Spectroscopic and Structural Analysis of Cu2+-Induced Fluorescence Quenching of ZsYellow
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Preparation
2.2. Spectroscopic Analysis
2.3. Crystallization and X-ray Diffraction Data
2.4. Structure Determination
3. Results
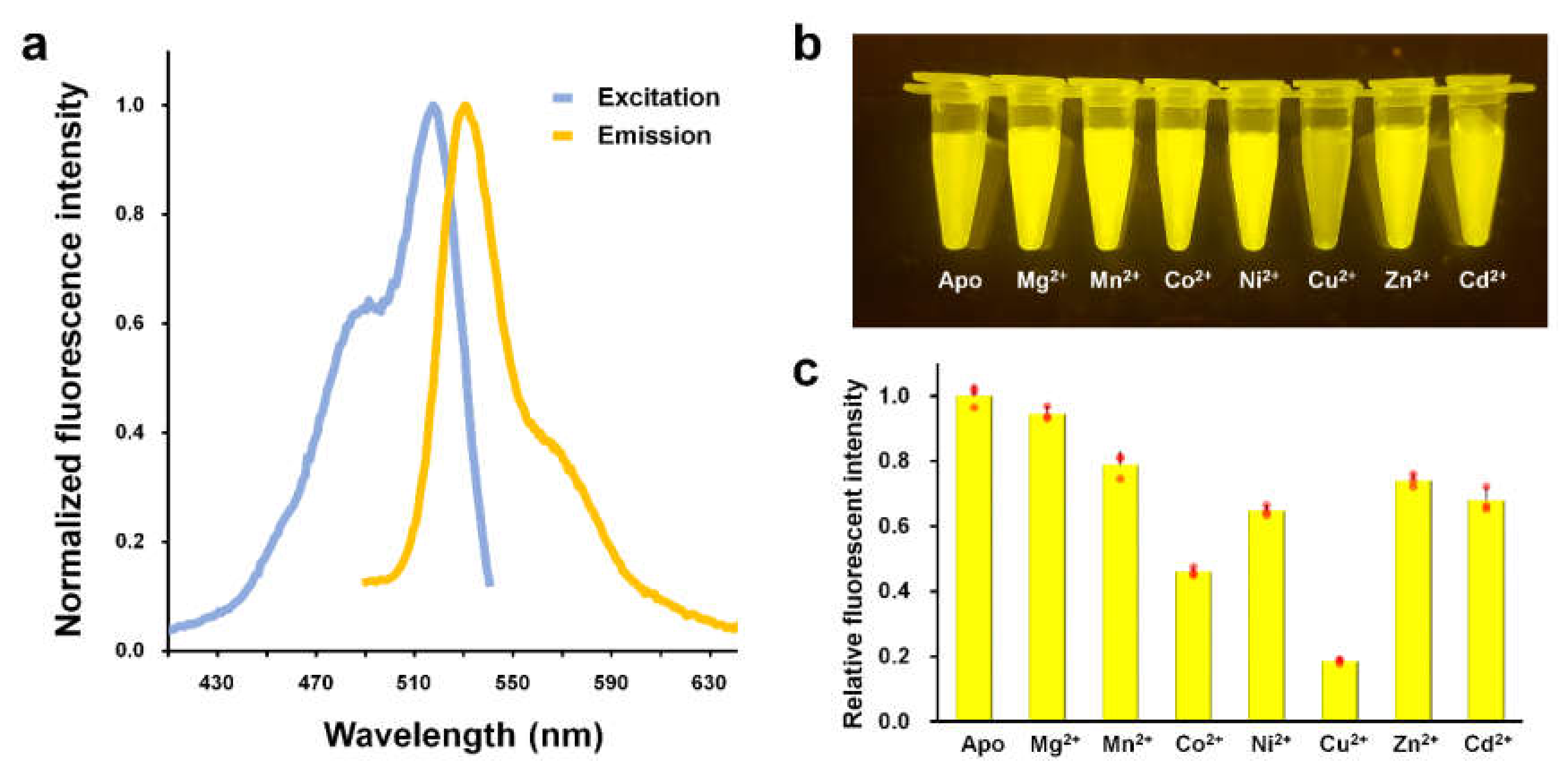
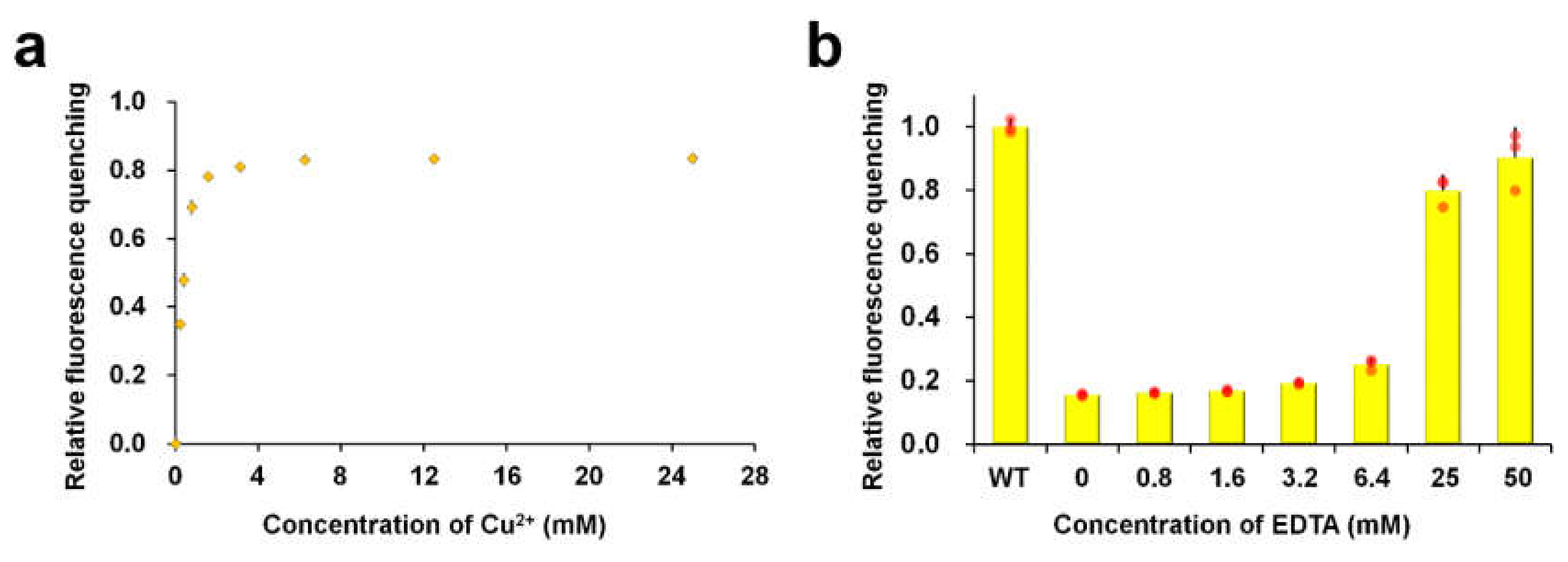
3.1. Cu2+-Induced Fluorescence Quenching
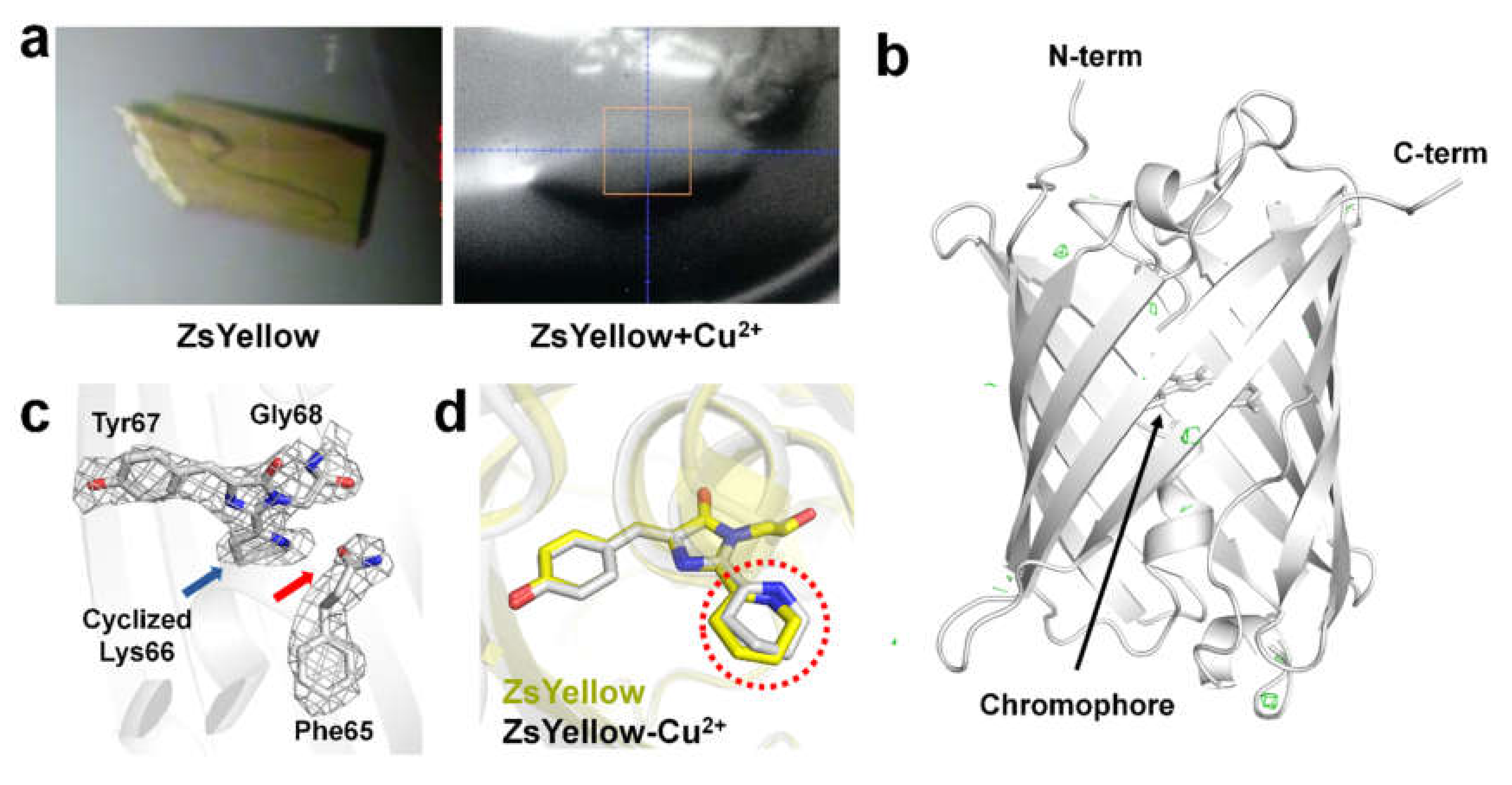
3.2. Structural Analysis of ZsYellow Soaked in Cu2+
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tsien, R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar] [CrossRef] [PubMed]
- Remington, S.J. Green fluorescent protein: A perspective. Protein Sci. 2011, 20, 1509–1519. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, M. Green fluorescent protein (GFP): Applications, structure, and related photophysical behavior. Chem. Rev. 2002, 102, 759–781. [Google Scholar] [CrossRef] [PubMed]
- Seward, H.E.; Bagshaw, C.R. The photochemistry of fluorescent proteins: Implications for their biological applications. Chem. Soc. Rev. 2009, 38, 2842–2851. [Google Scholar] [CrossRef]
- Sample, V.; Newman, R.H.; Zhang, J. The structure and function of fluorescent proteins. Chem. Soc. Rev. 2009, 38, 2852–2864. [Google Scholar] [CrossRef]
- Denay, G.; Schultz, P.; Hansch, S.; Weidtkamp-Peters, S.; Simon, R. Over the rainbow: A practical guide for fluorescent protein selection in plant FRET experiments. Plant Direct 2019, 3, e00189. [Google Scholar] [CrossRef]
- Bird, L.E.; Rada, H.; Verma, A.; Gasper, R.; Birch, J.; Jennions, M.; Lwe, J.; Moraes, I.; Owens, R.J. Green fluorescent protein-based expression screening of membrane proteins in Escherichia coli. J. Vis. Exp. 2015, 95, e52357. [Google Scholar] [CrossRef]
- Rana, M.S.; Wang, X.; Banerjee, A. An improved strategy for fluorescent tagging of membrane proteins for overexpression and purification in mammalian cells. Biochemistry 2018, 57, 6741–6751. [Google Scholar] [CrossRef]
- Xu, Y.; Hwang, K.Y.; Nam, K.H. Spectral and structural analysis of large Stokes shift fluorescent protein dKeima570. J. Microbiol. 2018, 56, 822–827. [Google Scholar] [CrossRef]
- Kim, S.E.; Hwang, K.Y.; Nam, K.H. Spectral and structural analysis of a red fluorescent protein from Acropora digitifera. Protein Sci. 2019, 28, 375–381. [Google Scholar] [CrossRef]
- Saeed, S.; Mehreen, H.; Gerlevik, U.; Tariq, A.; Manzoor, S.; Noreen, Z.; Sezerman, U.; Bokhari, H. HriGFP novel flourescent protein: Expression and applications. Mol. Biotechnol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Eli, P.; Chakrabartty, A. Variants of DsRed fluorescent protein: Development of a copper sensor. Protein Sci. 2006, 15, 2442–2447. [Google Scholar] [CrossRef] [PubMed]
- Sumner, J.P.; Westerberg, N.M.; Stoddard, A.K.; Hurst, T.K.; Cramer, M.; Thompson, R.B.; Fierke, C.A.; Kopelman, R. DsRed as a highly sensitive, selective, and reversible fluorescence-based biosensor for both Cu+ and Cu2+ ions. Biosens. Bioelectron. 2006, 21, 1302–1308. [Google Scholar] [CrossRef] [PubMed]
- Peterffy, J.P.; Szabo, M.; Szilagyi, L.; Lanyi, S.; Abraham, B. Fluorescence of a histidine-modified enhanced green fluorescent protein (EGFP) effectively quenched by copper(II) ions. part II. molecular determinants. J. Fluoresc. 2015, 25, 871–883. [Google Scholar] [CrossRef]
- Yu, X.; Strub, M.P.; Barnard, T.J.; Noinaj, N.; Piszczek, G.; Buchanan, S.K.; Taraska, J.W. An engineered palette of metal ion quenchable fluorescent proteins. PLoS ONE 2014, 9, e95808. [Google Scholar]
- Kim, I.J.; Kim, S.; Park, J.; Eom, I.; Kim, S.; Kim, J.H.; Ha, S.C.; Kim, Y.G.; Hwang, K.Y.; Nam, K.H. Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development. FEBS Lett. 2016, 590, 2982–2990. [Google Scholar] [CrossRef]
- Bae, J.E.; Kim, I.J.; Nam, K.H. Spectroscopic analysis of the Cu2+-induced fluorescence quenching of fluorescent proteins amcyan and morange2. Mol. Biotechnol. 2018, 60, 485–491. [Google Scholar] [CrossRef]
- Bae, J.E.; Kim, I.J.; Nam, K.H. Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein zsyellow by protonation of glu221. Biochem. Biophys. Res. Commun. 2017, 493, 562–567. [Google Scholar] [CrossRef]
- Sorenson, A.E.; Schaeffer, P.M. A new bivalent fluorescent fusion protein for differential Cu(II) and Zn(II) ion detection in aqueous solution. Anal. Chim. Acta 2020, 1101, 120–128. [Google Scholar] [CrossRef]
- Lee, W.; Kim, H.; Kang, Y.; Lee, Y.; Yoon, Y. A biosensor platform for metal detection based on enhanced green fluorescent protein. Sensors 2019, 19, 1846. [Google Scholar] [CrossRef]
- Martinez, A.R.; Heil, J.R.; Charles, T.C. An engineered GFP fluorescent bacterial biosensor for detecting and quantifying silver and copper ions. Biometals 2019, 32, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Murao, K.; Tanabe, Y.; Oda, M.; Tanaka, T. Metal-ion-dependent GFP emission in vivo by combining a circularly permutated green fluorescent protein with an engineered metal-ion-binding coiled-coil. J. Am. Chem. Soc. 2007, 129, 11378–11383. [Google Scholar] [CrossRef]
- Richmond, T.A.; Takahashi, T.T.; Shimkhada, R.; Bernsdorf, J. Engineered metal binding sites on green fluorescence protein. Biochem. Biophys. Res. Commun. 2000, 268, 462–465. [Google Scholar] [CrossRef] [PubMed]
- Isarankura-Na-Ayudhya, C.; Tantimongcolwat, T.; Galla, H.J.; Prachayasittikul, V. Fluorescent protein-based optical biosensor for copper ion quantitation. Biol. Trace Elem. Res. 2010, 134, 352–363. [Google Scholar] [CrossRef]
- Barondeau, D.P.; Kassmann, C.J.; Tainer, J.A.; Getzoff, E.D. Structural chemistry of a green fluorescent protein Zn biosensor. J. Am. Chem. Soc. 2002, 124, 3522–3524. [Google Scholar] [CrossRef]
- Park, S.Y.; Ha, S.C.; Kim, Y.G. The Protein Crystallography Beamlines at the Pohang Light Source II. Biodesign 2017, 5, 30–34. [Google Scholar]
- Nagelberg, D.; Wang, J.; Su, R.; Torres-Vazquez, J.; Targoff, K.L.; Poss, K.D.; Knaut, H. Origin, Specification, and Plasticity of the Great Vessels of the Heart. Curr. Biol. 2015, 25, 2099–2110. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.X.; Yelon, D. Cadm4 restricts the production of cardiac outflow tract progenitor cells. Cell Rep. 2014, 7, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar]
- Vagin, A.; Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D 2010, 66, 22–25. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Vagin, A.A.; Steiner, R.A.; Lebedev, A.A.; Potterton, L.; McNicholas, S.; Long, F.; Murshudov, G.N. REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2184–2195. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef] [PubMed]
- Remington, S.J.; Wachter, R.M.; Yarbrough, D.K.; Branchaud, B.; Anderson, D.C.; Kallio, K.; Lukyanov, K.A. zFP538, a yellow-fluorescent protein from Zoanthus, contains a novel three-ring chromophore. Biochemistry 2005, 44, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Pletneva, N.V.; Pletnev, S.V.; Chudakov, D.M.; Tikhonova, T.V.; Popov, V.O.; Martynov, V.I.; Wlodawer, A.; Dauter, Z.; Pletnev, V.Z. Three-dimensional structure of yellow fluorescent protein zYFP538 from Zoanthus sp. at the resolution 1.8 Å. Bioorg. Khim. 2007, 33, 421–430. [Google Scholar] [CrossRef] [PubMed]

| Data Collection | ZsYellow-Cu2+ |
|---|---|
| Space group | P21221 |
| Cell dimensions | |
| a, b, c (Å) | 48.638, 72.929, 124.189 |
| Resolution (Å) | 50.0–2.60 (2.64–2.60) |
| Completeness | 94.0 (90.8) |
| Redundancy | 2.8 (2.4) |
| I/σ(I) | 10.43 (2.06) |
| Rmerge(%) a | 0.107 (0.376) |
| Refinement statistics | |
| Resolution (Å) | 26.42–2.60 |
| Rwork (%) b | 20.60 |
| Rfree (%) c | 21.50 |
| B-factor (Averaged) | |
| Protein | 34.06 |
| Water | 24.23 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.645 |
| Ramachandran plot (%) | |
| favored | 98.2 |
| Allowed | 1.8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, I.J.; Xu, Y.; Nam, K.H. Spectroscopic and Structural Analysis of Cu2+-Induced Fluorescence Quenching of ZsYellow. Biosensors 2020, 10, 29. https://doi.org/10.3390/bios10030029
Kim IJ, Xu Y, Nam KH. Spectroscopic and Structural Analysis of Cu2+-Induced Fluorescence Quenching of ZsYellow. Biosensors. 2020; 10(3):29. https://doi.org/10.3390/bios10030029
Chicago/Turabian StyleKim, In Jung, Yongbin Xu, and Ki Hyun Nam. 2020. "Spectroscopic and Structural Analysis of Cu2+-Induced Fluorescence Quenching of ZsYellow" Biosensors 10, no. 3: 29. https://doi.org/10.3390/bios10030029
APA StyleKim, I. J., Xu, Y., & Nam, K. H. (2020). Spectroscopic and Structural Analysis of Cu2+-Induced Fluorescence Quenching of ZsYellow. Biosensors, 10(3), 29. https://doi.org/10.3390/bios10030029







