Label-Free Biosensors for Laboratory-Based Diagnostics of Infections: Current Achievements and New Trends
Abstract
1. Introduction
2. Lateral Flow Immunoassay (LFIA) as Simplified Formats of Modern Biosensors
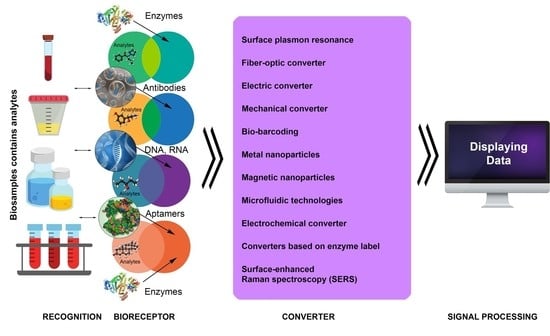
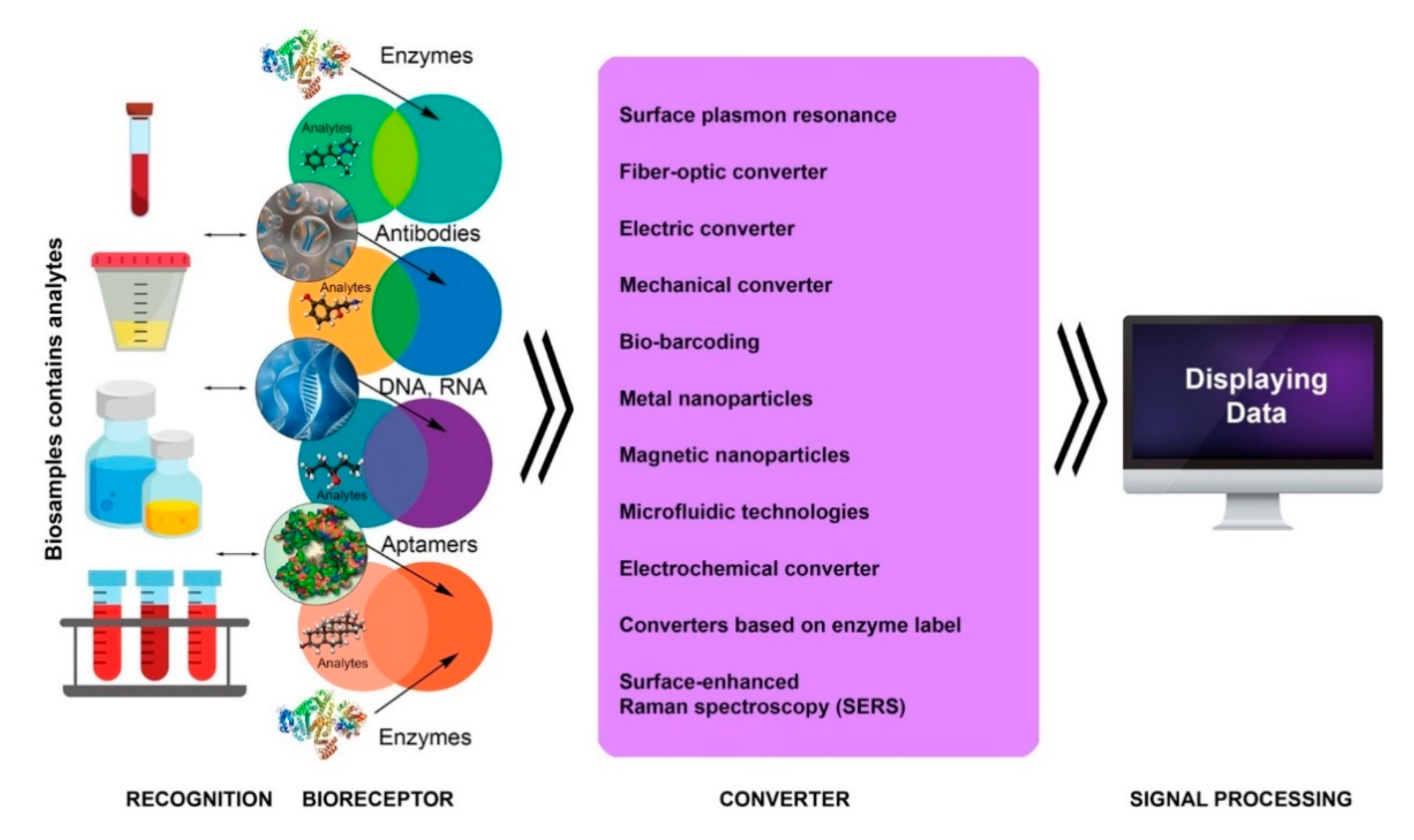
3. Introduction to Biosensor Technologies
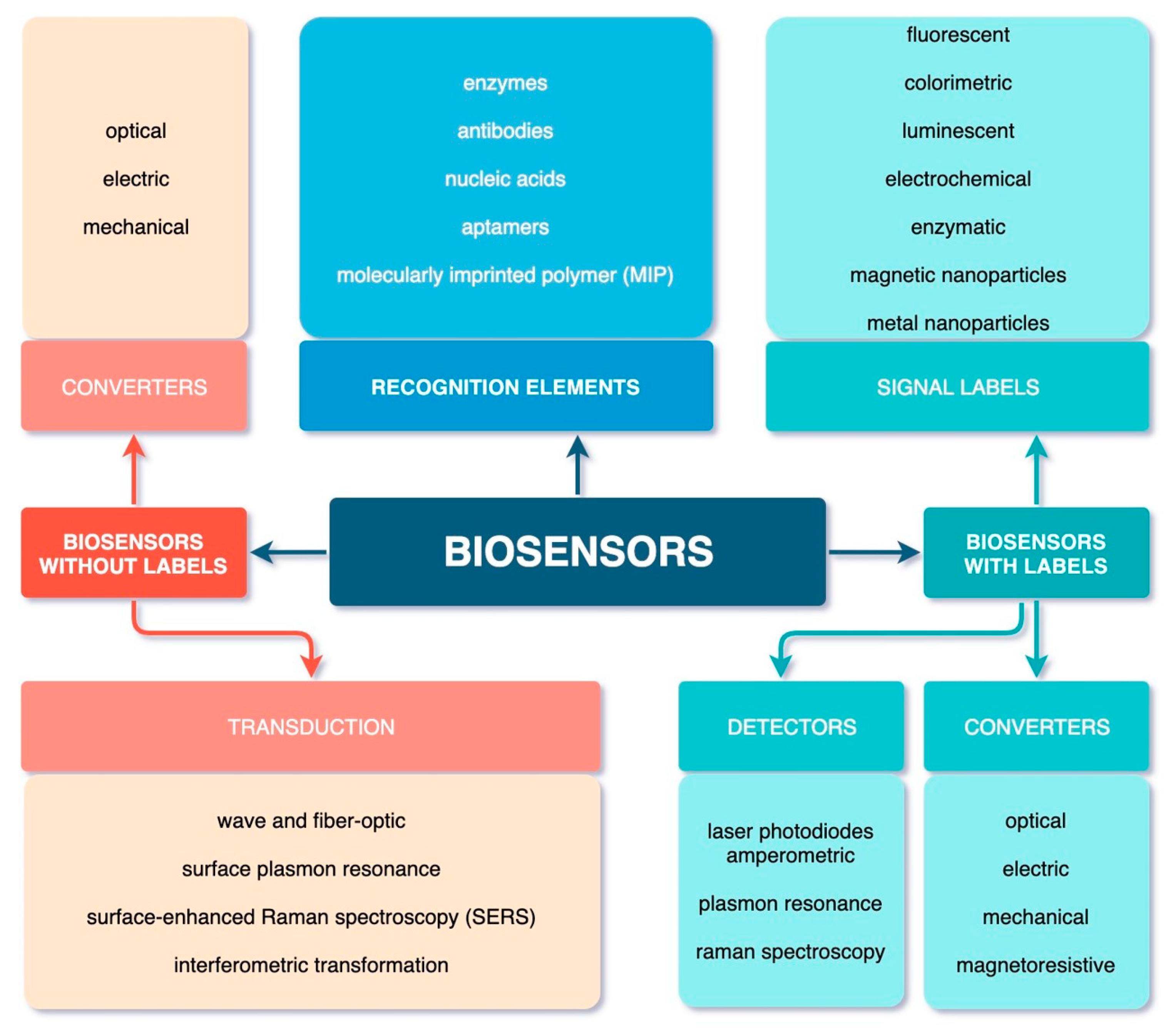
4. Main Types of Biosensors and Their Functions
4.1. Label-Free Biosensors
4.1.1. Label-Free Biosensors with Optical Converter
4.1.2. Electrochemical Label-Free Biosensors
4.1.3. Microwave Label-Free Biosensors
4.2. Mechanical Biosensors
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Havelaar, A.H.; Kirk, M.D.; Torgerson, P.R.; Gibb, H.J.; Hald, T.; Lake, R.J.; Praet, N.; Bellinger, D.C.; De Silva, N.R. World Health Organization Global Estimates and Regional Comparisons of the Burden of Foodborne Disease in 2010. PLOS Med. 2015, 12, e1001923. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO) Infectious Disease Newsletter. Available online: https://www.who.int/topics/infectious_diseases/factsheets/ru/ (accessed on 9 December 2019).
- Sheng, L.; Lu, Y.; Deng, S.; Liao, X.; Zhang, K.; Ding, T.; Gao, H.; Liu, D.; Deng, R.; Li, J. A transcription aptasensor: Amplified, label-free and culture-independent detection of foodborne pathogens via light-up RNA aptamers. Chem. Commun. (Camb.) 2019, 55, 10096–10099. [Google Scholar] [CrossRef] [PubMed]
- Clark, L.C., Jr.; Lyons, C. Electrode systems for continuous monitoring in cardiovascular surgery. Ann. N. Y. Acad. Sci. 1962, 102, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Jia, W.; Hou, C.; Lei, Y. Microbial biosensors: A review. Biosens. Bioelectron. 2011, 26, 1788–1799. [Google Scholar] [CrossRef] [PubMed]
- Sin, M.L.; Mach, K.E.; Wong, P.K.; Liao, J.C. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 2014, 14, 225–244. [Google Scholar] [CrossRef]
- Peltomaa, R.; Glahn-Martínez, B.; Benito-Peña, E.; Moreno-Bondi, M.C. Optical Biosensors for Label-Free Detection of Small Molecules. Sensors 2018, 18, 4126. [Google Scholar] [CrossRef]
- Zarei, M. Infectious pathogens meet point-of-care diagnostics. Biosens. Bioelectron. 2018, 106, 193–203. [Google Scholar] [CrossRef]
- Kozel, T.R.; Burnham-Marusich, A.R. Point-of-Care Testing for Infectious Diseases: Past, Present, and Future. J. Clin. Microbiol. 2017, 55, 2313–2320. [Google Scholar] [CrossRef]
- Kim, H.; Chung, D.R.; Kang, M. A new point-of-care test for the diagnosis of infectious diseases based on multiplex lateral flow immunoassays. Analyst 2019, 144, 2460–2466. [Google Scholar] [CrossRef]
- Fabri-Faja, N.; Calvo-Lozano, O.; Dey, P.; Terborg, R.A.; Estevez, M.C.; Belushkin, A.; Yesilköy, F.; Duempelmann, L.; Altug, H.; Pruneri, V.; et al. Early sepsis diagnosis via protein and miRNA biomarkers using a novel point-of-care photonic biosensor. Anal. Chim. Acta. 2019, 1077, 232–242. [Google Scholar] [CrossRef]
- Min, J.; Nothing, M.; Coble, B.; Zheng, H.; Park, J.; Im, H.; Weber, G.F.; Castro, C.M.; Swirski, F.K.; Weissleder, R.; et al. Integrated Biosensor for Rapid and Point-of-Care Sepsis Diagnosis. ACS Nano 2018, 12, 3378–3384. [Google Scholar] [CrossRef] [PubMed]
- Safenkova, I.V.; Panferov, V.G.; Panferova, N.A.; Varitsev, Y.A.; Zherdev, A.V.; Dzantiev, B.B. Alarm lateral flow immunoassay for detection of the total infection caused by the five viruses. Talanta 2019, 195, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, C.S.; Uldum, S.A.; Sørensen, J.F.; Skovsted, I.C.; Otte, S.; Elverdal, P.L. Evaluation of a new lateral flow test for detection of Streptococcus pneumoniae and Legionella pneumophila urinary antigen. J. Microbiol. Methods 2015, 116, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, Q.; Meng, Q.; Wu, F.; Zhang, L.; Tang, Y.; Guan, Y.; An, L. Quantum dots-based lateral flow immunoassay combined with image analysis for semiquantitative detection of IgE antibody to mite. Int. J. Nanomed. 2017, 12, 4805–4812. [Google Scholar] [CrossRef][Green Version]
- Boisen, M.L.; Oottamasathien, D.; Jones, A.B.; Millett, M.M.; Nelson, D.S.; Bornholdt, Z.A.; Fusco, M.L.; Abelson, D.M.; Oda, S.; Hartnett, J.N.; et al. Development of prototype filovirus recombinant antigen immunoassays. J. Infect. Dis. 2015, 212 (Suppl. 2), S359–S367. [Google Scholar] [CrossRef]
- Nielsen, K.; Yu, W.L.; Kelly, L.; Bermudez, R.; Renteria, T.; Dajer, A.; Fusco, M.L.; Abelson, D.M.; Oda, S.; Hartnett, J.N.; et al. Development of a lateral flow assay for rapid detection of bovine antibody to Anaplasma marginale. J. Immunoass. Immunochem. 2008, 29, 10–18. [Google Scholar] [CrossRef]
- Posthuma-Trumpie, G.A.; Korf, J.; van Amerongen, A. Lateral flow (immuno)assay: Its strengths, weaknesses, opportunities and threats. A literature survey. Anal. Bioanal. Chem. 2008, 393, 569–582. [Google Scholar] [CrossRef]
- Rohrman, B.A.; Leautaud, V.; Molyneux, E.; Richards–Kortum, R.R. A lateral flow assay for quantitative detection of amplified HIV-1 RNA. PLoS ONE 2012, 7, e45611. [Google Scholar] [CrossRef]
- Kamphee, H.; Chaiprasert, A.; Prammananan, T.; Wiriyachaiporn, N.; Kanchanatavee, A.; Dharakul, T. Rapid molecular detection of multidrug-resistant tuberculosis by PCR-nucleic acid lateral flow immunoassay. PLoS ONE 2015, 10, e0137791. [Google Scholar] [CrossRef]
- Pilavaki, E.; Demosthenous, A. Optimized Lateral Flow Immunoassay Reader for the Detection of Infectious Diseases in Developing Countries. Sensors (Basel) 2017, 17, 2673. [Google Scholar] [CrossRef]
- Zhaoa, S.; Wangb, S.; Zhanga, S.; Liua, J.; Dong, Y. State of the art: Lateral flow assay (LFA) biosensor for on-site rapid detection. Chin. Chem. Lett. 2018, 29, 1567–1577. [Google Scholar] [CrossRef]
- Espinosa, J.R.; Galván, M.; Quiñones, A.S.; Ayala, J.L.; Durón, S.M. DNA Biosensor Based on Double-Layer Discharge for the Detection of HPV Type 16. Sensors (Basel) 2019, 19, 3956. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.; Slaughter, G. Label-Free MicroRNA Optical Biosensors. Nanomaterials 2019, 9, 1573. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ragavan, K.V.; Kumar, S.; Swaraj, S.; Neethirajan, S. Advances in biosensors and optical assays for diagnosis and detection of malaria. Biosens. Bioelectron. 2018, 105, 188–210. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Nanda, R.; Sahoo, S.; Mohapatra, E. Biosensors in Health Care: The Milestones Achieved in Their Development towards Lab-On-Chip-Analysis. Biochem. Res. Int. 2016, 2016, 3130469. [Google Scholar] [CrossRef]
- IUPAC. Compendium of Chemical Terminology; McNaught, A.D., Wilkinson, A., Eds.; Blackwell Scientific Publications: Oxford, UK, 1997; Volume 2, ISBN 0-9678550-9-8. [Google Scholar] [CrossRef]
- Biomaterials Nanoarchitectonics; Ebara, M., Ed.; Elsevier Inc.: Tsukuba, Japan, 2016; 362p. [Google Scholar] [CrossRef]
- Russell, C.; Ward, A.C.; Vezza, V.; Hoskisson, P.; Alcorn, D.; Steenson, D.P.; Corrigan, D.K. Development of a needle shaped microelectrode for electrochemical detection of the sepsis biomarker interleukin-6 (IL-6) in real time. Biosens. Bioelectron. 2019, 126, 806–814. [Google Scholar] [CrossRef]
- Kumar, S.; Tripathy, S.; Jyoti, A.; Singh, S.G. Recent advances in biosensors for diagnosis and detection of sepsis: A comprehensive review. Biosens. Bioelectron. 2019, 124–125, 205–215. [Google Scholar] [CrossRef]
- Mazzaracchio, V.; Neagu, D.; Porchetta, A.; Marcoccio, E.; Pomponi, A.; Faggioni, G.; D’Amore, N.; Notargiacomo, A.; Pea, M.; Moscone, D.; et al. A label-free impedimetric aptasensor for the detection of Bacillus anthracis spore simulant. Biosens. Bioelectron. 2019, 126, 640–646. [Google Scholar] [CrossRef]
- Waller, D.F.; Hew, B.E.; Holdaway, C.; Jen, M.; Peckham, G.D. Rapid Detection of Bacillus anthracis Spores Using Immunomagnetic Separation and Amperometry. Biosensors (Basel) 2016, 6, 61. [Google Scholar] [CrossRef]
- Wynn, D.; Deo, S.; Daunert, S. Engineering Rugged Field Assays to Detect Hazardous Chemicals Using Spore-Based Bacterial Biosensors. Methods Enzymol. 2017, 589, 51–85. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Son, K.; Liu, Y.; Revzin, A. Biosensors for Cell Analysis. Ann. Rev. Biomed. Eng. 2015, 17, 165–190. [Google Scholar] [CrossRef] [PubMed]
- Biopolymer Composites in Electronics; Sadasivuni, K.K., Cabibihan, J.-J., Ponnamma, D., AlMaadeed, M.A., Kim, J., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2017; 544p. [Google Scholar] [CrossRef]
- Yagi, K. Applications of whole-cell bacterial sensors in biotechnology and environmental science. Appl. Microbiol. Biotechnol. 2007, 73, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Mangwani, N.; Dash, H.R.; Chauhan, A.; Das, S. Bacterial quorum sensing: Functional features and potential applications in biotechnology. J. Mol. Microb. Biotechnol. 2012, 22, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; White, I.M.; Shopova, S.I.; Zhu, H.; Suter, J.D.; Sun, Y. Sensitive optical biosensors for unlabeled targets: A review. Anal. Chim. Acta. 2008, 620, 8–26. [Google Scholar] [CrossRef] [PubMed]
- Sang, S.; Wang, Y.; Feng, Q.; Wei, Y.; Ji, J.; Zhang, W. Progress of new label-free techniques for biosensors: A review. Crit. Rev. Biotechnol. 2016, 36, 465–481. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, H.; Hu, Z.; Yu, G.; Yang, D.; Zhao, J. Label and label-free based surface-enhanced Raman scattering for pathogen bacteria detection: A review. Biosens. Bioelectron. 2017, 94, 131–140. [Google Scholar] [CrossRef]
- McLinden, T.; Sargeant, J.M.; Thomas, M.K.; Papadopoulos, A.; Fazil, A. Component costs of foodborne illness: A scoping review. BMC Public Health 2014, 14, 509. [Google Scholar] [CrossRef]
- Novick, A.; Weiner, M. Enzyme induction as an all-or-none phenomenon. Proc. Natl. Acad. Sci. USA 1957, 43, 553–566. [Google Scholar] [CrossRef]
- Jacob, F.; Monod, J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961, 3, 318–356. [Google Scholar] [CrossRef]
- Wu, X.; Xu, C.; Tripp, R.A.; Huang, Y.W.; Zhao, Y. Detection and differentiation of foodborne pathogenic bacteria in mung bean sprouts using field deployable label-free SERS devices. Analyst 2013, 138, 3005–3012. [Google Scholar] [CrossRef] [PubMed]
- Van der Meer, J.R.; Belkin, S. Where microbiology meets microengineering: Design and applications of reporter bacteria. Nat. Rev. Microbiol. 2010, 8, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Marks, H.; Schechinger, M.; Garza, J.; Locke, A.; Coté, G. Surface enhanced Raman spectroscopy (SERS) for in vitro diagnostic testing at the point of care. Nanophotonics 2017, 6, 681–701. [Google Scholar] [CrossRef]
- Lee, T.; Park, S.Y.; Jang, H.; Kim, G.H.; Lee, Y.; Park, C.; Mohammadniaei, M.; Lee, M.H.; Min, J. Fabrication of electrochemical biosensor consisted of multi-functional DNA structure/porous au nanoparticle for avian influenza virus (H5N1) in chicken serum. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 99, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Faria, H.A.M.; Zucolotto, V. Label-free electrochemical DNA biosensor for zika virus identification. Biosens. Bioelectron. 2019, 131, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Teengam, P.; Siangproh, W.; Tuantranont, A.; Vilaivan, T.; Chailapakul, O.; Henry, C.S. Electrochemical impedance-based DNA sensor using pyrrolidinyl peptide nucleic acids for tuberculosis detection. Anal. Chim. Acta. 2018, 1044, 102–109. [Google Scholar] [CrossRef]
- González-Pabón, M.J.; Figueredo, F.; Martínez-Casillas, D.C.; Cortón, E. Characterization of a new composite membrane for point of need paper-based micro-scale microbial fuel cell analytical devices. PLoS ONE 2019, 14, e0222538. [Google Scholar] [CrossRef]
- Lim, J.W.; Ha, D.; Lee, J.; Lee, S.K.; Kim, T. Review of Micro/Nanotechnologies for Microbial Biosensors. Front. Bioeng. Biotechnol. 2015, 3, 61. [Google Scholar] [CrossRef]
- Lei, Y.; Chen, W.; Mulchandani, A. Microbial biosensors. Anal. Chim. Acta 2006, 568, 200–210. [Google Scholar] [CrossRef]
- Urmann, K.; Reich, P.; Walter, J.G.; Beckmann, D.; Segal, E.; Scheper, T. Rapid and label-free detection of protein a by aptamer-tethered porous silicon nanostructures. J. Biotechnol. 2017, 257, 171–177. [Google Scholar] [CrossRef]
- Zhang, S.; Ma, L.; Ma, K.; Xu, B.; Liu, L.; Tian, W. Label-Free Aptamer-Based Biosensor for Specific Detection of Chloramphenicol Using AIE Probe and Graphene Oxide. ACS Omega 2018, 3, 12886–12892. [Google Scholar] [CrossRef] [PubMed]
- Gharatape, A.; Yari Khosroushahi, A. Optical Biomarker-based Biosensors for Cancer/Infectious Disease Medical Diagnoses. Appl. Immunohistochem. Mol. Morphol. 2019, 27, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Zhou, J. Application of biosensors to detection of epidemic diseases in animals. Res. Vet. Sci. 2018, 118, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Kahraman, M.; Mullen, E.R.; Korkmaz, A.; Wachsmann-Hogiu, S. Fundamentals and applications of SERS-based bioanalytical sensing. Nanophotonics 2017, 6, 831–852. [Google Scholar] [CrossRef]
- Qiu, C.; Zhai, H.; Hou, J. Biosensors Design in Yeast and Applications in Metabolic Engineering. FEMS Yeast Res. 2019, 19, foz082. [Google Scholar] [CrossRef]
- Zhang, F.Z.; Keasling, J. Biosensors and their applications in microbial metabolic engineering. Trends Microbiol. 2011, 19, 323–329. [Google Scholar] [CrossRef]
- Dietrich, J.A.; McKee, A.E.; Keasling, J.D. High-throughput metabolic engineering: Advances in small-molecule screening and selection. Ann. Rev. Biochem. 2010, 79, 563–590. [Google Scholar] [CrossRef]
- Durrieu, C.; Lagarde, F.; Jaffrezic-Renault, N. Nanotechnology assets in biosensors design for environmental monitoring. In Nanomaterials: A Danger or a Promise; Brayner, R., Fiévet, F., Coradin, T., Eds.; Springer: London, UK, 2013; pp. 189–229. [Google Scholar]
- Chang, H.J.; Voyvodic, P.L.; Zúñiga, A.; Bonnet, J. Microbially derived biosensors for diagnosis, monitoring and epidemiology. Microb. Biotechnol. 2017, 10, 1031–1035. [Google Scholar] [CrossRef]
- Renella, G.; Giagnoni, L. Light dazzles from the black box: Whole-cell biosensors are ready to inform on fundamental soil biological processes. Chem. Biol. Technol. Agric. 2016, 3, 8. [Google Scholar] [CrossRef]
- Park, M.; Tsai, S.L.; Chen, W. Microbial biosensors: Engineered microorganisms as the sensing machinery. Sensors (Basel) 2013, 13, 5777–5795. [Google Scholar] [CrossRef]
- Roy, V.; Adams, B.L.; Bentley, W.E. Developing next generation antimicrobials by intercepting AI-2 mediated quorum sensing. Enzym. Microb. Technol. 2011, 49, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Amine, A.; Arduini, F.; Moscone, D.; Palleschi, G. Recent advances in biosensors based on enzyme inhibition. Biosens. Bioelectron. 2016, 76, 180–194. [Google Scholar] [CrossRef] [PubMed]
- Barreiros dos Santos, M.; Agusil, J.P.; Prieto-Simón, B.; Sporer, C.; Teixeira, V.; Samitier, J. Highly sensitive detection of pathogen Escherichia coli O157:H7 by electrochemical impedance spectroscopy. Biosens. Bioelectron. 2013, 45, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Rocchitta, G.; Spanu, A.; Babudieri, S.; Latte, G.; Madeddu, G.; Galleri, G.; Nuvoli, S.; Bagella, P.; Demartis, M.I.; Fiore, V.; et al. Enzyme Biosensors for Biomedical Applications: Strategies for Safeguarding Analytical Performances in Biological Fluids. Sensors (Basel) 2016, 16, 780. [Google Scholar] [CrossRef]
- Liu, X.; Marrakchi, M.; Xu, D.; He, D.; Andreescu, S. Biosensors based on modularly designed synthetic peptides for recognition, detection and live/dead differentiation of pathogenic bacteria. Biosens. Bioelectron. 2016, 80, 9–16. [Google Scholar] [CrossRef]
- Golichenari, B.; Velonia, K.; Nosrati, R.; Nezami, A.; Farokhi-Fard, A.; Abnous, K.; Behravan, J.; Tsatsakis, A.M. Label-free nano-biosensing on the road to tuberculosis detection. Biosens. Bioelectron. 2018, 113, 124–135. [Google Scholar] [CrossRef]
- Golichenari, B.; Nosrati, R.; Farokhi-Fard, A.; Faal Maleki, M.; Gheibi Hayat, S.M.; Ghazvini, K.; Vaziri, F.; Behravan, J. Electrochemical-based biosensors for detection of Mycobacterium tuberculosis and tuberculosis biomarkers. Crit. Rev. Biotechnol. 2019, 39, 1056–1077. [Google Scholar] [CrossRef]
- Mowbray, S.E.; Amiri, A.M. A Brief Overview of Medical Fiber Optic Biosensors and Techniques in the Modification for Enhanced Sensing Ability. Diagnostics (Basel) 2019, 9, 23. [Google Scholar] [CrossRef]
- Abisado, R.G.; Benomar, S.; Klaus, J.R.; Dandekar, A.A.; Chandler, J.R. Bacterial Quorum Sensing and Microbial Community Interactions. mBio 2018, 9. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Bhardwaj, S.K.; Mehta, J.; Kim, K.H.; Deep, A. MOF-Bacteriophage Biosensor for Highly Sensitive and Specific Detection of Staphylococcus aureus. ACS Appl. Mater. Interfaces 2017, 9, 33589–33598. [Google Scholar] [CrossRef]
- Nasrin, F.; Chowdhury, A.D.; Takemura, K.; Lee, J.; Adegoke, O.; Deo, V.K.; Abe, F.; Suzuki, T.; Park, E.Y. Single-step detection of norovirus tuning localized surface plasmon resonance-induced optical signal between gold nanoparticles and quantum dots. Biosens. Bioelectron. 2018, 122, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Damborský, P.; Švitel, J.; Katrlík, J. Optical biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Nquyet, N.T.; Yen, L.T.H.; Doan, V.Y.; Hoang, N.L.; Van Thu, V.; Lan, H.; Trung, T.; Pham, V.H.; Tam, P.D. A label-free and highly sensitive DNA biosensor based on the core-shell structured CeO2-NR@Ppy nanocomposite for Salmonella detection. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 96, 790–797. [Google Scholar] [CrossRef]
- Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M. Molecular Biosensors for Electrochemical Detection of Infectious Pathogens in Liquid Biopsies: Current Trends and Challenges. Sensors (Basel) 2017, 17, 2533. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Oueslati, R.; Cheng, C.; Zhao, L.; Chen, J.; Almeida, R.; Wu, J. Rapid, highly sensitive detection of Gram-negative bacteria with lipopolysaccharide based disposable aptasensor. Biosens. Bioelectron. 2018, 112, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xu, J.; Liu, J.; Wang, X.; Chen, B. Disease-Related Detection with Electrochemical Biosensors: A Review. Sensors (Basel) 2017, 17, 2375. [Google Scholar] [CrossRef]
- Zanchetta, G.; Lanfranco, R.; Giavazzi, F.; Bellini, T.; Buscaglia, M. Emerging applications of label-free optical biosensors. Nanophotonics 2017, 6, 158. [Google Scholar] [CrossRef]
- Dudak, F.C.; Boyaci, I.H. Rapid and label-free bacteria detection by surface plasmon resonance (SPR) biosensors. Biotechnol. J. 2009, 4, 1003–1011. [Google Scholar] [CrossRef]
- Zhang, Y.; Tian, J.; Li, K.; Tian, H.; Xu, W. Label-free visual biosensor based on cascade amplification for the detection of Salmonella. Anal. Chim. Acta 2019, 1075, 144–151. [Google Scholar] [CrossRef]
- Yang, F.; Chang, T.L.; Liu, T.; Wu, D.; Du, H.; Liang, J.; Tian, F. Label-free detection of Staphylococcus aureus bacteria using long-period fiber gratings with functional polyelectrolyte coatings. Biosens. Bioelectron. 2019, 133, 147–153. [Google Scholar] [CrossRef]
- Wang, Z.; Yao, X.; Wang, R.; Ji, Y.; Yue, T.; Sun, J.; Li, T.; Wang, J.; Zhang, D. Label-free strip sensor based on surface positively charged nitrogen-rich carbon nanoparticles for rapid detection of Salmonella enteritidis. Biosens. Bioelectron. 2019, 132, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.H.; Huang, S.C.; Chen, K.P.; Li, B.R.; Li, Y.K. Effective Construction of a High-Capacity Boronic Acid Layer on a Quartz Crystal Microbalance Chip for High-Density Antibody Immobilization. Sensors (Basel) 2019, 19, 28. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Soylu, M.C.; Kirimli, C.E.; Wu, W.; Sen, B.; Joshi, S.G.; Emery, C.L.; Au, G.; Niu, X.; Hamilton, R.; et al. Rapid, label-free genetic detection of enteropathogens in stool without genetic isolation or amplification. Biosens. Bioelectron. 2019, 130, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.F.D.; Magalhães, J.M.C.S.; Barroso, M.F.; Oliva-Teles, T.; Freire, C.; Delerue-Matos, C. In situ formation of gold nanoparticles in polymer inclusion membrane: Application as platform in a label-free potentiometric immunosensor for Salmonella typhimurium detection. Talanta 2019, 194, 134–142. [Google Scholar] [CrossRef]
- Srinivasan, S.; Ranganathan, V.; DeRosa, M.C.; Murari, B.M. Label-free aptasensors based on fluorescent screening assays for the detection of Salmonella typhimurium. Anal. Biochem. 2018, 559, 17–23. [Google Scholar] [CrossRef]
- Rubab, M.; Shahbaz, H.M.; Olaimat, A.N.; Oh, D.H. Biosensors for rapid and sensitive detection of Staphylococcus aureus in food. Biosens. Bioelectron. 2018, 105, 49–57. [Google Scholar] [CrossRef]
- Wu, R.; Ma, Y.; Pan, J.; Lee, S.H.; Liu, J.; Zhu, H.; Gu, R.; Shea, K.J.; Pan, G. Efficient. Capture, Rapid Killing and Ultrasensitive Detection of Bacteria by a Nano-Decorated Multi-Functional Electrode Sensor. Biosens. Bioelectron. 2018, 101, 52–59. [Google Scholar] [CrossRef]
- Kimuda, S.G.; Biraro, I.A.; Bagaya, B.S.; Raynes, J.G.; Cose, S. Characterising antibody avidity in individuals of varied Mycobacterium tuberculosis infection status using surface plasmon resonance. PLoS ONE 2018, 13, e0205102. [Google Scholar] [CrossRef]
- Rebelo, R.; Barbosa, A.I.; Caballero, D.; Kwon, I.K.; Oliveira, J.M.; Kundu, S.C.; Reis, R.L.; Correlo, V.M. 3D biosensors in advanced medical diagnostics of high mortality diseases. Biosens. Bioelectron. 2019, 130, 20–39. [Google Scholar] [CrossRef]
- Erdem, Ö.; Saylan, Y.; Cihangir, N.; Denizli, A. Molecularly imprinted nanoparticles based plasmonic sensors for real-time Enterococcus faecalis detection. Biosens. Bioelectron. 2019, 126, 608–614. [Google Scholar] [CrossRef]
- Hoyos-Nogués, M.; Gil, F.J.; Mas-Moruno, C. Antimicrobial Peptides: Powerful Biorecognition Elements to Detect Bacteria in Biosensing Technologies. Molecules 2018, 23, 1683. [Google Scholar] [CrossRef] [PubMed]
- Liebana, S.; Brandao, D.; Alegret, S.; Pividori, M.I. Electrocehmical immunosensors, genosensors and phagosensors for Salmonela detection. Anal. Methods 2014, 6, 8858–8873. [Google Scholar] [CrossRef]
- Cui, F.; Xu, Y.; Wang, R.; Liu, H.; Chen, L.; Zhang, Q.; Mu, X. Label-free impedimetric glycan biosensor for quantitative evaluation interactions between pathogenic bacteria and mannose. Biosens. Bioelectron. 2018, 103, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Bu, T.; Huang, Q.; Yan, L.; Zhang, W.; Dou, L.; Huang, L.; Yang, Q.; Zhao, B.; Yang, B.; Li, T.; et al. Applicability of biological dye tracer in strip biosensor for ultrasensitive detection of pathogenic bacteria. Food Chem. 2019, 274, 816–821. [Google Scholar] [CrossRef]
- Zarei, S.S.; Soleimanian-Zad, S.; Ensafi, A.A. An impedimetric aptasensor for Shigella dysenteriae using a gold nanoparticle-modified glassy carbon electrode. Mikrochim. Acta. 2018, 185, 538. [Google Scholar] [CrossRef]
- Chuensirikulchai, K.; Laopajon, W.; Phunpae, P.; Apiratmateekul, N.; Surinkaew, S.; Tayapiwatana, C.; Pata, S.; Kasinrerk, W. Sandwich antibody-based biosensor system for identification of Mycobacterium tuberculosis complex and nontuberculous mycobacteria. J Immunoass. Immunochem. 2019, 29, 590–604. [Google Scholar] [CrossRef]
- Kravets, V.G.; Kabashin, A.V.; Barnes, W.L.; Grigorenko, A.N. Plasmonic Surface Lattice Resonances: A Review of Properties and Applications. Chem. Rev. 2018, 118, 12–5912. [Google Scholar] [CrossRef]
- Ermolaeva, T.N.; Kalmykova, E.N. Piezoelectric immunosensors: Analytical potentials and outlooks. Russ. Chem. Rev. 2006, 75, 397. [Google Scholar] [CrossRef]
- Muratsugu, M.; Ohta, F.; Miya, Y.; Hosokawa, T.; Kurosawa, S.; Kamo, N.; Ikeda, H. Quartz crystal microbalance for the detection of microgram quantities of human serum albumin: Relationship between the frequency change and the mass of protein adsorbed. Anal. Chem. 1993, 65, 2933–2937. [Google Scholar] [CrossRef]
- Savas, S.; Altintas, Z. Graphene Quantum Dots as Nanozymes for Electrochemical Sensing of Yersinia enterocolitica in Milk and Human Serum. Materials (Basel) 2019, 12, 2189. [Google Scholar] [CrossRef]
- Russel, M.; Sophocleous, M.; JiaJia, S.; Xu, W.; Xiao, L.; Maskow, T.; Alam, M.; Georgiou, J. High-frequency, dielectric spectroscopy for the detection of electrophysiological/biophysical differences in different bacteria types and concentrations. Anal. Chim. Acta 2018, 1028, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Piekarz, I.; Górska, S.; Odrobina, S.; Drab, M.; Wincza, K.; Gamian, A.; Gruszczynski, S. A microwave matrix sensor for multipoint label-free Escherichia coli detection. Biosens. Bioelectron. 2019, 147, 111784. [Google Scholar] [CrossRef] [PubMed]
- Biagi, M.C.; Fabregas, R.; Gramse, G.; van der Hofstadt, M.; Juárez, A.; Kienberger, F.; Fumagalli, L.; Gomila, G. Nanoscale Electric Permittivity of Single Bacterial Cells at Gigahertz Frequencies by Scanning Microwave Microscopy. ACS Nano 2016, 10, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Esteban-Ferrer, D.; Edwards, M.A.; Fumagalli, L.; Juárez, A.; Gomila, G. Electric Polarization Properties of Single Bacteria Measured with Electrostatic Force Microscopy. ACS Nano 2014, 8, 9843–9849. [Google Scholar] [CrossRef] [PubMed]
- Checa, M.; Millan-Solsona, R.; Blanco, N.; Torrents, E.; Fabregas, R.; Gomila, G. Mapping the dielectric constant of a single bacterial cell at the nanoscale with scanning dielectric force volume microscopy. Nanoscale. 2019, 11, 20809–20819. [Google Scholar] [CrossRef]
- Flores-Cosío, G.; Herrera-López, E.J.; Arellano-Plaza, M.; Gschaedler-Mathis, A.; Sanchez, A.; Amaya-Delgado, L. Dielectric property measurements as a method to determine the physiological state of Kluyveromyces marxianus and Saccharomyces cerevisiae stressed with furan aldehydes. Appl. Microbiol. Biotechnol. 2019, 103, 9633–9642. [Google Scholar] [CrossRef]
- Mehrotra, P.; Chatterjee, B.; Sen, S. EM-Wave Biosensors: A Review of RF, Microwave, mm-Wave and Optical Sensing. Sensors (Basel) 2019, 19, 1013. [Google Scholar] [CrossRef]
- Oberoi, K.S.; Daya, K.S.; Tirumalai, P.S. Microwave Sensor for Detection of E. Coli in Water. In Proceedings of the Sixth International Conference on Sensing Technology (ICST), Kolkata, India, 18–21 December 2012. [Google Scholar] [CrossRef]
- Narang, R.; Mohammadi, S.; Ashani, M.M.; Narang, R.; Mohammadi, S.; Ashani, M.M.; Sadabadi, H.; Hejazi, H.; Zarifi, M.H.; Sanati-Nezhad, A. Sensitive, Real-time and Non-Intrusive Detection of Concentration and Growth of Pathogenic Bacteria using Microfluidic-Microwave Ring Resonator Biosensor. Sci. Rep. 2018, 8, 15807. [Google Scholar] [CrossRef]
- Tigli, O.; Zaghloul, M. A Novel Circular SAW (Surface Acoustic Wave) Device in CMOS. Proc. IEEE Sens. 2007, 474–477. [Google Scholar] [CrossRef]
- Alvarez, M.; Lechuga, L.M. Microcantilever-based platforms as biosensing tools. Analyst 2010, 135, 827–836. [Google Scholar] [CrossRef]
- Wu, G.; Datar, R.H.; Hansen, K.M.; Thundat, T.; Cote, R.J.; Majumdar, A. Bioassay of prostate-specific antigen (PSA) using microcantilevers. Nat. Biotechnol. 2001, 19, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Sang, S.; Zhao, Y.; Zhang, W.; Li, P.; Hu, J.; Li, G. Surface stress-based biosensors. Biosens. Bioelectron. 2014, 51, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Dahlin, A.V. Plasmonic Biosensors: An Integrated View of Refractometric Detection; De Monfort University: Leicester, UK, 2012; Volume 4, 316p. [Google Scholar] [CrossRef]
- Ilica, B.; Yang, Y.; Craighead, H.G. Virus detection using nanoelectromechanical devices. Appl. Phys. Lett. 2004, 85, 2604. [Google Scholar] [CrossRef]
- Arlett, J.L.; Myers, E.B.; Roukes, M.L. Comparative advantages of mechanical biosensors. Nat. Nanotechnol. 2011, 6, 203–215. [Google Scholar] [CrossRef]
- Barton, R.A.; Ilic, B.; Verbridge, S.S.; Cipriany, B.R.; Parpia, J.M.; Craighead, H.G. Fabrication of a nanomechanical mass sensor containing a nanofluidic channel. Nano Lett. 2010, 10, 2058–2063. [Google Scholar] [CrossRef]
- Lee, J.; Moon, S.U.; Lee, Y.S.; Ali, B.A.; Al-Khedhairy, A.A.; Ali, D.; Ahmed, J.; Al Salem, A.M.; Kim, S. Quantum dot-based molecular beacon to monitor intracellular microRNAs. Sensors (Basel) 2015, 15, 12872–12883. [Google Scholar] [CrossRef]
- Shin, S.; Kim, J.P.; Sim, S.J.; Lee, J. A multisized piezoelectric microcantilever biosensor array for the quantitative analysis of mass and surface stress. Appl. Phys. Lett. 2008, 93, 102902. [Google Scholar] [CrossRef]
- Kuss, S.; Amin, H.M.A.; Compton, R.G. Electrochemical Detection of Pathogenic Bacteria-Recent Strategies, Advances and Challenges. Chem. Asian J. 2018, 13, 2758–2769. [Google Scholar] [CrossRef]
- Muniandy, S.; Teh, S.J.; Appaturi, J.N.; Thong, K.L.; Lai, C.W.; Ibrahim, F.; Leo, B.F. A reduced graphene oxide-titanium dioxide nanocomposite based electrochemical aptasensor for rapid and sensitive detection of Salmonella enterica. Bioelectrochemistry 2019, 127, 136–144. [Google Scholar] [CrossRef]
- Singh, R.; Mukherjee, M.D.; Sumana, G.; Gupta, R.K.; Sood, S.; Malhotra, B.D. Biosensors for pathogen detection: A smart approach towards clinical diagnosis. Sens. Actuators B Chem. 2014, 197, 385–404. [Google Scholar] [CrossRef]
- Gonzalez-Sapienza, G.; Rossotti, M.A.; Tabares-da Rosa, S. Single-Domain Antibodies as Versatile Affinity Reagents for Analytical and Diagnostic Applications. Front. Immunol. 2017, 8, 977. [Google Scholar] [CrossRef] [PubMed]
- Bever, C.S.; Dong, J.X.; Vasylieva, N.; Barnych, B.; Cui, Y.; Xu, Z.L.; Hammock, B.D.; Gee, S.J. VHH antibodies: Emerging reagents for the analysis of environmental chemicals. Anal. Bioanal. Chem. 2016, 408, 5985–6002. [Google Scholar] [CrossRef]
- Shriver-Lake, L.C.; Liu, J.L.; Zabetakis, D.; Sugiharto, V.A.; Lee, C.; Defang, G.N.; Wu, S.L.; Anderson, G.P.; Goldman, E.R. Selection and Characterization of Anti-Dengue NS1 Single Domain Antibodies. Sci. Rep. 2018, 8, 18086. [Google Scholar] [CrossRef] [PubMed]
- Goode, J.; Dillon, G.; Liu, P.A. The development and optimisation of nanobody based electrochemical immunosensors for IgG. Sens. Actuators B Chem. 2016, 234, 478–484. [Google Scholar] [CrossRef]
- Della Pia, E.A.; Martinez, K.L. Single domain antibodies as a powerful tool for high quality surface plasmon resonance studies. PLoS ONE 2015, 10, e0124303. [Google Scholar] [CrossRef] [PubMed]
- Manjavacas, A.; Zundel, L.; Sanders, S. Analysis of the Limits of the Near-Field Produced by Nanoparticle Arrays. ACS Nano 2019, 13, 10682–10693. [Google Scholar] [CrossRef]
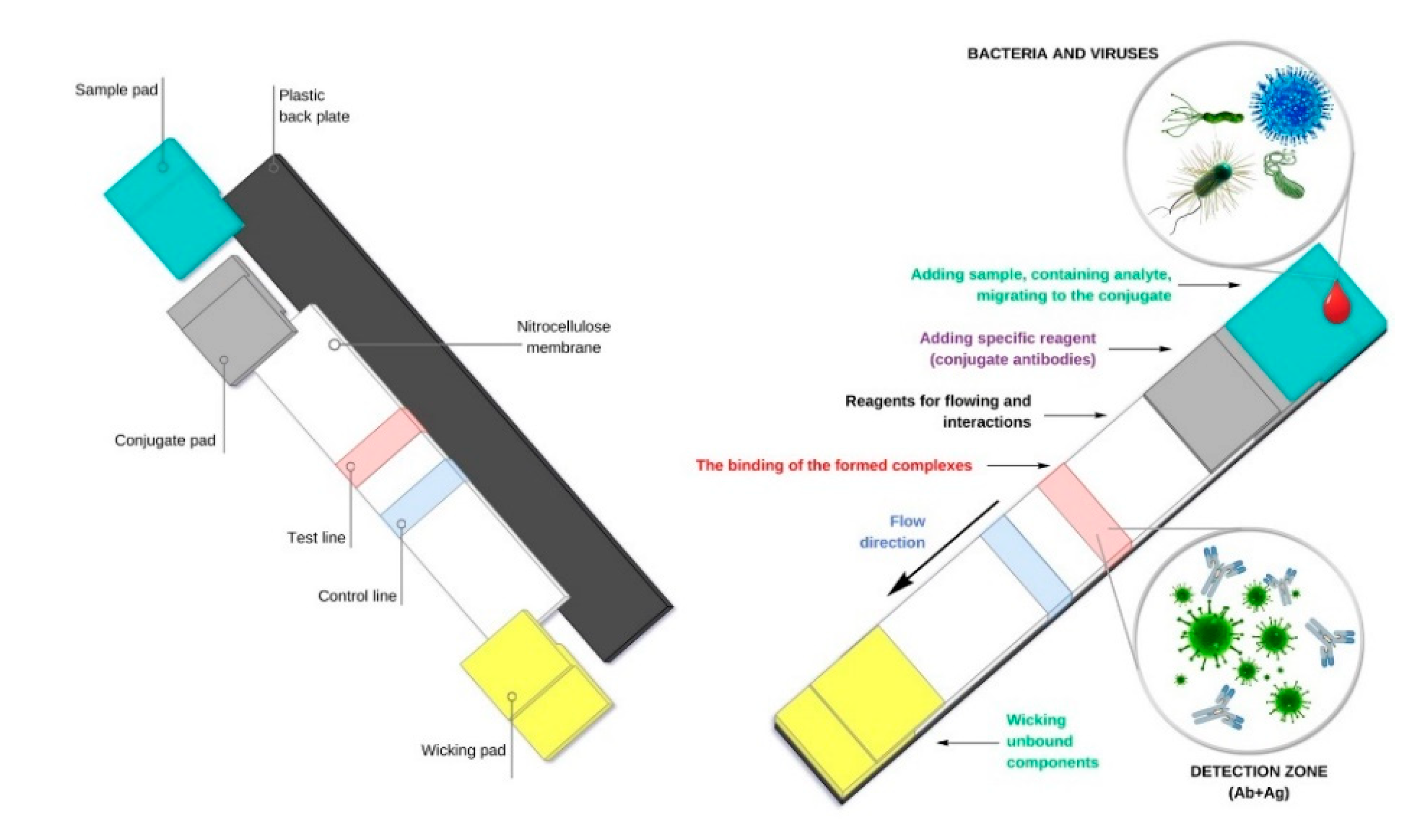
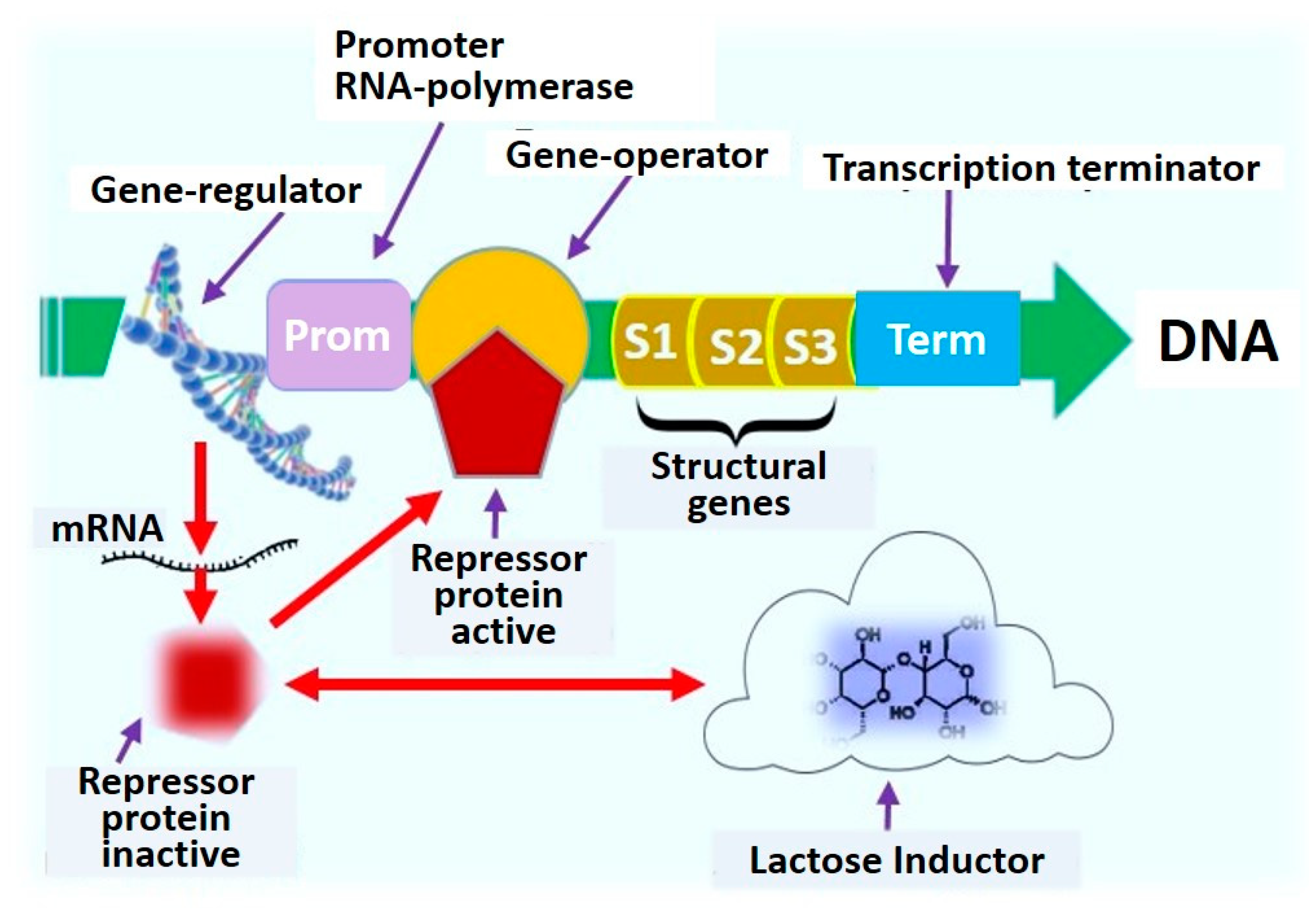
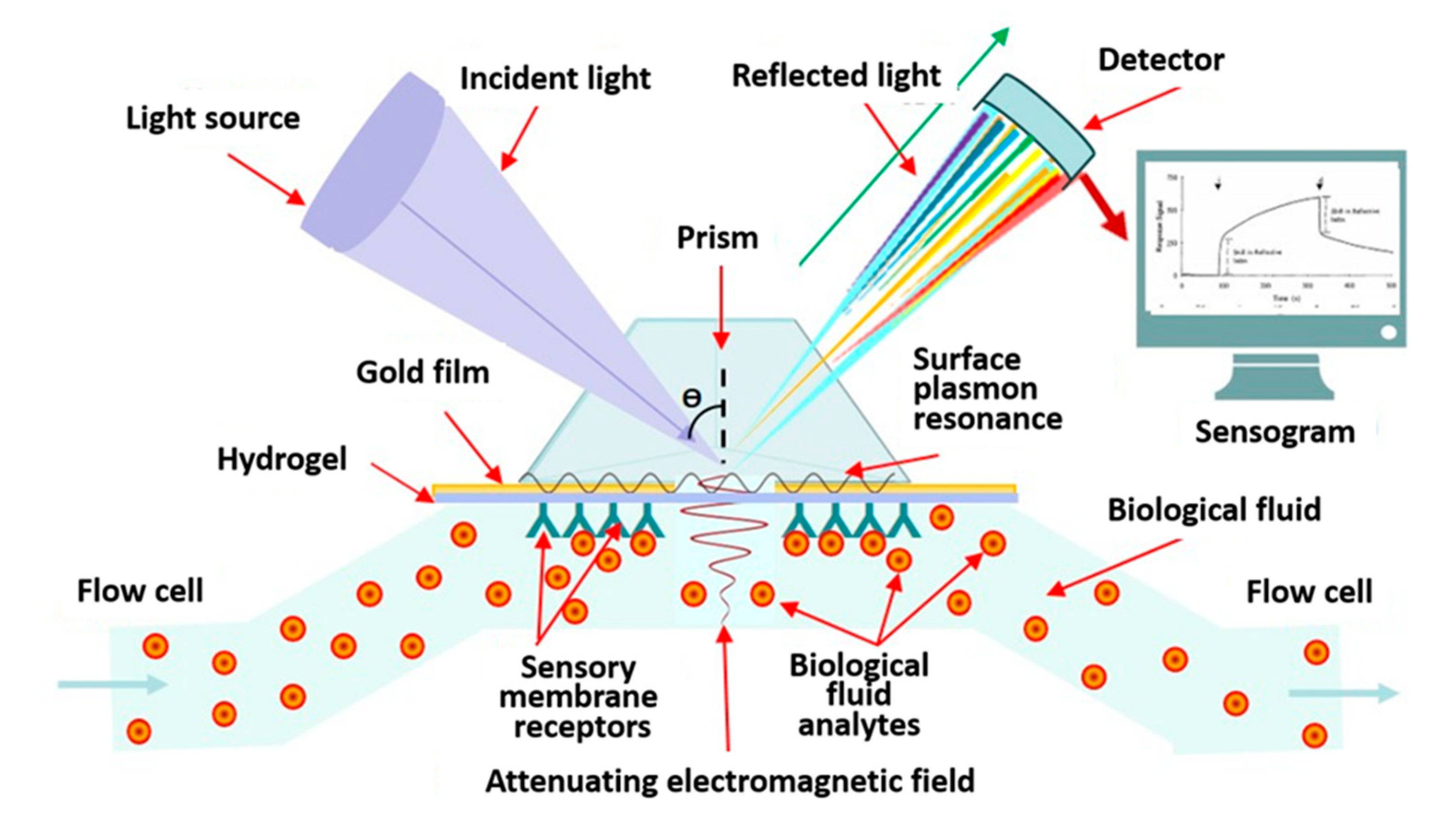
| Advantages | Disadvantages | References |
|---|---|---|
| ◾ Cheap, rapid, inexpensive, and easy to apply tests. ◾ Long shelf-life of test systems. ◾ Test systems do not require special temperature conditions for storage. ◾ No additional special equipment is required. ◾ They do not need qualified personnel. ◾ They can be used by general practice physicians or patients at home. ◾ Visual result is clear and easily distinguishable. ◾ Tests are usually sold as kits with a set of all the items needed to perform the test. ◾ Possible increase in sensitivity of test systems by the use of plasmon resonance, surface-enhanced Raman scattering (SERS), chemiluminescent or fluorescent labels. ◾ Possibility of multiplexed formats of test systems | ◾ Suitable only for primary screening and require confirmation of positive results by independent methods. ◾ Special equipment (scanners, reflectometers, CCD cameras) and software are required to obtain quantitative results. ◾ Technological improvement of the method increases cost and duration of the analysis. ◾ In a competitive format, response negatively correlates with concentration. ◾ Possible technical errors in application of specimen may affect the accuracy and reproducibility of result. ◾ Increase in sensitivity of tests is based on the use of gold, silver, or enzyme nanoparticles, which limits shelf-life, increases cost of analysis, and breaks the one-step rule of application. ◾ Tested specimen must be in the form of a solution. Preliminary dissolution of dry specimens is mandatory. ◾ When the analyte content in the solution is low, the specimen needs to be concentrated. | [14] [17,21] [15,19] [16,18,20] [18,19,20] [16,18,20] [13] [14,16,17] [18] [13,15,16] [18,19] |
| Advantages | References |
|---|---|
| ● A simplified pattern of analysis. | [3,29,49,51,81] |
| ● Reduced analysis time (rapid response time). | [7,29,82] |
| ● Lower cost of analysis. | [7,28,80] |
| ● Reduced consumption of organic solvents. | [33,64,78,83] |
| ● Portability and small dimensions. | [33,43,73] |
| ● No need in qualified medical personnel. | [3,7,39,64,83] |
| ● Opportunity to quantify biomolecules in real-time mode. | [25,26,78,84,85] |
| ● Target analytes are detected in natural forms, without. modifications and labels. | [22,33,73,80,82] |
| ● High sensitivity. | [22,25,26,43,64,85,86] |
| ● Direct measurement of analytes. | [43,51,64,80] |
| ● Opportunity to detect small molecules. | [3,7,25,26,43,79] |
| ● Opportunity of multiplexing. | [28,29,64,83] |
| ● Access to kinetic and thermodynamic parameters. | [22,26,39,80,86] |
| Recognizing Bioreceptor | Conversion Method | Test Models of Pathogens, Sensitivity | References |
|---|---|---|---|
| Bacteriophage | Photoluminescence | S. aureus 4 × 108 ufc/mL | [70] |
| Antimicrobial peptides | Impedancemetry | E. coli, S. aureus, P. aeruginosa, S. epidermidis, 102 ufc/mL | [75] |
| Antibacterial nanoparticles Zn-CuO and graphene oxide Man/MUA-MH/Au * | Impedancemetry and electrochemical impedance spectroscopy | E. coli, S. aureus 50 ufc/mL and antibacterial effect 100% (30 min) | [93] |
| Thiolated protein G on: - gold electrodes - gold nanoparticles | Cyclic voltammetry and electrochemical impedance spectroscopy | S. typhimurium, 2.16 × 106 ufc/mL E. coli, 50–103 ufc/mL | [102] |
| Enzymes | Electrochemical | E. coli O157:H7 150 ufc/mL | [68] |
| Nucleic acids (DNA, RNA) | Electrochemical | S. aureus, 140 ufc/mL S. typhimurium, 48 ufc/mL | [72] |
| Nucleic acids (DNA, RNA) | Electrochemical | S. aureus, M. tuberculosis | [73] |
| Aptamer on AuNP | Autofluorescence quenching | S. typhimurium, 48 ufc/mL | [92] |
| Monoclonal antibodies | Optical | S. enteritidis, 80 ufc/mL Listeria monocytogenes | [103] |
| Thiolated aptamer | Impedancemetry | Shigella dysenteriae | [104] |
| Nucleic acids (DNA, RNA) | Electrochemical impedance spectroscopy | M. tuberculosis | [50] |
| Monoclonal antibodies | Surface plasmon resonance | Enterococcus faecalis, 104–108 ufc/mL | [99] |
| Aptamer | Impedancemetry | Bacillus cereus, 104–106 ufc/mL Bacillus anthracis (spores) | [32] |
| Nucleic acids (DNA) | Cyclic voltammetry and electrochemical impedance spectroscopy | Salmonella spp. | [81] |
| Enzyme Simulator (Graphene Quantum Dots, GQD) | Electrochemical | Yersinia enterocolitica, 5 (milk)–30 (serum) ufc/mL | [105] |
| Monoclonal antibodies | Surface plasmon resonance | S. aureus, 224 ufc/mL, 30 min | [71] |
| Monoclonal antibodies | Visualization | Salmonella enteritidis, 102–108 ufc/mL | [88] |
| DNA, aptamer | Electrochemical | Bird flu virus H5N1 (AIV) | [48] |
| Nucleic acids (DNA) | Electrochemical impedance | Zika virus, 25.0 ± 1.7 hМ. | [49] |
| Aptamer (rGO-TiO2) | Electrochemical | S. enterica Typhimurium, 101–108 ufc/mL | [98] |
| Nucleic acids (DNA) | Piezoelectric | Clostridium difficile, sensitivity 95% and specificity 95% | [90] |
| Monoclonal antibodies | Surface plasmon resonance | M. tuberculosis, 102–106 ufc/mL | [93,101] |
| Aptamer | Fluorescent | S. enterica Typhimurium, 6–10 ufc/mL | [92] |
| Monoclonal antibodies | Potentiometry | S. enterica Typhimurium, 6 ufc/mL | [91] |
| Nucleic acids (DNA) | Electrochemical impedance | M. tuberculosis, 102–106 ufc/mL | [50] |
| Aptamer (RNA) | Fluorescent | S. aureus, 102–106 ufc/mL | [3] |
| Nicolson-Ross-Weir method | Dielectric spectroscopy | Bacillus Subtilis, 2.10–1.30 × 109 ufc/mL E. coli 1.60–1.00 × 109 ufc/mL | [106] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andryukov, B.G.; Besednova, N.N.; Romashko, R.V.; Zaporozhets, T.S.; Efimov, T.A. Label-Free Biosensors for Laboratory-Based Diagnostics of Infections: Current Achievements and New Trends. Biosensors 2020, 10, 11. https://doi.org/10.3390/bios10020011
Andryukov BG, Besednova NN, Romashko RV, Zaporozhets TS, Efimov TA. Label-Free Biosensors for Laboratory-Based Diagnostics of Infections: Current Achievements and New Trends. Biosensors. 2020; 10(2):11. https://doi.org/10.3390/bios10020011
Chicago/Turabian StyleAndryukov, Boris G., Natalya N. Besednova, Roman V. Romashko, Tatyana S. Zaporozhets, and Timofey A. Efimov. 2020. "Label-Free Biosensors for Laboratory-Based Diagnostics of Infections: Current Achievements and New Trends" Biosensors 10, no. 2: 11. https://doi.org/10.3390/bios10020011
APA StyleAndryukov, B. G., Besednova, N. N., Romashko, R. V., Zaporozhets, T. S., & Efimov, T. A. (2020). Label-Free Biosensors for Laboratory-Based Diagnostics of Infections: Current Achievements and New Trends. Biosensors, 10(2), 11. https://doi.org/10.3390/bios10020011