himananto, O.; Sutapun, B. Significant sensitivity improvement for camera-based lateral flow immunoassay readers. Sensors 2018, 18, 4026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, K.; Wang, K.; Qin, W.; Hou, Y.; Lu, W.; Xu, H.; Wo, Y.; Cui, D. Use of quantum dot beads-labeled monoclonal antibody to improve the sensitivity of a quantitative and simultaneous immunochromatographic assay for neuron specific enolase and carcinoembryonic antigen. Talanta 2017, 164, 463–469. [Google Scholar] [CrossRef]
- Lee, S.; Kim, G.; Moon, J. Development of a Smartphone-based reading system for lateral flow immunoassay. J. Nanosci. Nanotechnol. 2014, 14, 8453–8457. [Google Scholar] [CrossRef]
- Ruppert, C.; Phogat, N.; Laufer, S.; Kohl, M.; Deigner, H.-P. A smartphone readout system for gold nanoparticle-based lateral flow assays: Application to monitoring of digoxigenin. Microchim. Acta 2019, 186, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Spiked CEA (ng/mL) | Intra-Assay | Inter-Assay | ||||
|---|---|---|---|---|---|---|
| Mean ± SD | Recovery (%) | CV (%) | Mean ± SD | Recovery (%) | CV (%) | |
| 0.5 | 0.6 ± 0.06 | 120 | 10 | 0.6 ± 0.07 | 120 | 11.66 |
| 2 | 2.3 ± 0.15 | 115 | 6.5 | 2.4 ± 0.25 | 120 | 10.41 |
| 20 | 19 ± 1.2 | 95 | 6.3 | 18 ± 1.5 | 90 | 8.33 |
| 35 | 34 ± 2.6 | 97 | 7.6 | 35 ± 3.4 | 100 | 9.70 |
| 45 | 46 ± 3.4 | 102 | 7.4 | 46 ± 5.2 | 102 | 11.30 |
| Samples | Number of Positive Results | Number of Negative Results | Characteristic Parameter | |
|---|---|---|---|---|
| 12 (+) | 11 | 1 | Sensitivity | 91% |
| 14 (−) | 1 | 13 | Specificity | 93% |
| Detection Strategy | Used Tag | LR * (ng/mL) | LOD ** (ng/mL) | Detection Time (min) | REASSURED * Criteria | Ref. |
|---|---|---|---|---|---|---|
| Commercial magnetic strip reader | Magnetic particles | 1–100 | 0.045 | 30 | -EASS-R- - | [17] |
| Smartphone-based colorimetric image analysis | Magnetic NPs containing Ab and biotinylated DNA | 0.25–100 | 0.0375 | 15 | -EASS-R- - | [16] |
| Fluorescent handing system | Quantum dots | 1–100 | 5 | 20 | REASS-R-D | [15] |
| Fluorescent handing system | Quantum dot nanobeads | 1–50 | 0.049 | 15 | REASS-R-D | [32] |
| Commercial fluorescent reader | Quantum-dot-doped polystyrene nanoparticles | 2.8–680 | 0.35 | 15 | -EASSURED | [14] |
| Smartphone-based colorimetric image analysis | GNP@PDA | 0.05–50 | 0.05 | 15 | REASSURED | this work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahmoudi, T.; Pourhassan-Moghaddam, M.; Shirdel, B.; Baradaran, B.; Morales-Narváez, E.; Golmohammadi, H. On-Site Detection of Carcinoembryonic Antigen in Human Serum. Biosensors 2021, 11, 392. https://doi.org/10.3390/bios11100392
Mahmoudi T, Pourhassan-Moghaddam M, Shirdel B, Baradaran B, Morales-Narváez E, Golmohammadi H. On-Site Detection of Carcinoembryonic Antigen in Human Serum. Biosensors. 2021; 11(10):392. https://doi.org/10.3390/bios11100392
Chicago/Turabian StyleMahmoudi, Tohid, Mohammad Pourhassan-Moghaddam, Behnaz Shirdel, Behzad Baradaran, Eden Morales-Narváez, and Hamed Golmohammadi. 2021. "On-Site Detection of Carcinoembryonic Antigen in Human Serum" Biosensors 11, no. 10: 392. https://doi.org/10.3390/bios11100392
APA StyleMahmoudi, T., Pourhassan-Moghaddam, M., Shirdel, B., Baradaran, B., Morales-Narváez, E., & Golmohammadi, H. (2021). On-Site Detection of Carcinoembryonic Antigen in Human Serum. Biosensors, 11(10), 392. https://doi.org/10.3390/bios11100392







