Interaction between Copper Oxide Nanoparticles and Amino Acids: Influence on the Antibacterial Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Treatment of CuO NPs with Amino Acids
2.3. Synthesis of Cu-Amino Acid Complexes
2.4. FTIR and TGA-DSC
2.5. ICP-MS
2.6. SEM
2.7. DLS, ELS and CSA
2.8. Antibacterial Test
3. Results and Discussion
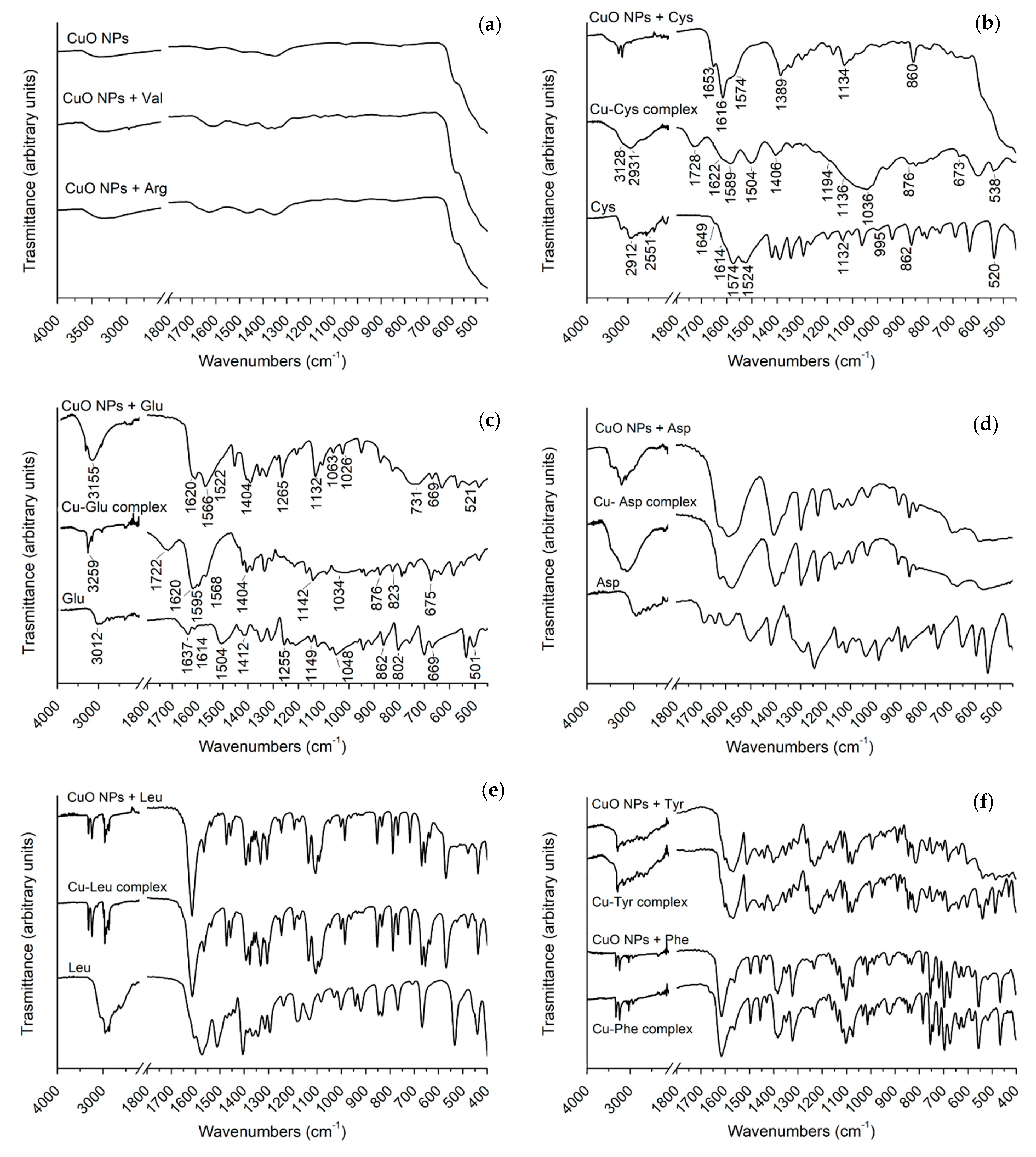
3.1. ATR-FTIR
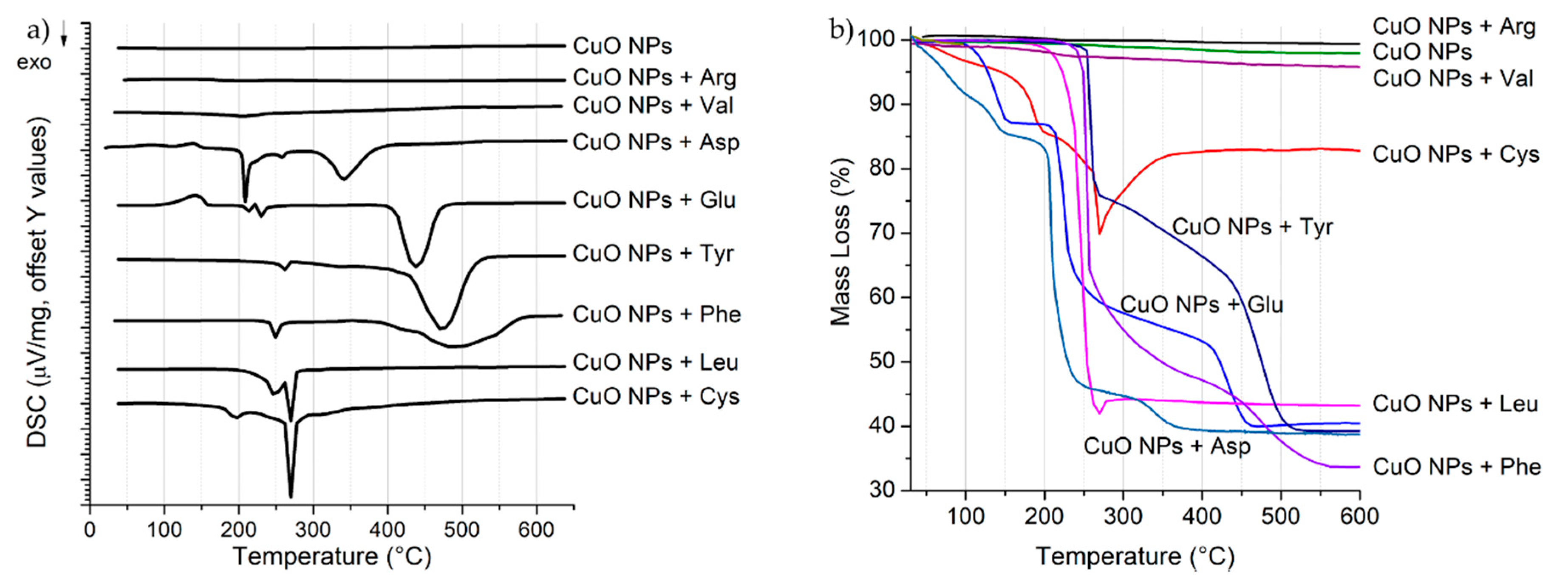
3.2. DSC-TGA
3.3. Total Copper Content in Pristine CuO NPs, Treated CuO NPs and Cu-Amino Acid Complexes
3.4. SEM
3.5. Colloidal Characterization
3.6. DLS
3.7. ELS
3.8. CSA
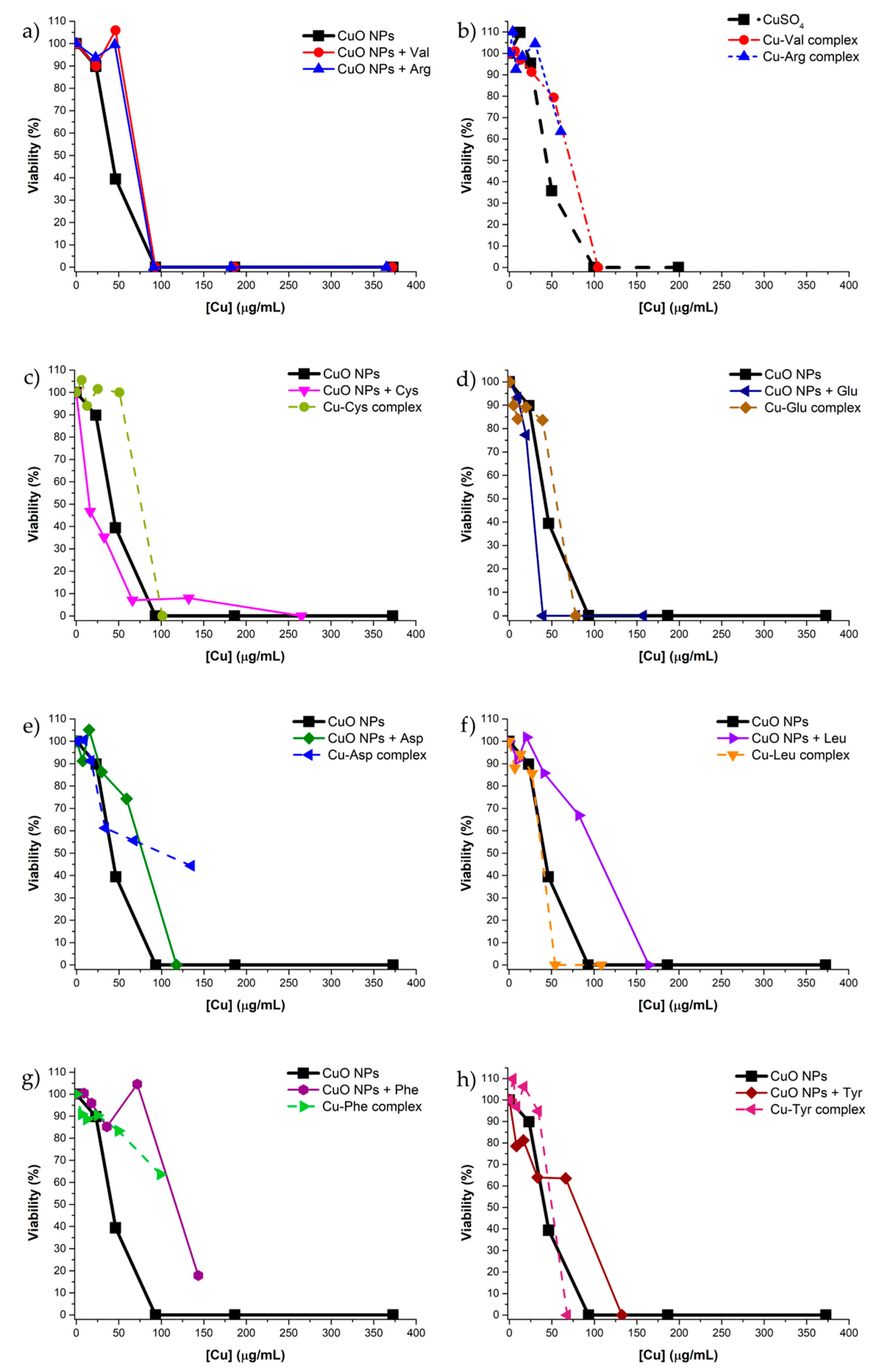
3.9. Antibacterial Test
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dizaj, S.M.; Lotfipour, F.; Barzegar-Jalali, M.; Zarrintan, M.H.; Adibkia, K. Antimicrobial activity of the metals and metal oxide nanoparticles. Mater. Sci. Eng. C 2014, 44, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Stankic, S.; Suman, S.; Haque, F.; Vidic, J. Pure and multi metal oxide nanoparticles: Synthesis, antibacterial and cytotoxic properties. J. Nanobiotechnol. 2016, 14, 73. [Google Scholar] [CrossRef] [PubMed]
- Raghunath, A.; Perumal, E. Metal oxide nanoparticles as antimicrobial agents: A promise for the future. Int. J. Antimicrob. Agents 2017, 49, 137–152. [Google Scholar] [CrossRef]
- Sikora, P.; Augustyniak, A.; Cendrowski, K.; Nawrotek, P.; Mijowska, E. Antimicrobial Activity of Al2O3, CuO, Fe3O4, and ZnO Nanoparticles in Scope of Their Further Application in Cement-Based Building Materials. Nanomaterials 2018, 8, 212. [Google Scholar] [CrossRef]
- Salvioni, L.; Galbiati, E.; Collico, V.; Alessio, G.; Avvakumova, S.; Corsi, F.; Tortora, P.; Prosperi, D.; Colombo, M. Negatively charged silver nanoparticles with potent antibacterial activity and reduced toxicity for pharmaceutical preparations. Int. J. Nanomed. 2017, 12, 2517–2530. [Google Scholar] [CrossRef]
- Phull, A.-R.; Abbas, Q.; Ali, A.; Raza, H.; Kim, S.J.; Zia, M.; Haq, I. Antioxidant, cytotoxic and antimicrobial activities of green synthesized silver nanoparticles from crude extract of Bergenia ciliata. Futur. J. Pharm. Sci. 2016, 2, 31–36. [Google Scholar] [CrossRef]
- Baptista, P.V.; McCusker, M.P.; Carvalho, A.; Ferreira, D.A.; Mohan, N.M.; Martins, M.; Fernandes, A.R. Nano-strategies to fight multidrug resistant bacteria—“A Battle of the Titans”. Front. Microbiol. 2018, 9, 1441. [Google Scholar] [CrossRef]
- Webster, T.J.; Seil, I. Antimicrobial applications of nanotechnology: Methods and literature. Int. J. Nanomed. 2012, 7, 2767. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.A.; Adeleye, A.S.; Conway, J.R.; Garner, K.L.; Zhao, L.; Cherr, G.N.; Hong, J.; Gardea-Torresdey, J.L.; Godwin, H.A.; Hanna, S.; et al. Comparative environmental fate and toxicity of copper nanomaterials. NanoImpact 2017, 7, 28–40. [Google Scholar] [CrossRef]
- Azam, A.; Ahmed, A.S.; Oves, M.; Khan, M.; Memic, A. Size-dependent antimicrobial properties of CuO nanoparticles against Gram-positive and -negative bacterial strains. Int. J. Nanomed. 2012, 7, 3527. [Google Scholar] [CrossRef] [PubMed]
- Ortelli, S.; Costa, A.L.; Blosi, M.; Brunelli, A.; Badetti, E.; Bonetto, A.; Hristozov, D.; Marcomini, A. Colloidal characterization of CuO nanoparticles in biological and environmental media. Environ. Sci. Nano 2017, 4, 1264–1272. [Google Scholar] [CrossRef]
- Pantano, D.; Neubauer, N.; Navratilova, J.; Scifo, L.; Civardi, C.; Stone, V.; Von Der Kammer, F.; Müller, P.; Sobrido, M.S.; Angeletti, B.; et al. Transformations of Nanoenabled Copper Formulations Govern Release, Antifungal Effectiveness, and Sustainability throughout the Wood Protection Lifecycle. Environ. Sci. Technol. 2018, 52, 1128–1138. [Google Scholar] [CrossRef]
- Radic, S. Biophysical Interaction Between Nanoparticles and Biomolecules. Ph.D. Thesis, Clemson University, Clemson, SC, USA, 2015. [Google Scholar]
- Zakharova, O.V.; Godymchuk, A.Y.; Gusev, A.A.; Gulchenko, S.I.; Vasyukova, I.A.; Kuznetsov, D.V. Considerable Variation of Antibacterial Activity of Cu Nanoparticles Suspensions Depending on the Storage Time, Dispersive Medium, and Particle Sizes. BioMed Res. Int. 2015, 2015, 412530. [Google Scholar] [CrossRef] [PubMed]
- Phogat, N.; Uddin, I.; Jahan, A. Interaction of Nanoparticles with Biomolecules, Protein, Enzymes, and Its Applications. Precis. Med. 2018, 253–276. [Google Scholar]
- Stanila, A.; Marcu, A.; Rusu, D.; Rusu, M.; David, L. Spectroscopic studies of some copper(II) complexes with amino acids. J. Mol. Struct. 2007, 834–836, 364–368. [Google Scholar] [CrossRef]
- Hu, R.; Yu, Q.; Liang, F.; Ma, L.; Chen, X.; Zhang, M.; Liang, H.; Yu, K. Syntheses and crystal structures of cis- and trans-copper(II) complexes of L-arginine. J. Coord. Chem. 2008, 61, 1265–1271. [Google Scholar] [CrossRef]
- Dokken, K.M.; Parsons, J.G.; McClure, J.; Gardea-Torresdey, J.L. Synthesis and structural analysis of copper(II) cysteine complexes. Inorg. Chim. Acta 2009, 362, 395–401. [Google Scholar] [CrossRef]
- Brunelli, A.; Badetti, E.; Basei, G.; Izzo, F.C.; Hristozov, D.; Marcomini, A. Effects of organic modifiers on the colloidal stability of TiO2 nanoparticles. A methodological approach for NPs categorization by multivariate statistical analysis. NanoImpact 2018, 9, 114–123. [Google Scholar] [CrossRef]
- Lerche, D. Dispersion stability and particle characterization by sedimentation kinetics in a centrifugal field. J. Dispers. Sci. Technol. 2002, 23, 699–709. [Google Scholar] [CrossRef]
- Doğan, A.; Köseoğlu, F.; Kılıç, E. The Stability Constants of Copper(II) Complexes with Some α-Amino Acids in Dioxan–Water Mixtures. Anal. Biochem. 2001, 295, 237–239. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Von Dem Bussche, A.; Kabadi, P.K.; Kane, A.B.; Hurt, R.H. Biological and environmental transformations of copper-based nanomaterials. ACS Nano 2013, 7, 8715–8727. [Google Scholar] [CrossRef]
- Deschamps, P.; Zerrouk, N.; Nicolis, I.; Martens, T.; Curis, E.; Charlot, M.-F.; Girerd, J.J.; Prangé, T.; Bénazeth, S.; Chaumeil, J.C.; et al. Copper(II)–l-glutamine complexation study in solid state and in aqueous solution. Inorg. Chim. Acta 2003, 353, 22–34. [Google Scholar] [CrossRef]
- Reddy, K.R. Green synthesis, morphological and optical studies of CuO nanoparticles. J. Mol. Struct. 2017, 1150, 553–557. [Google Scholar] [CrossRef]
- Berestova, T.V.; Kuzina, L.G.; Amineva, N.A.; Faizrakhmanov, I.S.; Massalimov, I.A.; Mustafin, A.G. ATR-FTIR spectroscopic investigation of the cis- and trans-bis-(α-amino acids) copper(II) complexes. J. Mol. Struct. 2017, 1137, 260–266. [Google Scholar] [CrossRef]
- Peng, C.; Shen, C.; Zheng, S.; Yang, W.; Hu, H.; Liu, J.; Shi, J. Transformation of CuO Nanoparticles in the Aquatic Environment: Influence of pH, Electrolytes and Natural Organic Matter. Nanomaterials 2017, 7, 326. [Google Scholar] [CrossRef] [PubMed]
- Fitts, J.P.; Persson, P.; Brown, G.E.; Parks, G.A. Structure and Bonding of Cu(II)–Glutamate Complexes at the γ-Al2O3–Water Interface. J. Colloid Interface Sci. 1999, 220, 133–147. [Google Scholar] [CrossRef]
- Herlinger, A.W.; Wenhold, S.L.; Long, T.V. Infrared spectra of amino acids and their metal complexes. II.Geometrical isomerism in bis(amino acidato)copper(II) complexes. J. Am. Chem. Soc. 1970, 92, 6474–6481. [Google Scholar] [CrossRef]
- De Jong, W.H.; De Rijk, E.; Bonetto, A.; Wohlleben, W.; Stone, V.; Brunelli, A.; Badetti, E.; Marcomini, A.; Gosens, I.; Cassee, F.R. Toxicity of copper oxide and basic copper carbonate nanoparticles after short-term oral exposure in rats. Nanotoxicology 2018, 13, 1–23. [Google Scholar] [CrossRef]
- Bondarenko, O.; Ivask, A.; Käkinen, A.; Kahru, A. Sub-toxic effects of CuO nanoparticles on bacteria: Kinetics, role of Cu ions and possible mechanisms of action. Environ. Pollut. 2012, 169, 81–89. [Google Scholar] [CrossRef]
- Malanovic, N.; Lohner, K. Gram-positive bacterial cell envelopes: The impact on the activity of antimicrobial peptides. Biochim. Biophys. Acta 2016, 1858, 936–946. [Google Scholar] [CrossRef] [PubMed]
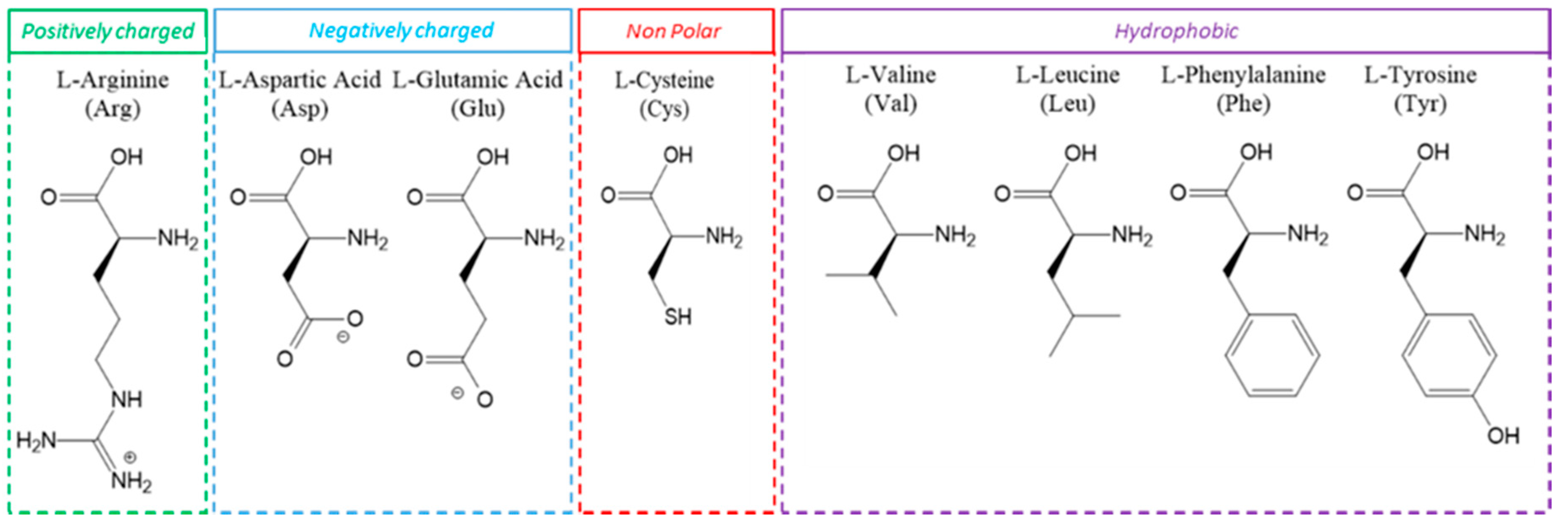
| Component/Parameter | Type/Value/Mode |
|---|---|
| Nebulizer | Meinhard quartz microconcentric |
| Spray Chamber | Quartz cyclonic |
| Triple Cone Interface Material | Nickel/Aluminum |
| Plasma Gas Flow | 18 L/min |
| Auxiliary Gas Flow | 1.2 L/min |
| Nebulizer Gas Flow | 0.96–1 L/min |
| Sample Uptake Rate | 200–250 µL/min |
| RF Power | 1600 W |
| Isotope | 63Cu |
| NPs | Complexes | ||
|---|---|---|---|
| ICP-MS Cu (%) | TGA Cu (%) | ICP-MS Cu (%) | |
| CuO | 74.6 ± 0.6 | 78 | - |
| Val | 74.4 ± 0.5 | 77 | 20.9 ± 0.5 |
| Arg | 73.0 ± 0.3 | 80 | 12.1 ± 0.7 |
| Cys | 53.0 ± 0.3 | 55 | 20.3 ± 0.6 |
| Asp | 23.5 ± 0.3 | 29 | 27.0 ± 0.6 |
| Glu | 31.5 ± 0.5 | 32 | 15.6 ± 0.5 |
| Leu | 32.9 ± 0.4 | 30 | 21.7 ± 1.6 |
| Phe | 28.7 ± 0.2 | 27 | 19.8 ± 0.5 |
| Tyr | 26.5 ± 0.7 | 31 | 13.6 ± 0.7 |
| NPs | Hydrodynamic Size (nm) | ξ (mV) | Sedimentation Velocity (μm/s) |
|---|---|---|---|
| CuO | 340 ± 206 | 15.4 ± 0.6 | 0.03 ± 0.01 |
| CuO + Val | 839 ± 182 | 13.1 ± 1.0 | 0.04 ± 0.01 |
| CuO + Arg | 518 ± 103 | 7.8 ± 0.9 | 0.07 ± 0.01 |
| CuO + Cys | 2630 ± 1031 | −4.3 ± 0.8 | 0.26 ± 0.01 |
| CuO + Asp | 2714 ± 1371 | −0.3 ± 0.7 | 0.33 ± 0.01 |
| CuO + Glu | 2084 ± 1053 | 1.3 ± 1.7 | 0.22 ± 0.03 |
| CuO + Leu | 2803 ± 1522 | −8.1 ± 1.9 | 0.34 ± 0.03 |
| CuO + Phe | 1491 ± 895 | −4.0 ± 0.5 | 0.22 ± 0.01 |
| CuO + Tyr | 4205 ± 1009 | −6.5 ± 1.4 | 0.28 ± 0.01 |
| Materials | MIC (μg Cu/mL) | Materials | MIC (μg Cu/mL) |
|---|---|---|---|
| CuO NPs | 94 | CuSO4 | 99 |
| CuO NPs + Val | 93 | Cu-Val complex | 104 |
| CuO NPs + Arg | 91 | Cu-Arg complex | >60 |
| CuO NPs + Cys | 265 | Cu-Cys complex | 101 |
| CuO NPs + Asp | 120 | Cu-Asp complex | >135 |
| CuO NPs + Glu | 39 | Cu-Glu complex | 78 |
| CuO NPs + Leu | 165 | Cu-Leu complex | 54 |
| CuO NPs + Phe | >143 | Cu-Phe complex | >99 |
| CuO NPs + Tyr | 135 | Cu-Tyr complex | 68 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Badetti, E.; Calgaro, L.; Falchi, L.; Bonetto, A.; Bettiol, C.; Leonetti, B.; Ambrosi, E.; Zendri, E.; Marcomini, A. Interaction between Copper Oxide Nanoparticles and Amino Acids: Influence on the Antibacterial Activity. Nanomaterials 2019, 9, 792. https://doi.org/10.3390/nano9050792
Badetti E, Calgaro L, Falchi L, Bonetto A, Bettiol C, Leonetti B, Ambrosi E, Zendri E, Marcomini A. Interaction between Copper Oxide Nanoparticles and Amino Acids: Influence on the Antibacterial Activity. Nanomaterials. 2019; 9(5):792. https://doi.org/10.3390/nano9050792
Chicago/Turabian StyleBadetti, Elena, Loris Calgaro, Laura Falchi, Alessandro Bonetto, Cinzia Bettiol, Benedetta Leonetti, Emmanuele Ambrosi, Elisabetta Zendri, and Antonio Marcomini. 2019. "Interaction between Copper Oxide Nanoparticles and Amino Acids: Influence on the Antibacterial Activity" Nanomaterials 9, no. 5: 792. https://doi.org/10.3390/nano9050792
APA StyleBadetti, E., Calgaro, L., Falchi, L., Bonetto, A., Bettiol, C., Leonetti, B., Ambrosi, E., Zendri, E., & Marcomini, A. (2019). Interaction between Copper Oxide Nanoparticles and Amino Acids: Influence on the Antibacterial Activity. Nanomaterials, 9(5), 792. https://doi.org/10.3390/nano9050792