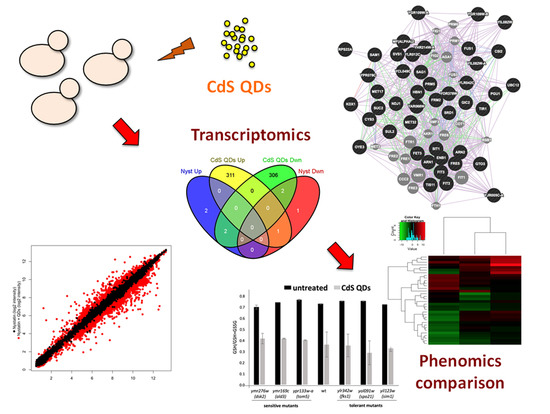
In Vivo-In Vitro Comparative Toxicology of Cadmium Sulphide Quantum Dots in the Model Organism Saccharomyces cerevisiae
Abstract
1. Introduction
2. Materials and Methods
2.1. Cadmium Sulfide Quantum Dots (CdS QDs) Characterization
2.2. Strains Used
2.3. Microarray Experiments
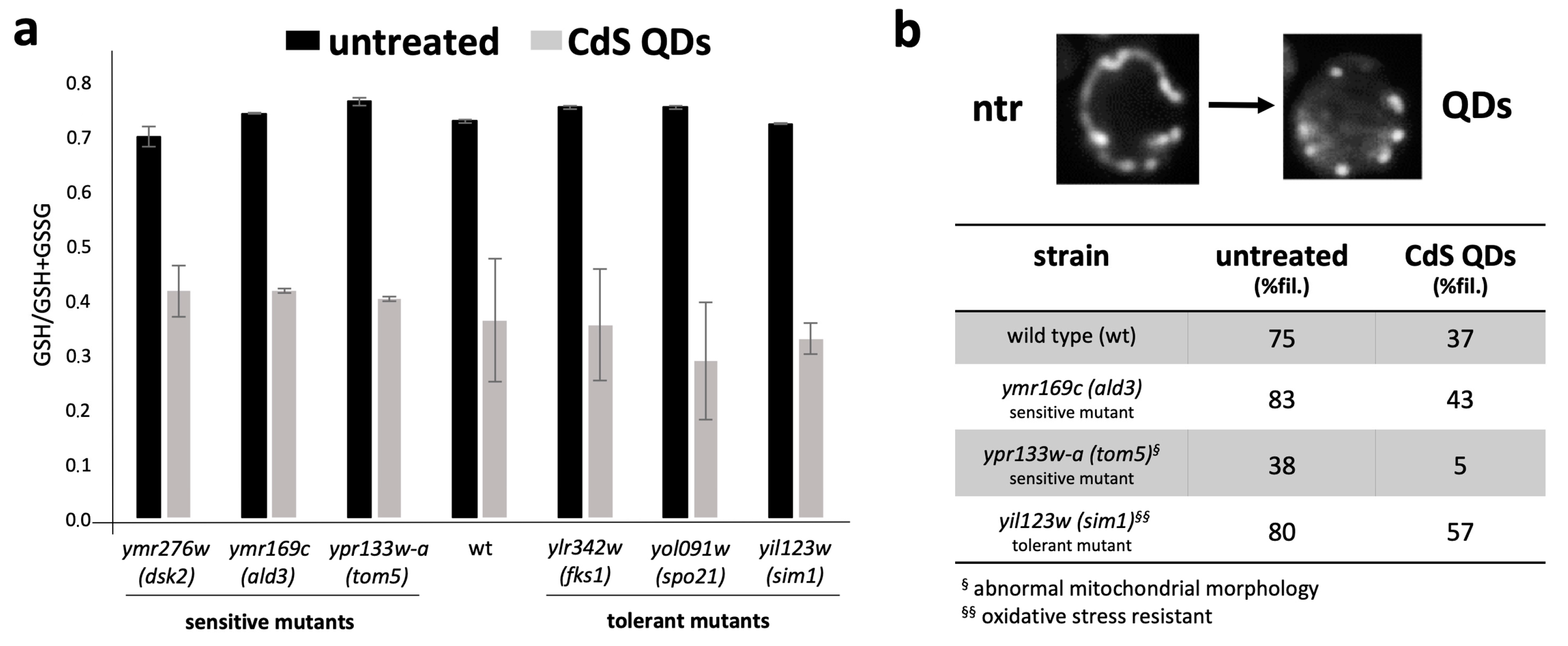
2.4. Determination of Glutathione Redox State and Mitochondrial Morphotype
2.5. Statistical Analysis
3. Results
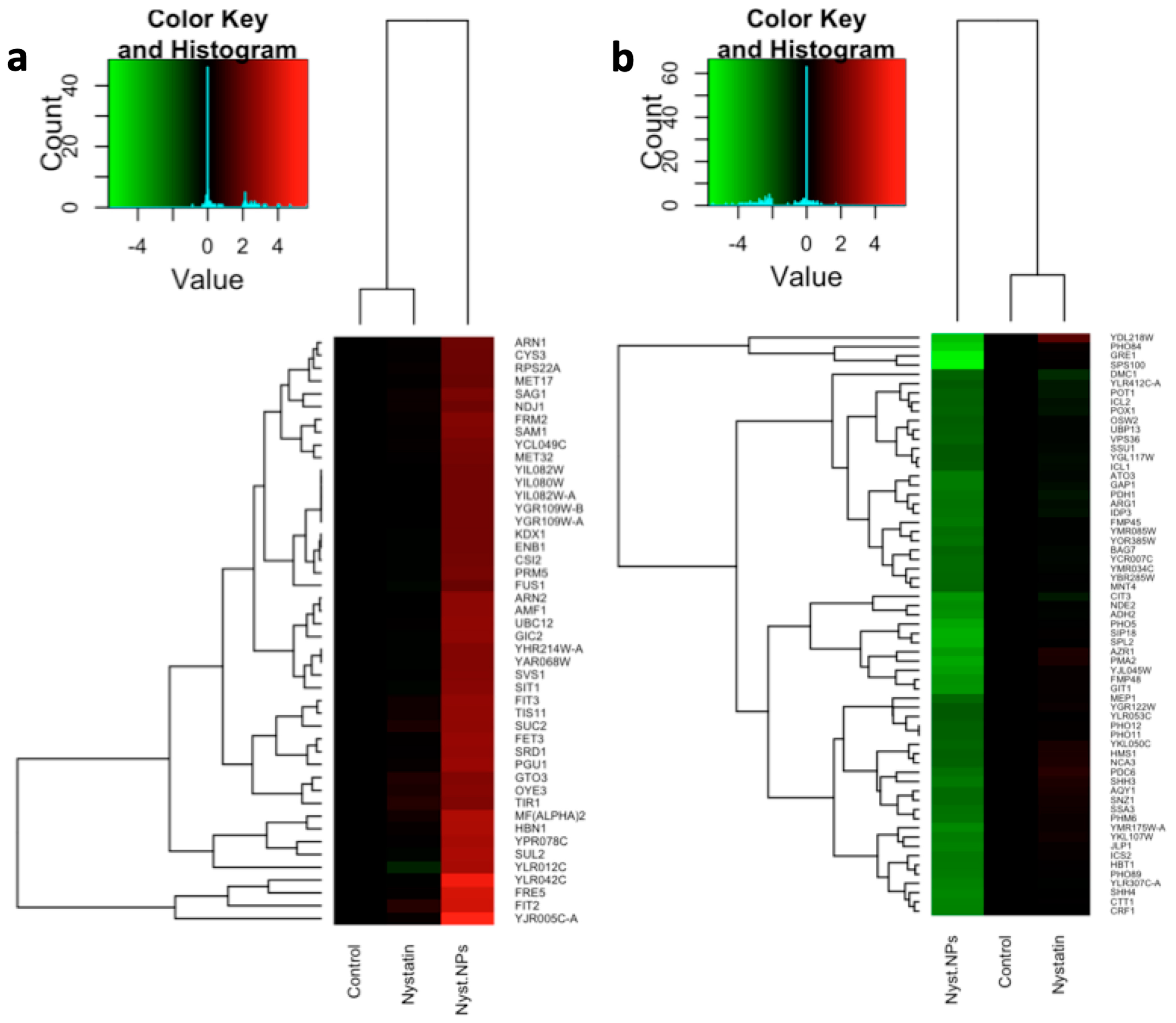
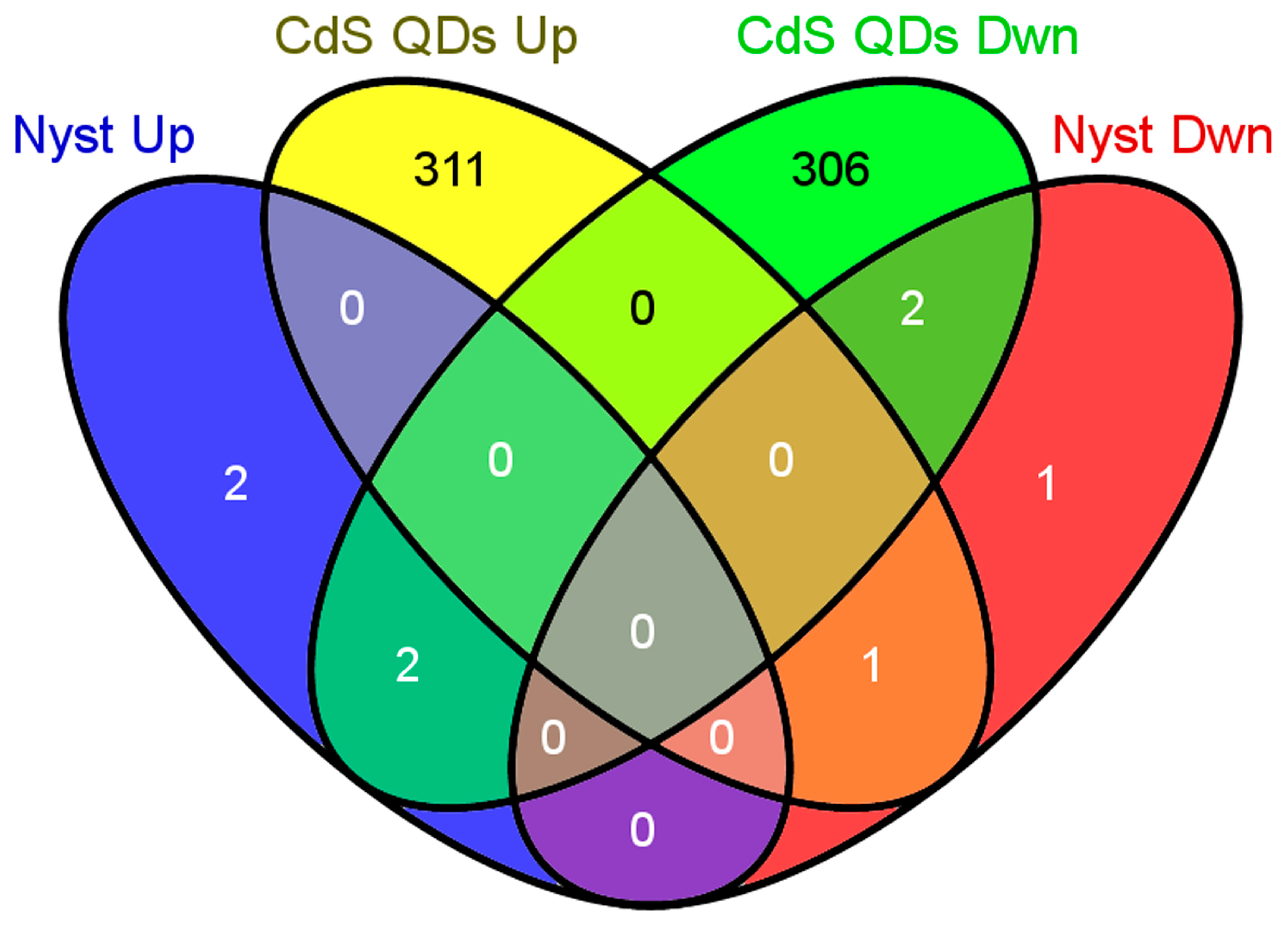
3.1. Microarray Data Analysis
3.2. Gene Ontology
3.3. Comparison of Two Yeast Inhibitors: Nystatin vs. CdS QDs
3.4. Comparison with Cadmium Ion Response
3.5. Comparison between Phenomics and Transcriptomics Data
3.6. Confirmation of Growth Characteristics in a Pool of Selected Mutants
3.7. Physiological and Molecular Tests on a Pool of Selected Tolerant and Sensitive Mutants
4. Discussion
4.1. Sub-Inhibitory Concentration of Nystatin Did Not Influence Yeast Response to CdS QDs
4.2. Cadmium Ion Response Comparison
4.3. Biological Function of the Genes Involved in the Response of Yeast to CdS QDs
4.4. Comparison between Phenomic and Transcriptomic Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stark, W.J. Nanoparticles in biological systems. Angew. Chem. Int. Ed. 2011, 50, 1242–1258. [Google Scholar] [CrossRef]
- Inshakova, E.; Inshakov, O. World market for nanomaterials: Structure and trends. MATEC Web Confer. 2017, 129, 02013. [Google Scholar] [CrossRef]
- Choi, J.Y.; Ramachandran, G.; Kandlikar, M. The impact of toxicity testing costs on nanomaterial regulation. Environ. Sci. Technol. 2009, 43, 3030–3034. [Google Scholar] [CrossRef]
- Caballero-Guzman, A.; Nowack, B. A critical review of engineered nanomaterial release data: Are current data useful for material flow modeling? Environ. Pollut. 2016, 213, 502–517. [Google Scholar] [CrossRef]
- Iavicoli, I.; Leso, V.; Schulte, P.A. Biomarkers of susceptibility: State of the art and implications for occupational exposure to engineered nanomaterials. Toxicol. Appl. Pharmacol. 2016, 299, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Avellan, A.; Simonin, M.; McGivney, E.; Bossa, N.; Spielman-Sun, E.; Rocca, J.D.; Bernhardt, E.S.; Geitner, N.K.; Unrine, J.M.; Wiesner, M.R.; et al. Gold nanoparticle biodissolution by a freshwater macrophyte and its associated microbiome. Nat. Nanotech. 2018, 13, 1072–1077. [Google Scholar] [CrossRef] [PubMed]
- Gardea-Torresdey, J.L.; Rico, C.M.; White, J.C. Trophic Transfer, Transformation, and Impact of Engineered Nanomaterials in Terrestrial Environments. Environ. Sci. Technol. 2014, 48, 2526–2540. [Google Scholar] [CrossRef]
- Kahru, A.; Dubourguier, H.C. From ecotoxicology to nanoecotoxicology. Toxicology 2010, 269, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Petersen, E.J.; Henry, T.B.; Zhao, J.; MacCuspie, R.I.; Kirschling, T.L.; Dobrovolskaia, M.A.; Hackley, V.; Xing, B.; White, J.C. Identification and avoidance of potential artifacts and misinterpretations in nanomaterial ecotoxicity measurements. Environ. Sci. Technol. 2014, 48, 4226–4246. [Google Scholar] [CrossRef]
- Krewski, D.; Acosta, D.; Andersen, M.; Anderson, H.; Bailar, J.C.; Boekelheide, K.; Brent, R.; Charnley, G.; Cheung, V.G.; Green, S.; et al. Toxicity testing in the 21st century: A vision and a strategy. J. Toxicol. Environ. Health B Crit. Rev. 2010, 13, 51–138. [Google Scholar] [CrossRef]
- Favero, P.P.; de Souza-Parise, M.; Fernandez, J.L.R.; Miotto, R.; Ferraz, A.C. Surface properties of CdS nanoparticles. Br. J. Phys. 2006, 36, 1032–1034. [Google Scholar] [CrossRef]
- Zhai, T.; Fang, X.; Li, L.; Bando, Y.; Golberg, D. One-dimensional CdS nanostructures: Synthesis, properties, and applications. Nanoscale 2010, 2, 168–187. [Google Scholar] [CrossRef] [PubMed]
- Winnik, F.M.; Maysinger, D. Quantum dot cytotoxicity and ways to reduce it. Acc. Chem. Res. 2013, 46, 672–680. [Google Scholar] [CrossRef]
- Duina, A.A.; Miller, M.E.; Keeney, J.B. Budding yeast for budding geneticists: A primer on the Saccharomyces cerevisiae model system. Genetics 2014, 197, 33–48. [Google Scholar] [CrossRef]
- Dos Santos, S.C.; Saá-Correia, I. Yeast toxicogenomics: Lessons from a eukaryotic cell model and cell factory. Curr. Opin. Biotechnol. 2015, 33, 183–191. [Google Scholar] [CrossRef]
- Marmiroli, M.; Pagano, L.; Pasquali, F.; Zappettini, A.; Tosato, V.; Bruschi, C.V.; Marmiroli, N. A genome-wide nanotoxicology screen of Saccharomyces cerevisiae mutants reveals the basis for cadmium sulphide quantum dot tolerance and sensitivity. Nanotoxicology 2016, 10, 84–93. [Google Scholar]
- Bao, S. Assessment of the toxicity of CuO nanoparticles by using Saccharomyces cerevisiae mutants with multiple genes deleted. Appl. Environ. Microbiol. 2015, 81, 8098–8107. [Google Scholar] [CrossRef] [PubMed]
- Suppi, S.; Kasemets, K.; Ivask, A.; Künnis-Beres, K.; Sihtmäe, M.; Kurvet, I.; Aruoja, V.; Kahru, A. A novel method for comparison of biocidal properties of nanomaterials to bacteria, yeasts and algae. J. Haz. Mater. 2015, 286, 75–84. [Google Scholar] [CrossRef]
- Oh, E.; Liu, R.; Nel, A.; Gemill, K.B.; Bilal, M.; Cohen, Y.; Medintz, I.L. Meta- analysis of cellular toxicity for cadmium-containing quantum dots. Nat. Nanotechnol. 2016, 11, 479–486. [Google Scholar] [CrossRef]
- Pasquali, F.; Agrimonti, C.; Pagano, L.; Zappettini, A.; Villani, M.; Marmiroli, M.; White, J.C.; Marmiroli, N. Nucleo-mitochondrial interaction of yeast in response to cadmium sulfide quantum dot exposure. J. Hazard. Mater. 2017, 324, 744–752. [Google Scholar] [CrossRef]
- Paesano, L.; Perotti, A.; Buschini, A.; Carubbi, C.; Marmiroli, M.; Maestri, E.; Iannotta, S.; Marmiroli, N. Markers for toxicity to HepG2 exposed to cadmium sulphide quantum dots; damage to mitochondria. Toxicology 2016, 374, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Kasemets, K.; Suppi, S.; Kunnis-Beres, K.; Kahru, A. Toxicity of CuO nanoparticles to yeast Saccharomyces cerevisiae BY4741 wild-type and its nine isogenic single-gene deletion mutants. Chem. Res. Toxicol. 2012, 26, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.R.; Boenzli, M.G.; Hindagolla, V.; Ding, J.; Miller, J.M.; Hutchison, J.E.; Greenwood, J.A.; Abeliovich, H.; Bakalinsky, A.T. Identification of gold nanoparticle-resistant mutants of Saccharomyces cerevisiae suggests a role for respiratory metabolism in mediating toxicity. Appl. Environ. Microbiol. 2013, 79, 728–733. [Google Scholar] [CrossRef] [PubMed]
- Nunnari, J.; Suomalainen, A. Mitochondria: In sickness and in health. Cell 2012, 148, 6, 1145–1159. [Google Scholar] [CrossRef]
- Laurent, J.M.; Young, J.H.; Kachroo, A.H.; Marcotte, E.M. Efforts to make and apply humanized yeast. Brief. Funct. Genom. 2016, 15, 155–163. [Google Scholar] [CrossRef]
- Marmiroli, M.; Pagano, L.; Savo Sardaro, M.L.; Villani, M.; Marmiroli, N. Genome-wide approach in Arabidopsis thaliana to assess the toxicity of cadmium sulfide quantum dots. Environ. Sci. Technol. 2014, 48, 5902–5909. [Google Scholar] [CrossRef]
- Villani, M.; Calestani, D.; Lazzarini, L.; Zanotti, L.; Mosca, R.; Zappettini, A. Extended functionality of ZnO nanotetrapods by solution-based coupling with CdS nanoparticles. J. Mater. Chem. 2012, 22, 5694–5699. [Google Scholar] [CrossRef]
- Pagano, L.; Pasquali, F.; Majumdar, S.; De La Torre-Roche, R.; Zuverza-Mena, N.; Villani, M.; Zappettini, A.; Marra, R.E.; Isch, S.M.; Marmiroli, M.; et al. Exposure of Cucurbita pepo to binary combinations of engineered nanomaterials: Physiological and molecular response. Environ. Sci.: Nano 2017, 4, 1579–1590. [Google Scholar] [CrossRef]
- Bilinski, C.A.; Marmiroli, N. Classical approaches to yeast strain selection. In Yeast Strain Selection; Marcel Dekker: New York, NY, USA, 1990; pp. 43–64. [Google Scholar]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols 2008, 4, 44–57. [Google Scholar] [CrossRef]
- Mostafavi, S.; Ray, D.; Warde-Farley, D.; Grouios, C.; Morris, Q. GeneMANIA: A real-time multiple association network integration algorithm for predicting gene function. Genom. Biol. 2008, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Philpott, C.C.; Protchenko, O.; Kim, Y.W.; Boretsky, Y.; Shakoury-Elizeh, M. The response to iron deprivation in Saccharomyces cerevisiae: Expression of siderophore-based systems of iron uptake. Biochem. Soc. Trans. 2002, 30, 698–702. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Iwai, Y.; Dancis, A.; Klausner, R.D. AFT1: A mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 1995, 14, 1231–1239. [Google Scholar] [CrossRef]
- Garay-Arroyo, A.; Covarrubias, A.A. Three genes whose expression is induced by stress in Saccharomyces cerevisiae. Yeast 1999, 15, 879–892. [Google Scholar] [CrossRef]
- Graziano, S.; Gullì, M.; Maestri, E.; Marmiroli, M. The global effect of exposing bakers’ yeast to 5-fluoruracil and nystatin; a view to Toxichip. Chemosphere 2016, 145, 470–479. [Google Scholar] [CrossRef]
- Bishop, D.K.; Park, D.; Xu, L.; Kleckner, N. DMC1: A meiosis-specific yeast homolog of, E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 1992, 69, 439–456. [Google Scholar] [CrossRef]
- Lutfiyya, L.L.; Johnston, M. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol. Cell. Biol. 1996, 16, 4790–4797. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.H.; Dunlap, P.E.; McBride, S.J.; Al-Refai, H.; Bushel, P.R.; Freedman, J.H. Global transcriptome and deletome profiles of yeast exposed to transition metals. PLoS Genet. 2008, 4, e1000053. [Google Scholar] [CrossRef]
- Cherest, H.; Davidian, J.C.; Thomas, D.; Benes, V.; Ansorge, W.; Surdin-Kerjan, Y. Molecular characterization of two high affinity sulfate transporters in Saccharomyces cerevisiae. Genetics 1997, 145, 627–635. [Google Scholar] [PubMed]
- Mouassite, M.; Guérin, M.G.; Camougrand, N.M. The SUN family of Saccharomyces cerevisiae: The double knock-out of UTH1 and SIM1 promotes defects in nucleus migration and increased drug sensitivity. FEMS Microbiol. Lett. 2000, 182, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Niazi, J.H.; Sang, B.-I.; Kim, Y.S.; Gu, M.B. Global Gene Response in Saccharomyces cerevisiae exposed to Silver Nanoparticles. Appl. Biochem. Biotechnol. 2011, 164, 1278–1291. [Google Scholar] [CrossRef]
- Pagano, L.; Maestri, E.; White, J.C.; Marmiroli, N.; Marmiroli, M. Quantum dots exposure in plants: Minimizing the adverse response. COESH 2018, 6, 71–76. [Google Scholar] [CrossRef]
- Pagano, L.; Maestri, E.; Caldara, M.; White, J.C.; Marmiroli, N.; Marmiroli, M. Engineered nanomaterial activity at the organelle level: Impacts on the chloroplasts and mitochondria. ACS Sustain. Chem. Eng. 2018, 6, 12562–12579. [Google Scholar] [CrossRef]
- Käosaar, S.; Kahrua, A.; Mantecca, P.; Kasemets, K. Profiling of the toxicity mechanisms of coated and uncoated silver nanoparticles to yeast Saccharomyces cerevisiae BY4741 using a set of its 9 single-gene deletion mutants defective in oxidative stress response, cell wall or membrane integrity and endocytosis. Toxicol. In Vitro 2016, 35, 149–162. [Google Scholar]
- Galván Márquez, I.; Ghiyasvand, M.; Massarsky, A.; Babu, M.; Samanfar, B.; Omidi, K.; Moon, T.W.; Smith, M.L.; Golshani, A. Zinc oxide and silver nanoparticles toxicity in the baker’s yeast Saccharomyces cerevisiae. PLoS ONE 2018, 13, e0193111. [Google Scholar]
- Kasemets, K.; Angela Ivask, A.; Dubourguier, H.C.; Kahru, A. Toxicity of nanoparticles of ZnO, CuO and TiO2 to yeast Saccharomyces cerevisiae. Toxicol. In Vitro 2009, 23, 1116–1122. [Google Scholar] [CrossRef]
- Piekarska, I.; Rytka, J.; Rempola, B. Regulation of sporulation in the yeast Saccharomyces cerevisiae. Acta Biochim. Pol. 2010, 57, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Horak, C.E.; Luscombe, N.M.; Qian, J.; Bertone, P.; Piccirrillo, S.; Mark Gerstein, M.; Snyder, M. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 2002, 16, 3017–3033. [Google Scholar] [CrossRef] [PubMed]
- Ruotolo, R.; Maestri, E.; Pagano, L.; Marmiroli, M.; White, J.C.; Marmiroli, N. Plant response to metal-containing engineered nanomaterials: An omics-based perspective. Environ. Sci. Technol. 2018, 52, 2451–2467. [Google Scholar] [CrossRef] [PubMed]
- Miller-Fleming, L.; Olin-Sandoval, V.; Campbell, K.; Ralser, M. Remaining Mysteries of Molecular Biology: The Role of Polyamines in the Cell. J. Mol. Biol. 2015, 427, 3389–3406. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; McIntosh, K.B.; Rudra, D.; Schawalder, S.; Shore, D.; Warner, J.R. Fine-Structure Analysis of Ribosomal Protein Gene Transcription. Mol. Cell. Biol. 2006, 26, 4853–4862. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, M. Review of in vitro toxicological research of quantum dot and potentially involved mechanisms. Sci. Tot. Environ. 2018, 625, 940–962. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Vishwakarma, K.; Ramawat, N.; Rai, P.; Singh, V.K.; Mishra, R.K.; Kumar, V.; Tripathi, D.K.; Sharma, S. Nanomaterials and microbes’ interactions: A contemporary overview. 3Biotech 2019, 9, 68. [Google Scholar] [CrossRef] [PubMed]
- Hardman, R. A Toxicologic Review of Quantum Dots: Toxicity Depends on Physicochemical and Environmental Factors. Environ Health Perspect. 2006, 114, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Holden, P.A.; Nisbet, R.M.; Lenihan, H.S.; Miller, R.J.; Cherr, G.N.; Schimel, J.P.; Gardea-Torresdey, J.L. Ecological Nanotoxicology: Integrating Nanomaterial Hazard Considerations Across the Subcellular, Population, Community, and Ecosystems Levels. Acc. Chem. Res. 2013, 46, 813–822. [Google Scholar] [CrossRef]
- Van Houten, B.; Hunter, S.E.; Meyer, J.N. Mitochondrial DNA damage induced autophagy, cell death, and disease. Front. Biosci. 2016, 21, 42–54. [Google Scholar] [CrossRef]
- Pimentel, C.; Batista-Nascimento, L.; Rodrigues-Pousada, C.; Menezes, R.A. Oxidative stress in Alzheimer’s and Parkinson’s diseases: Insights from the yeast Saccharomyces cerevisiae. Oxid. Med. Cell Longev. 2012. [Google Scholar] [CrossRef] [PubMed]

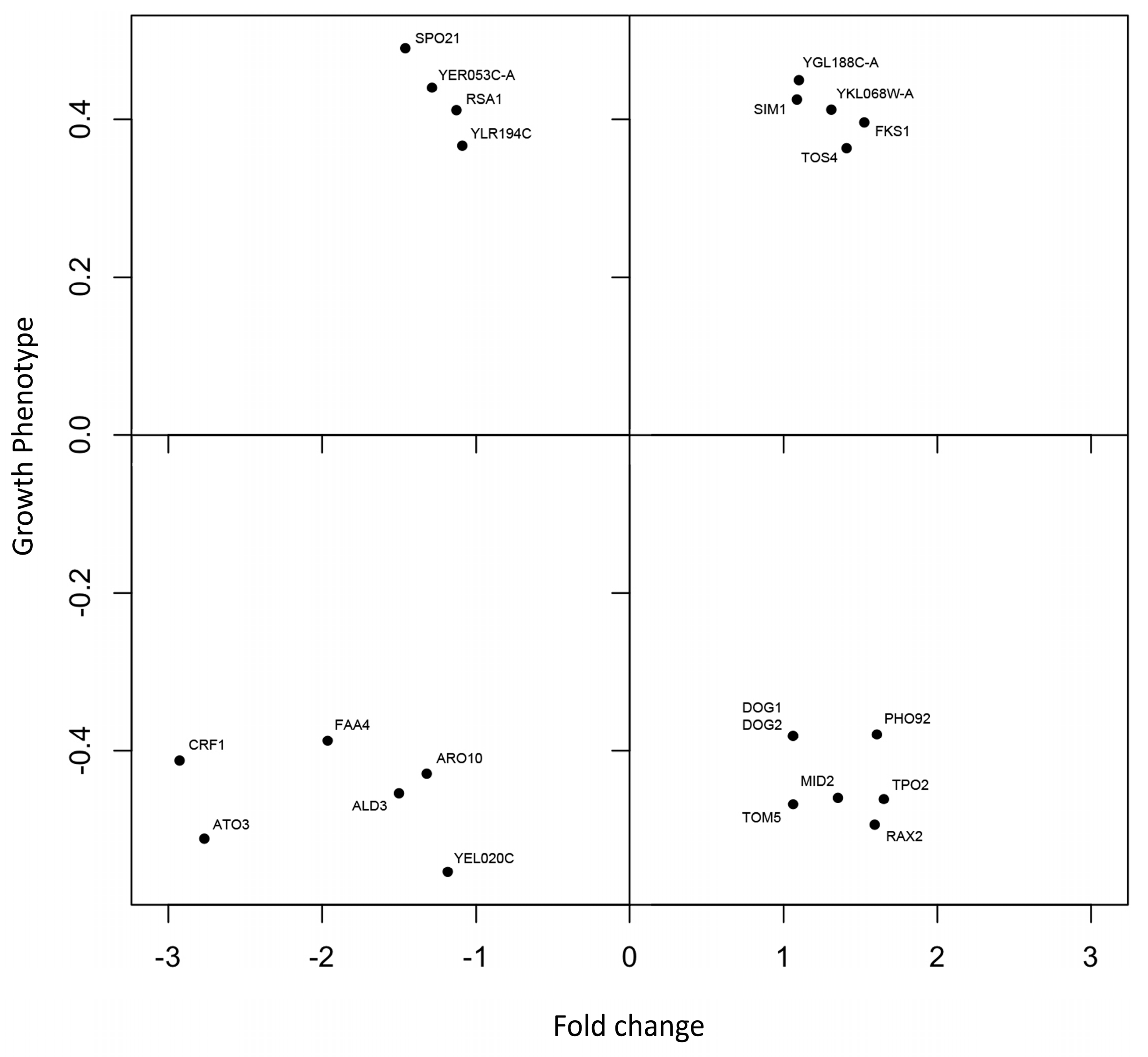
| Gene ID | Function |
|---|---|
| + + | |
| FKS1 | Catalytic subunit of 1,3-beta-D-glucan synthase; functionally redundant with alternate catalytic subunit Gsc2p; binds to regulatory subunit Rho1p; involved in cell wall synthesis and maintenance; localizes to sites of cell wall remodeling; FKS1 has a paralog, GSC2, that arose from the whole genome duplication |
| SIM1 | Protein of the SUN family (Sim1p, Uth1p, Nca3p, Sun4p); may participate in DNA replication; promoter contains SCB regulation box at −300 bp indicating that expression may be cell cycle-regulated; SIM1 has a paralog, SUN4, that arose from the whole genome duplication |
| TOS4 | Putative transcription factor, contains Forkhead Associated domain; found associated with chromatin; target of SBF transcription factor; expression is periodic and peaks in G1; involved in DNA replication checkpoint response; interacts with Rpd3 and Set3 histone deacetylase (HDAC) complexes; APCC(Cdh1) substrate; relative distribution to the nucleus increases upon DNA replication stress; TOS4 has a paralog, PLM2, that arose from the whole genome duplication |
| YGL188C-A | Unknown function |
| YKL068W-A | Unknown function |
| + − | |
| RSA1 | Protein involved in the assembly of 60S ribosomal subunits; functionally interacts with Dbp6p; functions in a late nucleoplasmic step of the assembly |
| SPO21 | Component of the meiotic outer plaque of the spindle pole body; involved in modifying the meiotic outer plaque that is required prior to the prospore membrane formation; SPO21 has a paralog, YSW1, that arose from the whole genome duplication |
| YER053C-A | Unknown function; green fluorescent protein (GFP)-fusion protein localizes to the endoplasmic reticulum; protein abundance increases in response to DNA replication stress |
| YLR194C | Structural constituent of the cell wall; attached to the plasma membrane by a GPI-anchor; expression is up-regulated in response to cell wall stress |
| − + | |
| DOG1 | 2-deoxyglucose-6-phosphate phosphatase; member of a family of low molecular weight phosphatases; confers 2-deoxyglucose resistance when overexpressed, in vivo substrate has not yet been identified; DOG1 has a paralog, DOG2, that arose from a single-locus duplication |
| DOG2 | 2-deoxyglucose-6-phosphate phosphatase; member of a family of low molecular weight phosphatases, induced by oxidative and osmotic stress, confers 2-deoxyglucose resistance when overexpressed; DOG2 has a paralog, DOG1, that arose from a single-locus duplication |
| MID2 | O-glycosylated plasma membrane protein; acts as a sensor for cell wall integrity signaling and activates the pathway; interacts with Rom2p, a guanine nucleotide exchange factor for Rho1p, and with the cell integrity pathway protein Zeo1p; MID2 has a paralog, MTL1, that arose from the whole genome duplication |
| PHO92 | Posttranscriptional regulator of phosphate metabolism; facilitates PHO4 mRNA degradation by interacting with Pop2p; regulates PHO4 mRNA stability by binding to PHO4’s 3’UTR in a phosphate-dependent manner; contains highly conserved YTH (YT521-B Homology) domain that exhibits RNA-binding activity; functional homolog of human YTHDF2 |
| RAX2 | N-glycosylated protein; involved in the maintenance of bud site selection during bipolar budding; localization requires Rax1p; RAX2 mRNA stability is regulated by Mpt5p |
| TOM5 | Component of the TOM (translocase of outer membrane) complex; responsible for the recognition and initial import of all mitochondrially directed proteins; involved in the etransfer of precursors from the Tom70p and Tom20p receptors to the Tom40p pore |
| TPO2 | Polyamine transporter of the major facilitator superfamily; member of the 12-spanner drug:H(+) antiporter DHA1 family; specific for spermine; localizes to the plasma membrane; transcription of TPO2 is regulated by Haa1p; TPO2 has a paralog, TPO3, that arose from the whole genome duplication |
| − − | |
| ALD3 | Cytoplasmic aldehyde dehydrogenase; involved in the beta-alanine synthesis; uses NAD+ as the preferred coenzyme; very similar to Ald2p; expression is induced by stress and repressed by glucose |
| ARO10 | Phenylpyruvate decarboxylase; catalyzes decarboxylation of phenylpyruvate to phenylacetaldehyde, which is the first specific step in the Ehrlich pathway; involved in protein N-terminal Met and Ala catabolism |
| ATO3 | Plasma membrane protein, putative ammonium transporter; regulation pattern suggests a possible role in the export of ammonia from the cell; phosphorylated in mitochondria; member of the TC 9.B.33 YaaH family of putative transporters |
| CRF1 | Transcriptional corepressor; involved in the repression of ribosomal protein (RP) gene transcription via the TOR signaling pathway which promotes the accumulation of Crf1p in the nucleus; role in the repression of RP genes varies by strain; CRF1 has a paralog, IFH1, that arose from the whole genome duplication |
| FAA4 | Long chain fatty acyl-CoA synthetase; activates imported fatty acids with a preference for C12:0-C16:0 chain lengths; functions in long chain fatty acid import; important for survival during stationary phase; localized to lipid particles; involved in the sphingolipid-to-glycerolipid metabolism; forms cytoplasmic foci upon DNA replication stress; FAA4 has a paralog, FAA1, that arose from the whole genome duplication |
| YEL020C | Protein of unknown function with low sequence identity to Pdc1p |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pagano, L.; Caldara, M.; Villani, M.; Zappettini, A.; Marmiroli, N.; Marmiroli, M. In Vivo-In Vitro Comparative Toxicology of Cadmium Sulphide Quantum Dots in the Model Organism Saccharomyces cerevisiae. Nanomaterials 2019, 9, 512. https://doi.org/10.3390/nano9040512
Pagano L, Caldara M, Villani M, Zappettini A, Marmiroli N, Marmiroli M. In Vivo-In Vitro Comparative Toxicology of Cadmium Sulphide Quantum Dots in the Model Organism Saccharomyces cerevisiae. Nanomaterials. 2019; 9(4):512. https://doi.org/10.3390/nano9040512
Chicago/Turabian StylePagano, Luca, Marina Caldara, Marco Villani, Andrea Zappettini, Nelson Marmiroli, and Marta Marmiroli. 2019. "In Vivo-In Vitro Comparative Toxicology of Cadmium Sulphide Quantum Dots in the Model Organism Saccharomyces cerevisiae" Nanomaterials 9, no. 4: 512. https://doi.org/10.3390/nano9040512
APA StylePagano, L., Caldara, M., Villani, M., Zappettini, A., Marmiroli, N., & Marmiroli, M. (2019). In Vivo-In Vitro Comparative Toxicology of Cadmium Sulphide Quantum Dots in the Model Organism Saccharomyces cerevisiae. Nanomaterials, 9(4), 512. https://doi.org/10.3390/nano9040512