3.1. Crafting a Collection of Diverse, Logic-Based Nucleic Acid Systems Geared Towards Therapeutic Development
Multiple nucleic acid systems were designed to specifically adhere to the ideal structural and functional criteria for an RNA-based conditional therapeutic that were outlined above. Each individual assembly is composed of only RNA and DNA oligonucleotides, with no additional 2′-modified nucleotides. Every initial-state assembly was designed to contain RNA/DNA hybrid duplexes to minimize potential ribonuclease cleavage of the RNA payloads, and the diagnostic components of each system are composed entirely of DNA to prevent its processing by ribonucleases, which would likely compromise its conditional function.
The conditional systems that follow display a continuous increase in design complexity, starting with a simple bimolecular switch that is able to detect a single input biomarker and release an ssRNA oligo when the RNA biomarker is present. From there, several pairs of cognate RNA/DNA hybrid constructs are characterized that perform conditional dsRNA release though differing diagnostic mechanisms. The first utilizes a diagnostic method whereby the two cognate constructs recognize neighboring sequence regions of a single input trigger to induce dsRNA release. The subsequent RNA/DNA hybrid pairs were all designed such that the diagnostic component responsible for the RNA biomarker that governed conditional function was completely contained within one of the two hybrids, while the cognate partner hybrid recognized a biomarker-dependent structural change of the first hybrid. Using this strategy, it is first demonstrated that dsRNA release can be induced by a single input, but then also that dsRNA release can be repressed following slight alterations to the design of the diagnostic component. Ultimately, a single cognate pair of RNA/DNA hybrid constructs are coupled that are able to detect multiple RNA triggers, with the release of dsRNA being dependent on the presence of one biomarker and the absence of a second. As a whole, the suite of conditional systems provides diversity in terms of the oligonucleotide output that can be generated, and the ability to either induce or repress oligonucleotide release based on the presence or absence of a specific RNA of interest.
3.2. A Beacon-Derived Conditional Switch Releases a Single-Stranded Oligonucleotide in the Presence of an RNA Trigger
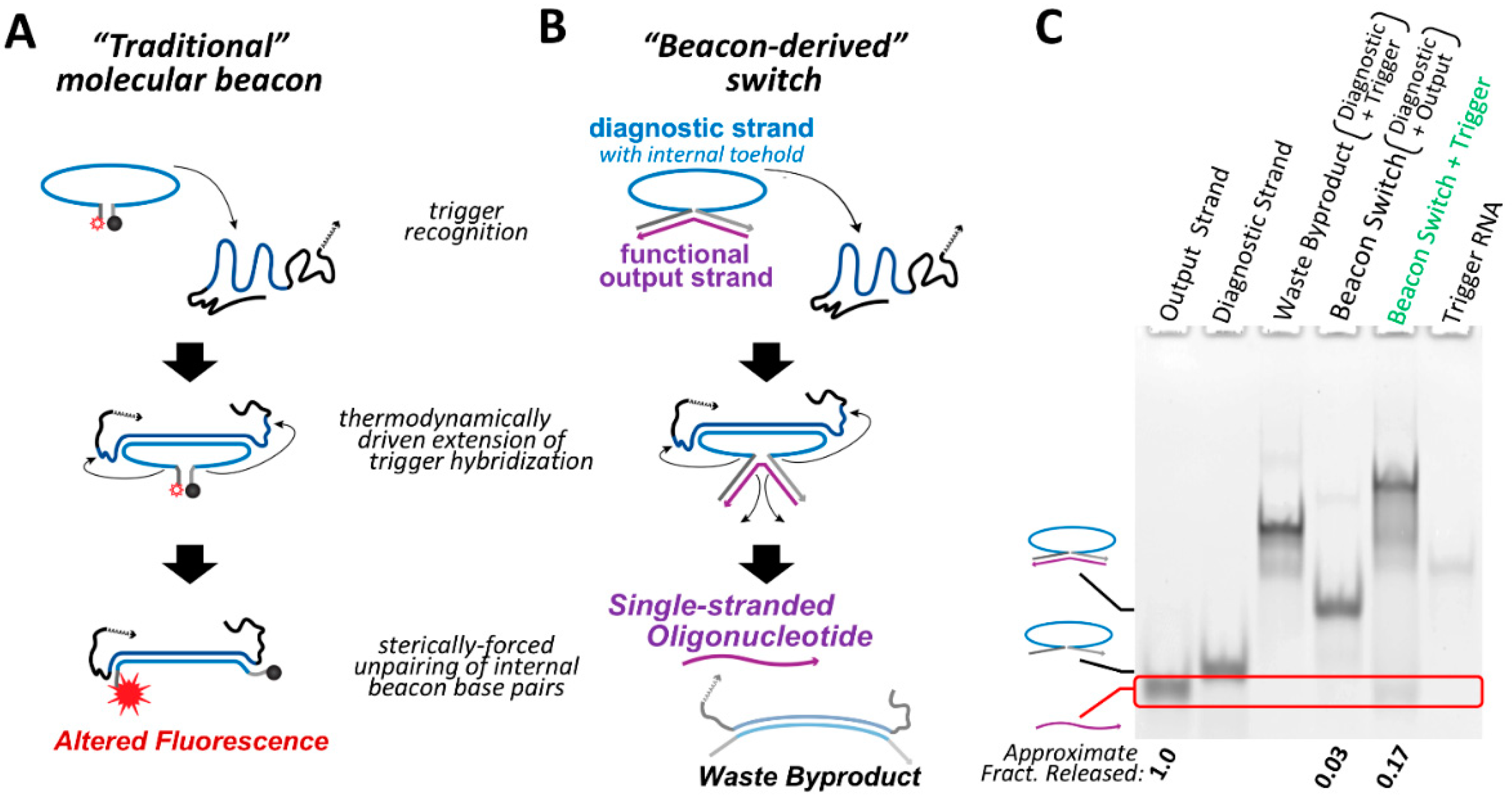
Traditional molecular beacons act as a unimolecular diagnostic tool, giving a fluorescent output signal that changes as a result of the presence of a specific oligonucleotide trigger [
21] (
Figure 1A). Rather than use fluorescence as an output signal, we have re-engineered the beacon system as a bimolecular switch construct that is able to release a single-stranded oligonucleotide upon recognition of a specific trigger sequence. Whereas traditional molecular beacons contain complementary regions at the 5′ and 3′ ends resulting in a hairpin structure (
Figure 1A), the beacon switch is designed such that the output oligonucleotide is complementary across its length to the 5′ and 3′ ends of the diagnostic strand, generating a structure that resembles the shape of a horseshoe (
Figure 1B). As with traditional molecular beacons, the diagnostic strand contains a large loop that is complementary to the trigger and serves as an internal toehold. Hybridization between the internal toehold and the trigger RNA acts as a thermodynamic driver that is intended to disrupt the pairing between the output strand and the diagnostic strand, resulting in the release of the single-stranded output.
Since the internal toehold of the diagnostic strand does not need to overlap with the 5′ and 3′ regions that are bound to the output oligonucleotide, essentially any set of trigger and target sequences can be implemented. The single-stranded output of the beacon switch could be composed of RNA or DNA depending on the desired function of the output strand. This conditional system could find application in instances where an irregular or diseased cellular state can be identified by a high copy number of a specific endogenous RNA, and the use of an AON, antagomir or other short single-stranded RNA would have a significant impact on rectifying the irregular state or inducing cell death.
For proof-of-principle illustration, a beacon switch was designed to respond to a fragment of the Kirsten rat sarcoma proto-oncogene (KRAS) mRNA as a trigger and release an RNA antagomir output strand in a conditional fashion. Analysis of beacon switch assembly and conditional output release was performed by non-denaturing polyacrylamide gel electrophoresis (PAGE) (
Figure 1C). Assembly between the diagnostic strand and output strand to form the beacon switch is extremely efficient based on non-denaturing PAGE and total nucleic acid staining, with only trace amounts of single-stranded output strand observed after assembly. Co-incubation of the assembled beacon switch with the KRAS trigger at 37 °C results in the release of the output strand and the appearance of a band corresponding to the expected waste product. A higher migrating band also appears in this lane, which is presumed to be a trinary molecular complex of the assembled beacon switch bound to the trigger. The amount of output strand observed to be released is likely a lower limit of the amount of single-stranded oligo that is actually newly accessible following interaction with the trigger RNA. This is because only one end of the single-stranded output needs to be released by the diagnostic strand to allow complete hybridization with the trigger oligonucleotide (
Figure S1). However, even a partially released single-stranded output oligo should be accessible to hybridize to a target RNA and still be able to perform its intended regulatory function.
3.3. Strand Exchange between RNA/DNA Hybrid Duplexes Can Be Facilitated by Toeholds That Target Adjacent Sequence Regions of an RNA Trigger
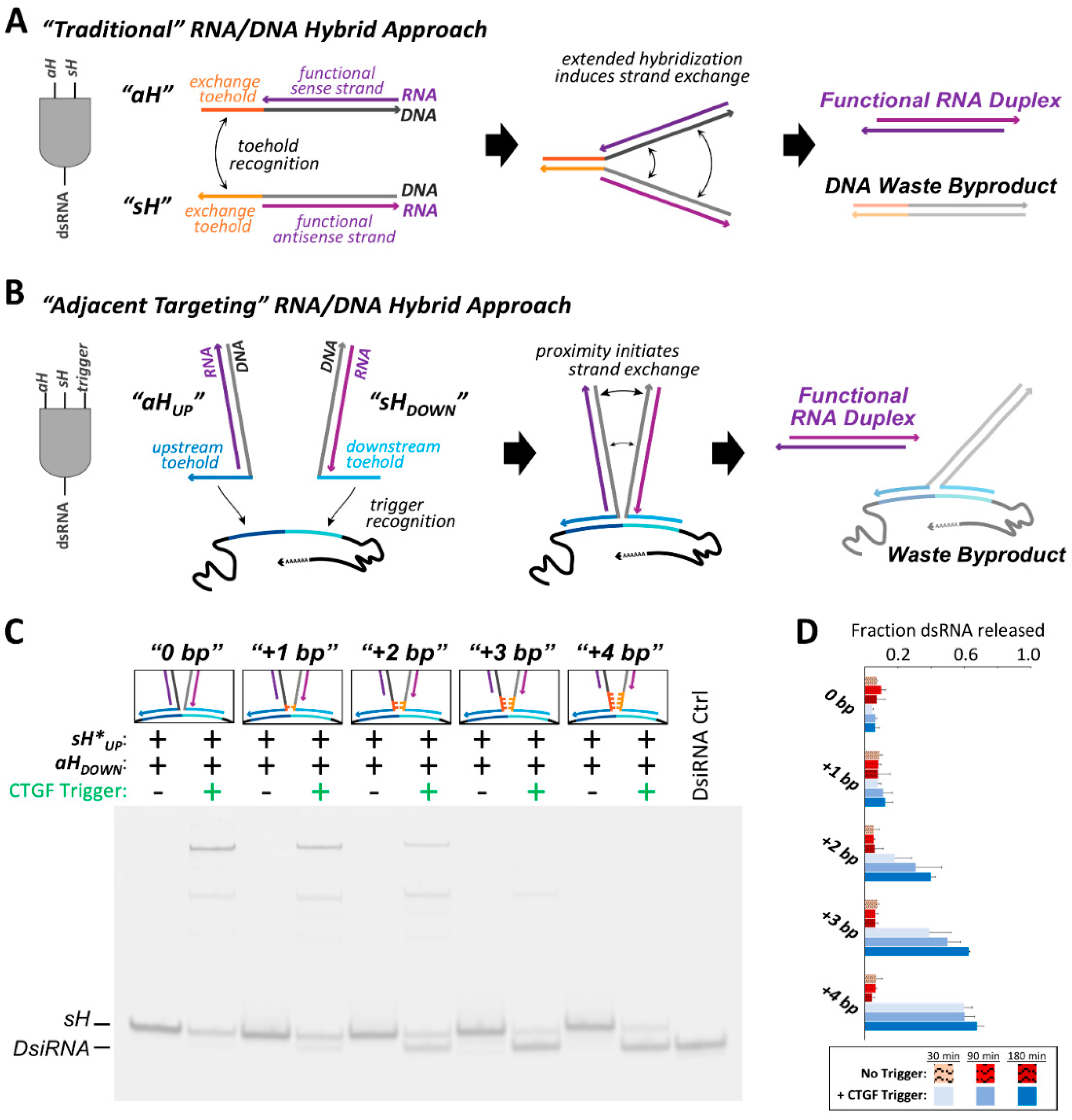
Cognate RNA/DNA hybrid pairs were previously designed that harbor split functional RNAs, devised to release a recombined functional dsRNA through recognition of complementary single stranded toeholds (
Figure 2A) [
31,
37,
38]. The separated single strands composing the functional duplex can be referred to as the sense strand and the antisense strand, and each of these RNA strands were annealed to a complementary DNA oligo. These assembled RNA/DNA hybrids are denoted as the sense hybrid (
sH) and the antisense hybrid (
aH), respectively. The “traditional” approach to cognate hybrid design utilized complementary single stranded toeholds emanating from
sH and
aH, with hybridization of these toeholds to one another initiating RNA/DNA strand exchange. Here, we have redesigned the toeholds to be complementary to adjacent regions of an RNA trigger sequence, rather than complementary to one another. As the toeholds can no longer drive strand exchange by hybridization to one another, release of the dsRNA product is conditional on the presence of the RNA trigger molecule.
In this “adjacent targeting” incarnation of the RNA/DNA hybrid system, a fragment of the connective tissue growth factor (CTGF) mRNA was used as the RNA trigger sequence, acting as a template for DNA toehold binding which in turn initiates strand exchange (
Figure 2B). Since the antisense hybrid binds upstream on the RNA trigger, it was termed
aHUP. Similarly, the sense hybrid is referred to as
sHDOWN. Binding of the cognate hybrid pair to the trigger RNA positions the two RNA/DNA hybrid regions adjacent to one another in space. The close proximity of the trigger-bound cognate hybrids will induce strand exchange through progressive hybridization of the trigger-bound DNA strands to one another, forming a three-way junction with the RNA trigger, and leading to formation and release of a dsRNA product. Like the beacon-derived switch, this activatable RNA/DNA hybrid system could find use in instances where a cell population of interest can be distinguished by the high relative expression level of an endogenous RNA. However, this RNA/DNA hybrid system (and those that follow) could be of use in cases where conditional generation of a double-stranded RNA is desirable, which could take the form of an RNA interference substrate, saRNA, aptamer, or another functionally relevant dsRNA. In this instance, the dsRNA product was designed as a 25/27-mer DsiRNA.
Formation of the dsRNA product was visualized by non-denaturing PAGE. The initial
aHUP/
sHDOWN cognate pair did not induce strand exchange and dsRNA release when co-incubated with the CTGF trigger for 180 min (
Figure 2C, “0 bp”). In the presence of the RNA trigger, a large fraction of the hybrid constructs appear to be stuck in an intermediate complex displaying slow electrophoretic mobility. Presumably, this observed band corresponds to a state in which both RNA/DNA hybrids are bound to the trigger through their respective toeholds, but strand exchange in not stimulated. Despite no observed dsRNA release from this system, the strand exchange reaction is predicted to be thermodynamically favored (
Figure S2). In an attempt to provide a greater driving force for strand exchange, additional sets of cognate hybrids pairs were designed in which additional complementary DNA nucleotides were inserted between the toehold region and the RNA/DNA hybrid region of each hybrid construct. These complementary nts were inserted to essentially serve as a nucleation site for strand exchange between the cognate partners once bound to the RNA trigger. In total, four additional hybrid pairs were designed which contained between 1 and 4 additional bps to seed the strand exchange (
Figure 2C).
Increasing the number of complementary DNA bps inserted immediately prior to the RNA/DNA hybrid regions resulted in increased DsiRNA release (
Figure 2C,D). Insertion of at least 2 DNA bps was needed to observe significant increases in DsiRNA release in the presence of the trigger RNA, as compared to background in the absence of the trigger after three hours (
Table S1). Insertion of 3 bps appears to be enough to achieve close to the maximal degree of product duplex release, as increasing to 4 inserted bps results in negligible further increases in DsiRNA release after 180 min. However, the gel electrophoresis experiments suggest that insertion of additional bps does seem to speed up the rate at which this plateau of apparent maximal possible product release is reached, as the +4 bp pair releases significantly more dsRNA after 30 min than the +3 bp system, and likewise the +3 bp system shows greater release than the +2 bp hybrid pair (
Figure 2D and
Table S2). Despite the +3 bp and +4 bp hybrid pairs eventually reaching a similar level of dsRNA release after three hours, their differences in the fraction of dsRNA released at early time points suggests that the initiation of strand exchange within the adjacent targeting system may be impeded by slow kinetics. Interestingly, despite these systems containing complementary DNA nts that could potentially serve as toeholds to promote strand exchange in the absence of the trigger RNA, increasing the number of inserted seed base pairs up to four did not result in significant differences in the degree of non-triggered dsRNA release when co-incubated over the longest duration examined (
Table 1 and
Table S1).
3.4. A Responsive Structural Element Can Act to Conditionally Induce Strand Exchange between RNA/DNA Hybrids
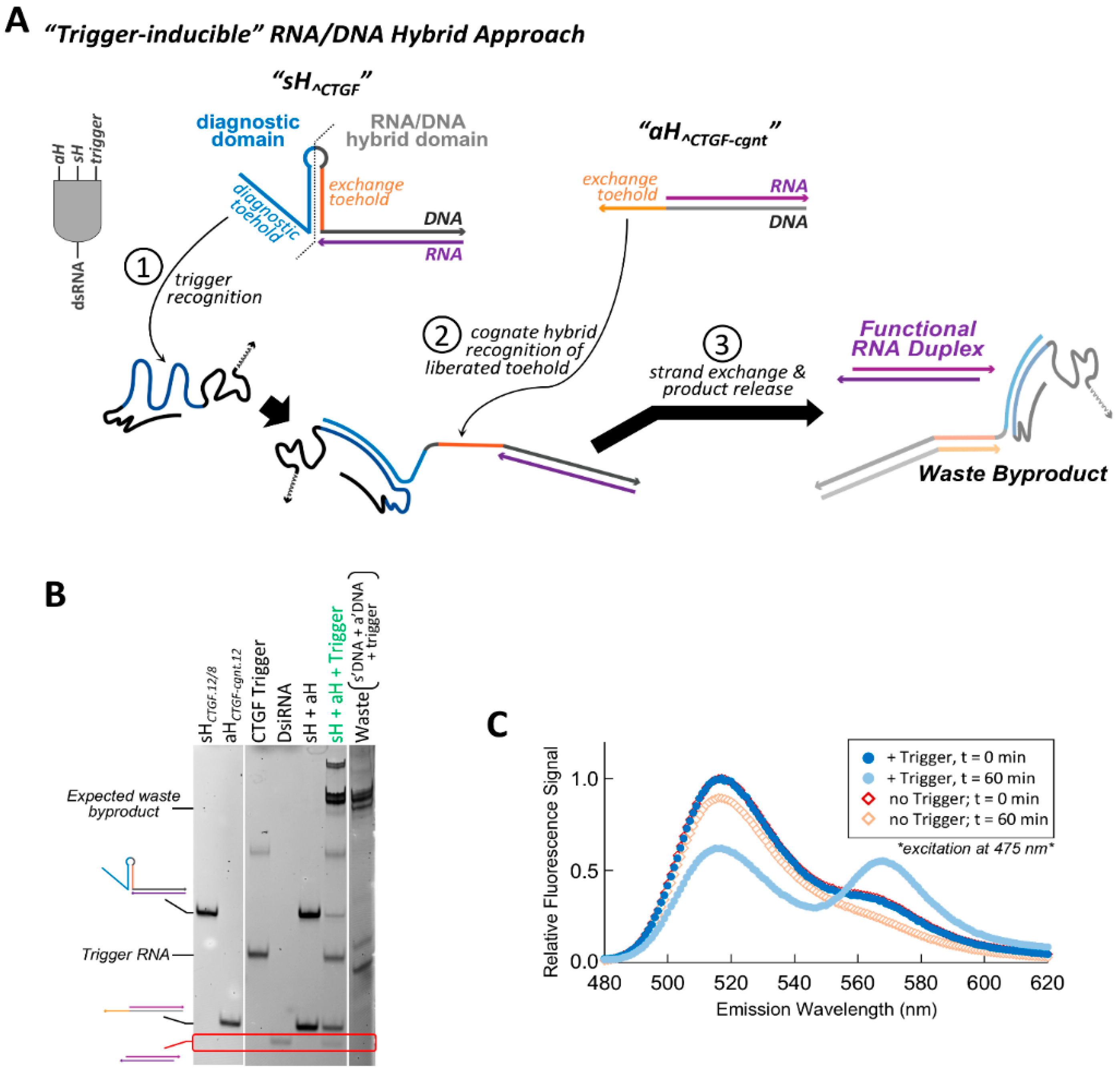
In an alternative approach for the implementation of conditional function within an RNA/DNA hybrid system, hybrid pairs were designed in which the accessibility of the toehold(s) needed to facilitate strand exchange was altered based on the presence or absence of a specific RNA trigger sequence. Although the adjacent targeting hybrid system described above performs its designed conditional function to release dsRNA, the fraction of dsRNA release for the best performing hybrid pair topped out at 0.67 after three hours. This second approach was pursued in an attempt to improve the efficiency of strand exchange and increase conditional dsRNA release. The “traditional” RNA/DNA hybrid methodology requiring the hybridization of complementary toeholds to one another for strand exchange serves as the basis of the conditional activation. We designate these single stranded toeholds as “exchange toeholds”, since they are required to initiate strand exchange. To create a hybrid system responsive to conditional activation, a structured hairpin element was incorporated in the DNA strand immediately adjacent to the RNA/DNA hybrid duplex region of the sense hybrid (
Figure 3A). This DNA hairpin ultimately controls the reassembly fate of the split functional RNA. In its initial folded state, the DNA hairpin is designed to sequester the entire length of the exchange toehold sequence within its helical stem, preventing the toehold from readily interacting with the complementary exchange toehold of the cognate antisense hybrid. The resulting hybrid pair initially exists in an “off” state that is unable to initiate strand exchange.
A new single stranded toehold, termed the “diagnostic toehold”, is then implemented as a means to control the conditional activation of the hybrid by altering the accessibility of the exchange toehold imbedded within the DNA hairpin upon recognition of a specific RNA trigger sequence (
Figure 3A). This single-stranded diagnostic toehold within the sense hybrid is positioned at the 5′ end of the DNA strand adjacent to the DNA hairpin (at the side opposite, the RNA/DNA hybrid region). By designing the sequence of the diagnostic toehold and the adjacent 5′ side of the DNA hairpin to be fully complementary to a region of an RNA trigger, hybridization of the trigger to the diagnostic toehold unzips the adjacent DNA hairpin and exposes the exchange toehold. Once the exchange toehold has been liberated, the complementary exchange toeholds of the hybrid pair can facilitate a strand exchange event and release a dsRNA output (
Figure 3A). It is intended that this method of exchange toehold recognition, whereby the hybridization of complementary toeholds to one another forms a single duplex that can be directly extended by stacking additional DNA bps formed during RNA/DNA hybrid strand exchange, will exert a greater kinetic and/or thermodynamic drive than the three-way junction dependent method employed within the adjacent targeting system.
To illustrate the function of this “trigger-inducible” hybrid system, conditional hybrid constructs were designed to release a 25/27-mer DsiRNA when triggered by a fragment of the CTGF mRNA. The DNA strand of the sense hybrid was designed to contain a central hairpin with a 12 bp stem and 8 nt loop. This sense hybrid is referred to as sH^CTGF.12/8, as the hybrid is designed to stimulate dsRNA release in the presence of CTGF (“^CTGF”) and contains a DNA hairpin composed of a 12 bp stem and 8 nt loop (“12/8”). The exchange toehold within sH^CTGF.12/8 is 12 nt in length and is initially completely sequestered within the DNA hairpin stem. The cognate partner hybrid is composed of an RNA/DNA hybrid duplex containing the DsiRNA antisense strand, with a 12 nt extension of the DNA strand at its 3′ end to encode the complementary exchange toehold. This hybrid is referred to as aH^CTGF-cgnt.12 to reflect that it contains a 12 nt exchange toehold (“12”) and is the cognate partner (“cgnt”) to the CTGF-triggered sH hybrid (“^CTGF”).
Non-denaturing PAGE and total nucleic acid staining was used to examine interactions occurring between the cognate hybrids, as well as between the hybrids and the trigger RNA (
Figure 3B). While not quantitative, initial analysis using a nucleic acid stain allowed for surveillance of all molecular species and products. As expected, no changes to the hybrids’ electrophoretic mobility is observed when incubated together at 37 °C in the absence of the trigger RNA, indicating that no interaction occurs between the hybrids and no dsRNA is released. Introduction of the RNA trigger activates
sH^CTGF.12/8 and induces the release of a dsRNA product when
aH^CTGF-cognt12 is also present. Higher migrating species are also observed when both hybrids are co-incubated with the trigger RNA. One of the high migrating bands corresponds to the expected waste product as indicated by similar migration of a control assembled from the RNA trigger and two DNA strands. An even slower migrating band is also observed and is likely to be a 5-molecule intermediate complex. Förster resonance energy transfer (FRET) experiments were performed to further verify the generation of the expected double stranded RNA product in the presence of the trigger molecule (
Figure 3C). The cognate hybrids used for these FRET studies had a 3′ donor fluorophore on the RNA sense strand, and a 5′ acceptor fluorophore on the RNA antisense strand. In the absence of the RNA trigger, the FRET-labeled hybrid pair show no significant change in their emission spectrum after one hour at 37 °C. However, one hour after the introduction of the CTGF trigger a large decrease in donor emission (~515 nm) and increase in acceptor fluorescence (~565 nm) is observed, indicating formation of the DsiRNA duplex product.
3.5. Alteration of Structural Elements Was Explored as a Means to Optimize dsRNA Release from Cognate RNA/DNA Hybrids
Within the inducible hybrid system, the accessibility of one exchange toehold is impeded by being sequestered within a responsive DNA hairpin. This toehold becomes liberated upon opening of the hairpin in the presence of an RNA trigger and allows for a strand exchange to proceed. As such, altering the stability of this responsive hairpin structure, as well as the length and accessibility of the liberated exchange toehold once the hairpin is open, could potentially modulate the degree of strand exchange between a cognate hybrid pair. The initially characterized
sH^CTGF.12/8 hybrid contained a 12 bp DNA hairpin stem capped by an 8 nt loop. Three additional CTGF-triggered
sH hybrids were designed to investigate how changing the structure of the responsive DNA hairpin affects strand exchange and dsRNA release (
Figure 4A). The first of these variants maintains a 12 bp DNA hairpin stem but expands the hairpin loop from 8 to 12 nts. This hybrid is denoted sH
^CTGF.12/12. The two additional
sH variants maintain the original 8 nt hairpin loop, but contain hairpin stems of 16 and 20 bps in length. These hybrids are named
sH^CTGF.16/8 and
sH^CTGF.20/8, respectively.
Each of the four
sH^CTGF constructs were assembled with a fluorescently labeled RNA sense strand to quantitatively examine their ability to liberate a dsRNA duplex following strand exchange with
aH^CTGF-cgnt.12 in the presence and absence of the CTGF trigger RNA (
Figure 4B). Interestingly, analysis using fluorescently labeled constructs revealed that the various
sH^CTGF /
aH^CTGF-cgnt.12 hybrid pairs release a small fraction of dsRNA when incubated together in the absence of the trigger RNA. This was not originally observed in the initial qualitative experiments that utilized staining with ethidium bromide. The degree of non-triggered release among pairs of hybrid constructs was relatively minor after 30 min (~2–5% of signal) and was observed to marginally increase over time for each variant
sH^CTGF construct paired with
aH^CTGF-cgnt.12 (
Figure 4C).
sH^CTGF.20/8, which is predicted to contain the most stable hairpin stem (
Figure S3), exhibited the smallest degree of non-triggered DsiRNA release compared to other hybrids pairs after 30 min. Likewise, s
H^CTGF.12/12 was predicted to have the weakest hairpin structure and displayed the greatest extent of non-triggered DsiRNA release after 30 min. This trend persists at longer time points; however, differences in non-triggered DsiRNA release among variant hybrids pairs were not all statistically significant, especially at longer time points (
Table S3).
Structural changes to the responsive DNA hairpin of the
sH^CTGF hybrids resulted in negligible differences in trigger-induced dsRNA release between the four
sH^CTGF/
aH^CTGF-cgnt.12 pairs assayed (
Figure 4C). However, these constructs did show a 12–18% improvement in conditional dsRNA release over the best performing adjacent targeting hybrid pair after three-hour incubations with the CTGF trigger (
Table 1). The lack of differences in triggered DsiRNA release among the variant
sH^CTGF constructs was somewhat surprising based on the predicted change in free energy (ΔΔG) between the unbound and CTGF trigger-bound states for each hybrid’s responsive DNA element (
Figure S3). However, it may be that the favorable change in free energy for each construct upon trigger binding is so great (ΔΔG < −25 kcal mol
−1 for each) that the comparatively small differences in ΔΔG between the various
sH^CTGF hybrids becomes inconsequential. Alternatively, differences in the ΔΔG of trigger binding could be offset by differences in steric accessibility of the newly liberated exchange toehold once the DNA hairpin has opened. Increasing the loop size or length of the hairpin stem increases the distance between the exchange toehold and the region bound by the RNA trigger once hybridized (
Figure S3). This could in turn alter the accessibility of the liberated exchange toehold to the incoming cognate hybrid. The
sH^CTGF.12/8/
aH^CTGF-cgnt.12 hybrid pair has the shortest nucleotide distance between the region bound by the trigger and its exchange toehold, and time course FRET experiments indicate the observed rate constant of dsRNA release is slower for this hybrid pairing than for any of the other three
sH^CTGF hybrids paired with
aH^CTGF-cgnt.12 (
Figure S4).
Extending the length of the exchange toehold was also explored as a means to boost triggered dsRNA release within the CTGF-inducible hybrid system. A variant
aH^CTGF-cgnt hybrid was designed containing a 16 nt toehold and was termed
aH^CTGF-cgnt.16. The toehold of
aH^CTGF-cgnt.16 was designed to encode the same 12 nt sequence as the
aH^CTGF-cgnt.12 toehold, with four additional nucleotides appended to the toehold’s distal end. These four additional nucleotides are complementary to corresponding regions within
sH^CTGF.16/8 and
sH^CTGF.20/8 and result in complete pairing of the 16 nt exchange toeholds between these cognate hybrids. However, these four added nucleotides at the distal end of the
aH^CTGF-cgnt.16 toehold do not have complementary sequences in
sH^CTGF.12/8 and
sH^CTGF.12/12 (
Figure 4A), leaving the distal end of the
aH^CTGF-cgnt.16 toehold unpaired.
Increasing the toehold length of the cognate antisense hybrid from 12 to 16 nucleotides was observed to have a negative impact on DsiRNA release when paired with any of the
sH^CTGF variants (
Figure 4D,E). The use of
aH^CTGF-cgnt.16 in place of
aH^CTGF-cgnt.12 had a negligible effect on the degree of non-triggered release but presented a large significant impediment to CTGF-triggered release in nearly all instances (
Table S4). The extent of diminished triggered-release was most pronounced when
aH^CTGF-cgnt.16 was paired with
sH^CTGF.12/8 and
sH^CTGF.12/12, suggesting that having non-complementary nucleotides at the distal end of the
aH^CTGF-cgnt.16 toehold interferes in some manner with the ability of the hybrids to promote the strand exchange reaction. As a way to compare the overall performance of the each conditionally-active hybrid pair, an “efficiency score” was determined for each time point examined. This efficiency score metric was calculated as the product of the fraction of triggered dsRNA release and the signal-to-noise ratio (triggered/non-triggered release). Larger scores indicated greater efficiency of conditional dsRNA release. Out of the eight pairs of CTGF-inducible hybrids and the five sets of adjacent-targeting hybrids, the
sH^CTGF.20/8/
aH^CTGF-cgnt.12 pairing displays the highest efficiency score for each time interval that was examined (
Table 1).
3.6. Redesigned Responsive Structural Elements Can Be Used to Inhibit Strand Exchange and Repress dsRNA Release
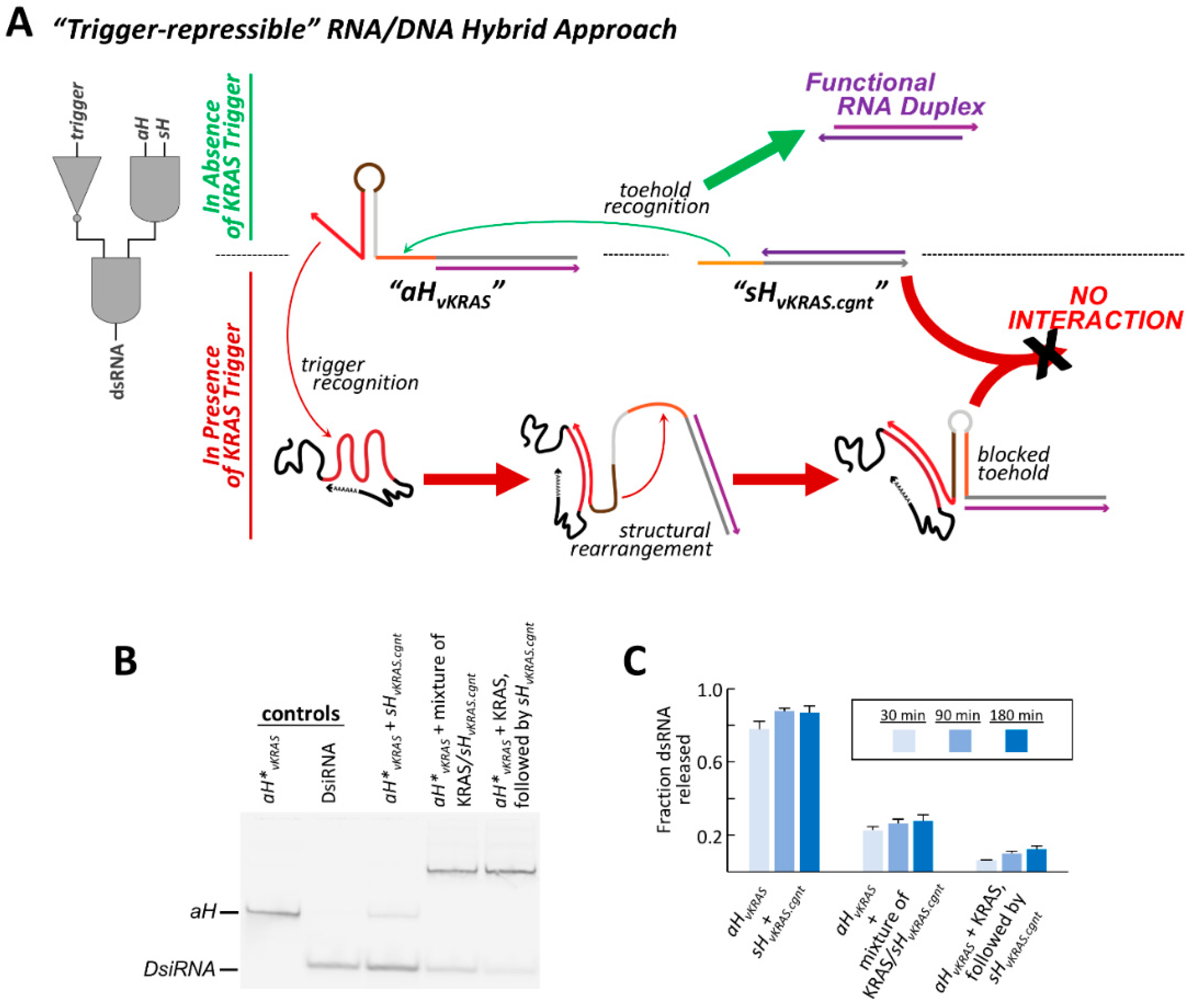
The concept and method of toehold sequestration used to impart conditional function within the trigger-inducible RNA/DNA hybrid system can be modified and redesigned to instead allow for the repression of strand exchange in the presence of a specific RNA trigger and thereby expands the degree of control over dsRNA release. In this embodiment, both exchange toeholds are initially free to undergo strand exchange, but one becomes sequestered into a DNA hairpin when interaction with an RNA trigger facilitates a structural rearrangement of that hybrid’s responsive structural element (
Figure 5A). Such a system would be of interest in situations where a cellular state of interest cannot be identified by the high expression of a particular RNA, but rather by a significant under expression of a specific RNA relative to the normal population.
Whereas the previously described inducible hybrid pairs contain a responsive hairpin element within the DNA strand of
sH that is triggered by CTGF, the repressible hybrid pair contains a responsive DNA element within
aH that is responsive to the KRAS mRNA-derived trigger. This new hybrid is termed “
aH∨
KRAS” to indicate that dsRNA release from the hybrid is negatively impacted by the KRAS trigger. In the absence of the cognate RNA trigger, the most stable DNA fold of
aH∨
KRAS is that which results in a single stranded exchange toehold and a 14 bp DNA hairpin (
Figure S5). When the trigger is present, however, it can bind to the 3′ diagnostic toehold present in
aH∨
KRAS and proceed to unzip the 14 bp hairpin, as the trigger is complementary to the entire 3′ side of the hairpin stem. As the initial 14 bp hairpin can no longer form, a structural rearrangement can occur where the exchange toehold pairs to the 12 nts that compose the apical loop of the original hairpin. This new hairpin structure makes the exchange toehold inaccessible to the cognate hybrid and represses the ability for the hybrid pair to release a dsRNA duplex (
Figure 5A).
The ability to repress hybrid strand exchange was examined for
aH∨
KRAS with its cognate hybrid, “
sH∨
KRAS-cgnt”, that contains a complementary 12 nt DNA exchange toehold extending from its RNA/DNA hybrid region. Analysis by non-denaturing PAGE at several time points illustrates that the cognate hybrids successfully undergo strand exchange and release dsRNA in the absence of the KRAS trigger (
Figure 5B,C). However, when the KRAS trigger and
sH∨
KRAS-cgnt hybrid are premixed and added simultaneously to
aH∨
KRAS, DsiRNA release is repressed more than 3-fold compared to in the absence of KRAS. A second context was also examined, where
aH∨
KRAS was permitted to interact with the KRAS trigger for five minutes prior to the addition of the cognate
sH∨
KRAS-cgnt hybrid. This scenario allowed the responsive DNA hairpin to rearrange and adopt its alternative “off”-state structure before the cognate exchange toehold was present in the reaction mix. In this context, DsiRNA release is reduced 12-fold after 30 min at 37 °C compared to in the absence of trigger, and maintains more than 7-fold repression after 3 h (
Figure 5C). FRET experiments further illustrate that strand exchange occurs quickly and efficiently in the absence of the KRAS trigger, but is severely impeded upon introduction of KRAS (
Figure S6).
To illustrate that repression of dsRNA release is dependent on the presence of a trigger RNA with a specific nucleotide sequence, additional non-cognate trigger molecules were co-incubated with the repressive hybrid pair. Neither of the non-cognate trigger molecules tested resulted in a reduction in dsRNA release (
Figure S7). This same degree of trigger specificity is observed for the CTGF-inducible hybrid system, as the
sH^CTGF.20/8/
aH^CTGF-cgnt.12 hybrid pair are only observed to initiate dsRNA release in the presence of the CTGF trigger, and not when co-incubated with non-cognate trigger molecules (
Figure S7). An orthogonal trigger-repressible system was also designed that is responsive to CTGF rather than KRAS, as a means to demonstrate versatility in accommodating various trigger sequence inputs, as well as an ability to position the response element at different locations within this generalized conditional system. In this system, the CTGF responsive DNA element was added to the sense hybrid rather than the antisense hybrid. Nonetheless, this cognate hybrid pair (
sH∨
CTGF/
aH∨
CTGF-cgnt) displays a repressed ability to generate dsRNA in the presence of the CTGF trigger, as intended (
Figure S8).
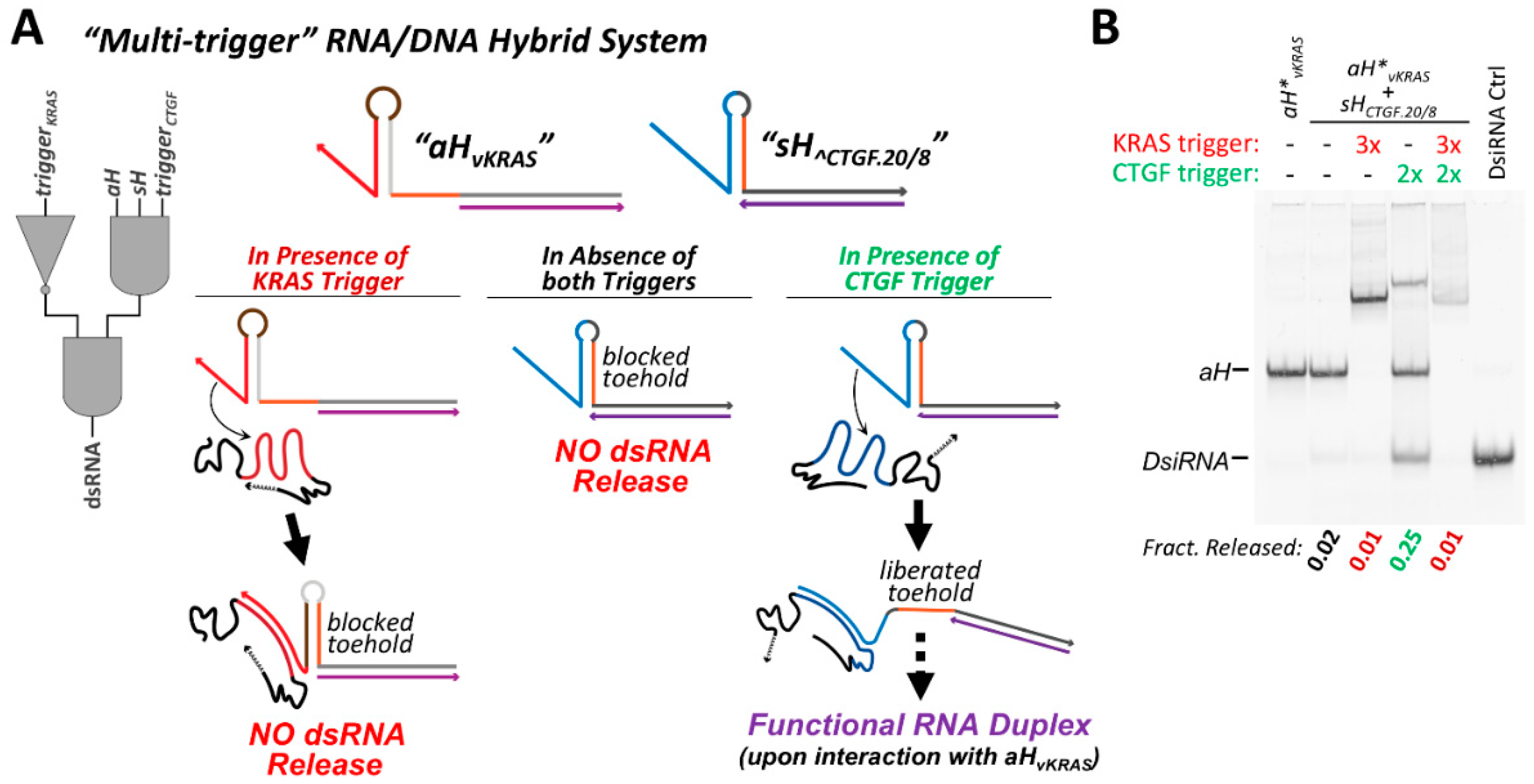
3.7. Cognate Hybrids Pairs with Multiple Responsive Elements Allow for Multi-Trigger Regulation
Because the strand exchange reaction between cognate hybrid partners is dependent on the accessibility of a specific toehold sequence (exchange toehold) present on each of the two hybrids, it is possible to generate a system in which the accessibility of each toehold is under the control of a different RNA trigger sequence. In the case of the trigger-repressible hybrids, such as aH∨KRAS, the trigger RNA imparts no sequence constraints on the exchange toehold and allows the exchange toehold to be any sequence that permits proper folding. With this in mind, the exchange toehold of construct aH∨KRAS was designed to be complementary to the exchange toehold of the sH^CTGF hybrids characterized previously.
Hybrid construct
sH^CTGF.20/8 was partnered with
aH∨
KRAS to generate a pair of conditional RNA/DNA hybrids whose function is dependent on the presence or absence of two RNA triggers, CTGF and KRAS (
Figure 6A). The strand exchange reaction between these two hybrids is initially inhibited, as
sH^CTGF.20/8 initially exists in an “off” state and requires interaction with the CTGF trigger to promote strand exchange.
aH∨
KRAS is initially in an active state; however, the exchange toehold of
aH∨
KRAS becomes inaccessible upon interaction with the KRAS trigger. For efficient strand exchange to occur between this hybrid pair, the presence of the CTGF trigger is required, as well as the absence of the KRAS trigger.
The degree to which dsRNA could be conditionally released from this cognate hybrid pair was assessed by non-denaturing PAGE (
Figure 6B) and FRET (
Figure S9). In the absence of any trigger molecules, the
sH^CTGF.20/8/
aH∨
KRAS hybrid pair releases very small amounts of dsRNA when co-incubated. The addition of the KRAS trigger to the hybrid pair reduces the degree of dsRNA release close to zero. However, if the CTGF trigger is added rather than the KRAS trigger, substantial release of dsRNA product occurs, as expected. Sequential addition of the KRAS trigger followed by the CTGF trigger to the hybrid pair results in very little dsRNA generation, suggesting that
aH∨
KRAS inactivation by the KRAS trigger occurs relatively quickly. Additional characterization was performed to examine how differences in the relative concentration of the two triggers affect dsRNA release. Various ratios of CTGF and KRAS trigger molecules were premixed and added to the co-incubating
sH^CTGF.20/8/
aH∨
KRAS hybrid pair. As might be expected, increasing the relative amount of CTGF trigger (activating) to KRAS trigger (deactivating) increases the extent of dsRNA release (
Figure S10). When equal amounts of the KRAS and CTGF triggers are added to the hybrid pair, the degree of dsRNA release is about 60% of the maximum amount of dsRNA released when an excess of CTGF trigger is added to the hybrids in the absence of the KRAS trigger. However, when the ratio of CTGF/KRAS triggers is varied away from 1:1, induction/repression of dsRNA release disproportionately favors the trigger that is present in a greater amount, beyond what would be predicted based on the trigger stoichiometry (i.e.,: when a 3:2 ratio of KRAS/CTGF is present, the fraction of dsRNA is less than 40% of the maximal dsRNA released in the absence of any KRAS).
3.8. Three-Strand RNA/DNA Hybrid Constructs Allow “Activated” Hybrids to Dissociate from Their Cognate Trigger
With both of the inducible and repressible conditional systems described above, the entirety of the hybrid construct containing the diagnostic toehold remains bound to the RNA trigger molecule following recognition and hybridization of the diagnostic toehold. However, one can imagine that there may be instances where the function of the conditional hybrid systems may benefit from allowing their RNA/DNA hybrid domains to freely diffuse away from their cognate trigger following hybridization through their diagnostic toehold/domain. A three-strand design approach was used to create an inducible hybrid that separates from the RNA trigger after hybridization. The design is based on that of the
sH^CTGF.20/8 hybrid. The 8 nt hairpin loop is removed, splitting the 20 bp hairpin into a duplex that assembles from two distinct DNA strands. One DNA strand retained the 5′ diagnostic toehold, while the other maintained the RNA/DNA hybrid region (
Figure S11). This new three-strand hybrid was termed “
sH^CTGF.20split” and works in conjunction with
aH^CTGF-cgnt.12. Analysis by non-denaturing PAGE illustrates that the three-piece hybrid
sH^CTGF.20split appears to function very similarly to that of
sH^CTGF.20/8, although the three-piece hybrid seems to have a slight increase in its degree of non-triggered dsRNA release (
Figure S11).
A similar approach was used to investigate a three-strand repressible hybrid construct based on the
aH∨
KRAS hybrid. A nick was positioned within the 5′ strand of the DNA hairpin, aiming to maintain stable formation of the initial 14 bp hairpin and allow strand exchange in the absence of the KRAS trigger. Four different variants were designed to identify a nick position that retained the greatest conditional function. The function of the four variants partnered with
sH∨
KRAS-cgnt was examined by non-denaturing PAGE (
Figure S12). Each of the three-strand repressible systems tested show a diminished ability to promote desirable dsRNA release in the absence of the KRAS trigger compared to the original design. This may stem from the possibility that a larger fraction of the three-strand hybrids initially adopt their “off” state when assembled. However, some three-strand systems did retain their repressible function. “
aH∨
KRAS.nick14”, where the nick was placed immediately below the hairpin loop and preserves the entire 14bp stem, displayed the greatest degree of conditional function. Progressively moving the nick down the stem resulted in continued loss of the responsive function to the KRAS trigger.