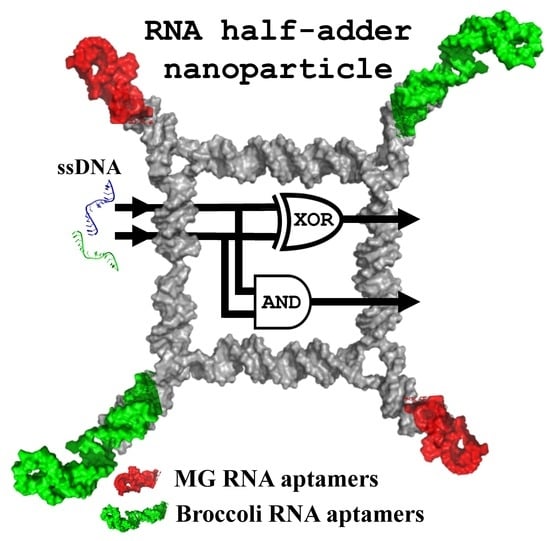
Fluorogenic RNA Aptamers: A Nano-platform for Fabrication of Simple and Combinatorial Logic Gates
Abstract
1. Introduction
2. Results and Discussion
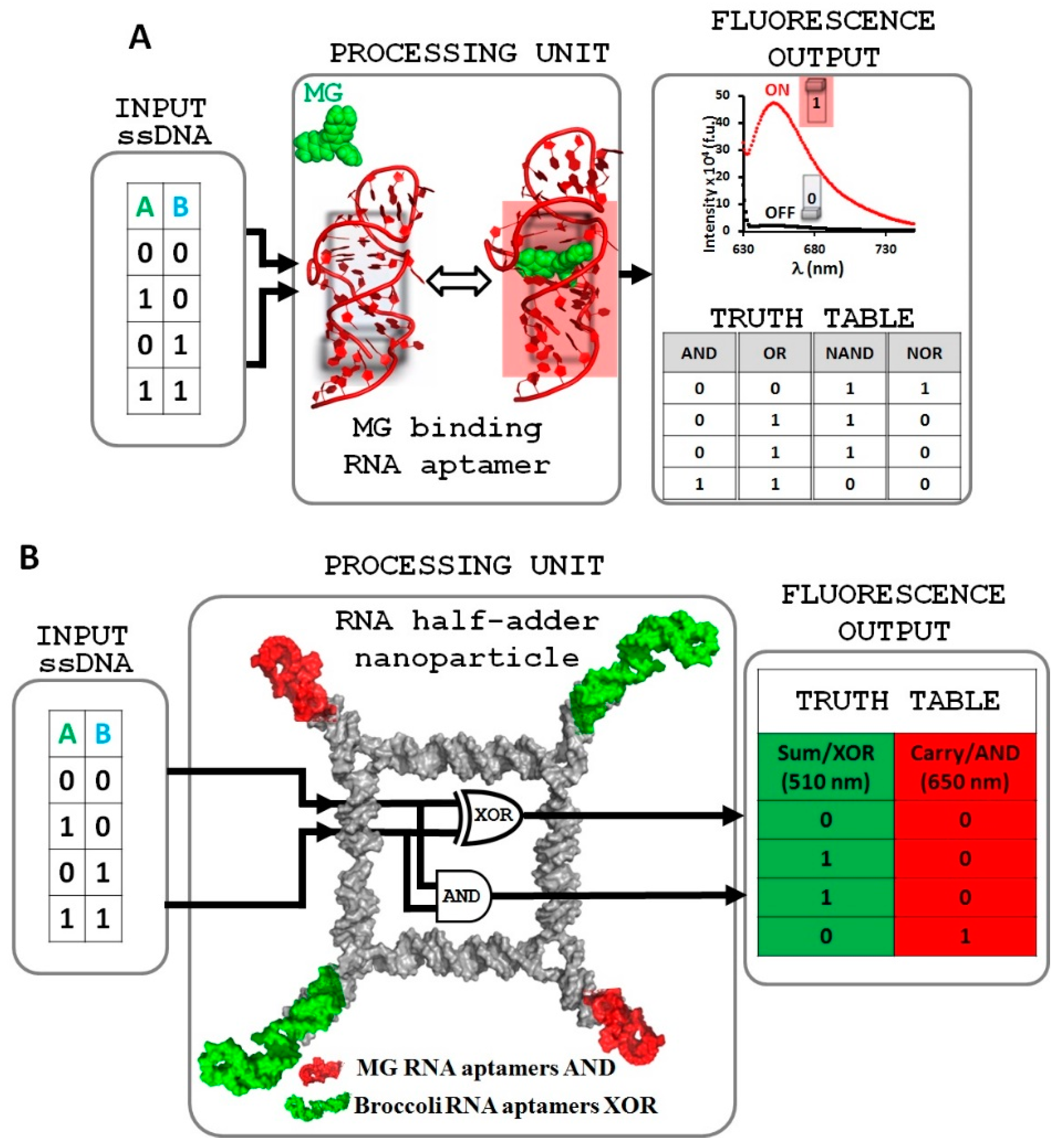
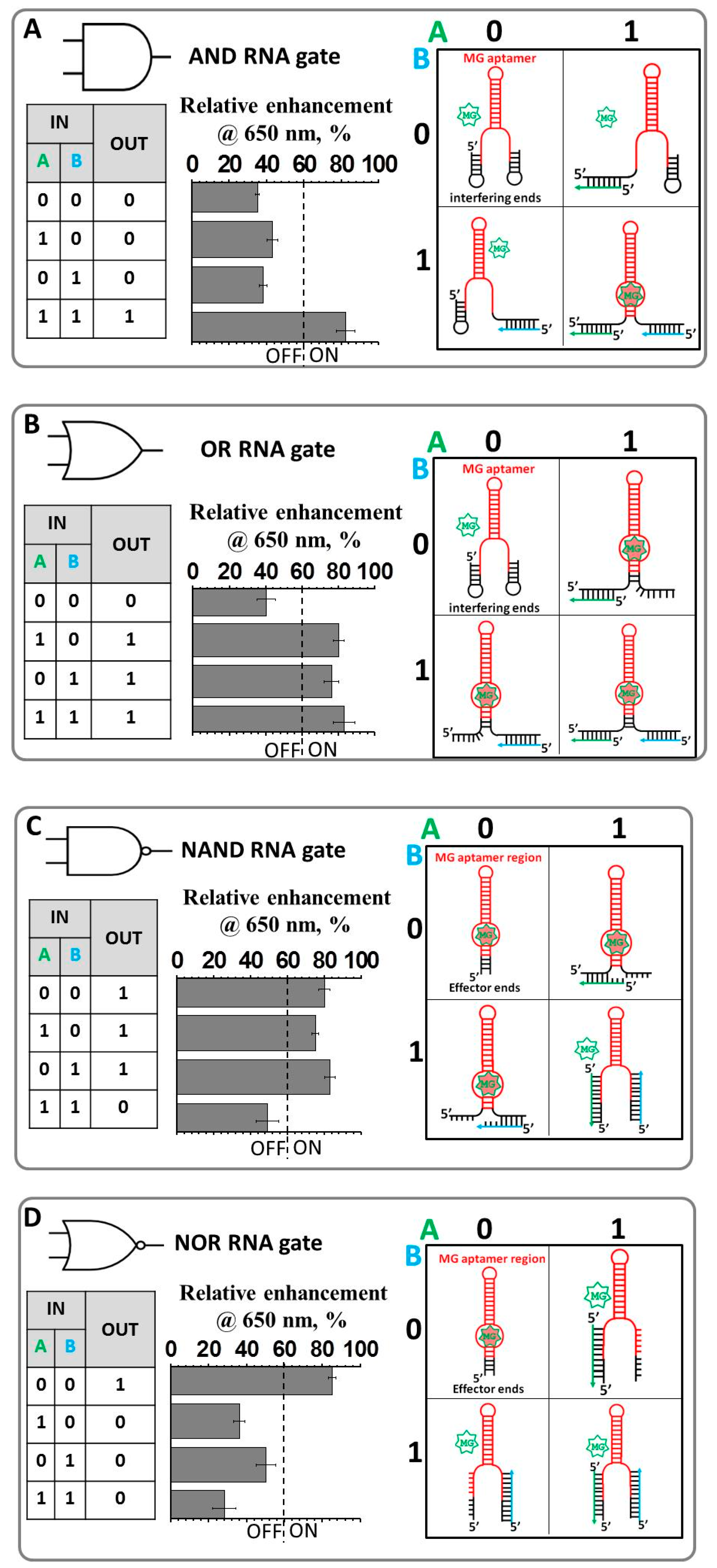
2.1. Design and Fabrication of a Logic Gate Possessing AND, OR, NAND and NOR Boolean Functions
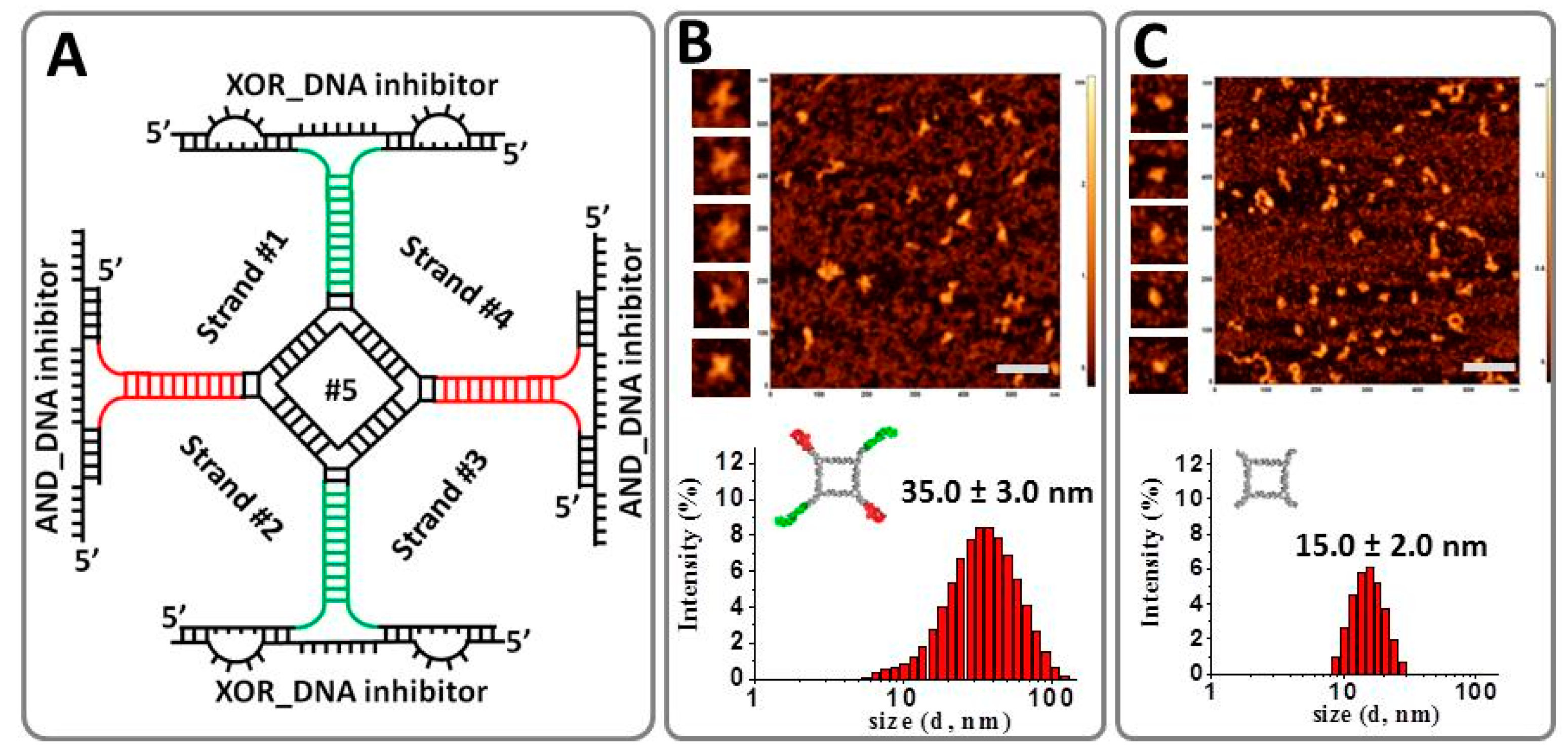
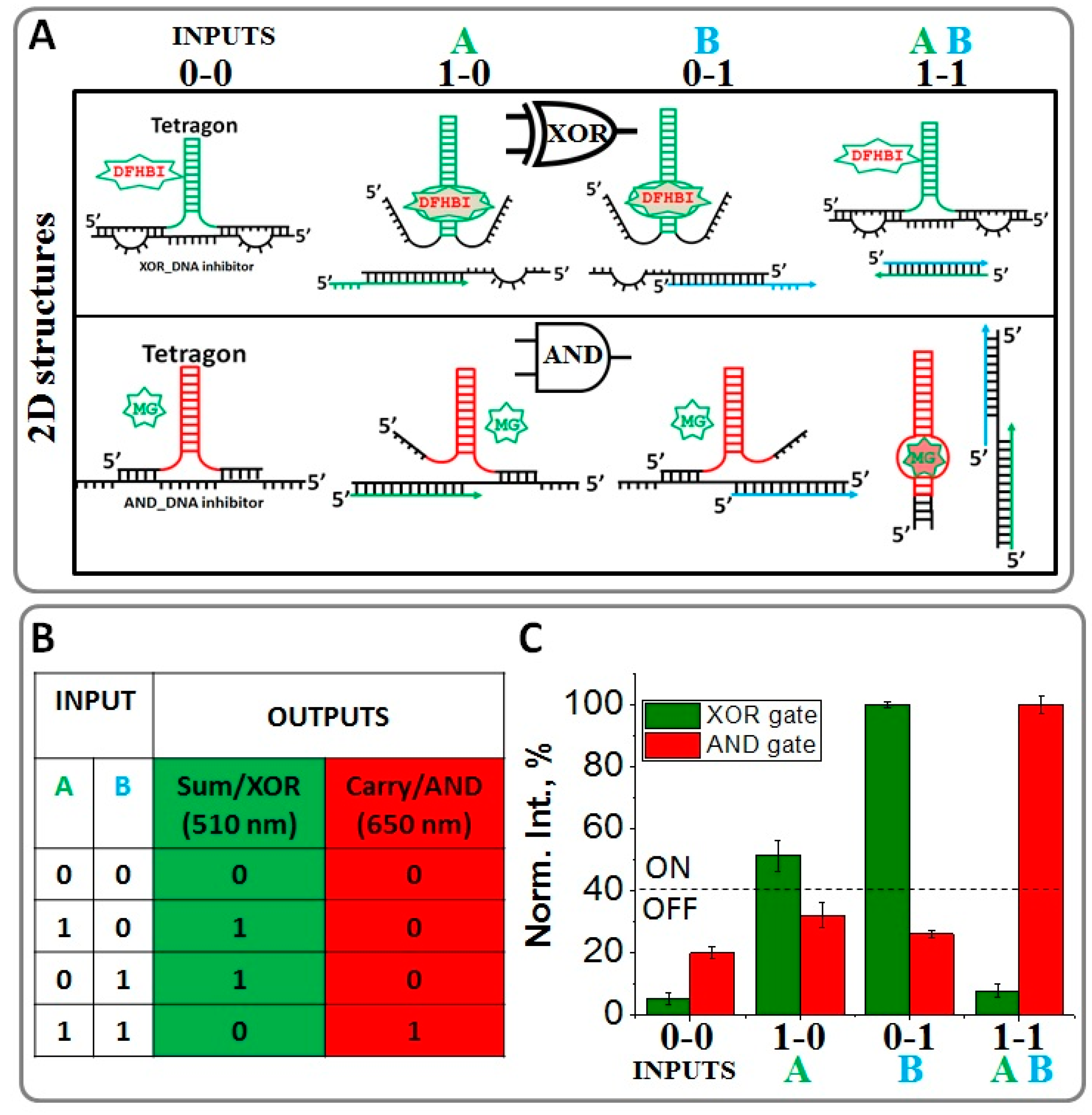
2.2. Implementing Logic Gates to Construct a Half-Adder Logic Circuit
3. Materials and Methods
3.1. Nucleic Acid Sequence Design, Synthesis, and Assembly
3.2. Fluorescence Measurements
3.3. Dynamic Light Scattering
3.4. Atomic Force Microscopy Imaging
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Guo, P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 2010, 5, 833–842. [Google Scholar] [CrossRef] [PubMed]
- Shukla, G.C.; Haque, F.; Tor, Y.; Wilhelmsson, L.M.; Toulme, J.J.; Isambert, H.; Guo, P.; Rossi, J.J.; Tenenbaum, S.A.; Shapiro, B.A. A boost for the emerging field of RNA nanotechnology. ACS Nano 2011, 5, 3405–3418. [Google Scholar] [CrossRef] [PubMed]
- Parlea, L.G.; Sweeney, B.A.; Hosseini-Asanjan, M.; Zirbel, C.L.; Leontis, N.B. The RNA 3D Motif Atlas: Computational methods for extraction, organization and evaluation of RNA motifs. Methods 2016, 103, 99–119. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, B.A.; Roy, P.; Leontis, N.B. An introduction to recurrent nucleotide interactions in RNA. Wiley Interdiscip. Rev. RNA 2015, 6, 17–45. [Google Scholar] [CrossRef] [PubMed]
- Jenner, L.; Melnikov, S.; Garreau de Loubresse, N.; Ben-Shem, A.; Iskakova, M.; Urzhumtsev, A.; Meskauskas, A.; Dinman, J.; Yusupova, G.; Yusupov, M. Crystal structure of the 80S yeast ribosome. Curr. Opin. Struct. Biol. 2012, 22, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Rupert, P.B.; Ferre-D’Amare, A.R. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature 2001, 410, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Scott, W.G.; Finch, J.T.; Klug, A. The Crystal-Structure of an All-Rna Hammerhead Ribozyme—A Proposed Mechanism for Rna Catalytic Cleavage. Cell 1995, 81, 991–1002. [Google Scholar] [CrossRef]
- Bui, M.N.; Brittany Johnson, M.; Viard, M.; Satterwhite, E.; Martins, A.N.; Li, Z.; Marriott, I.; Afonin, K.A.; Khisamutdinov, E.F. Versatile RNA tetra-U helix linking motif as a toolkit for nucleic acid nanotechnology. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.B.; Halman, J.R.; Satterwhite, E.; Zakharov, A.V.; Bui, M.N.; Benkato, K.; Goldsworthy, V.; Kim, T.; Hong, E.; Dobrovolskaia, M.A.; et al. Programmable Nucleic Acid Based Polygons with Controlled Neuroimmunomodulatory Properties for Predictive QSAR Modeling. Small 2017, 13. [Google Scholar] [CrossRef] [PubMed]
- Khisamutdinov, E.F.; Bui, M.N.; Jasinski, D.; Zhao, Z.; Cui, Z.; Guo, P. Simple Method for Constructing RNA Triangle, Square, Pentagon by Tuning Interior RNA 3WJ Angle from 60 degrees to 90 degrees or 108 degrees. MIMB 2015, 1316, 181–193. [Google Scholar] [CrossRef]
- Severcan, I.; Geary, C.; Verzemnieks, E.; Chworos, A.; Jaeger, L. Square-shaped RNA particles from different RNA folds. Nano Lett. 2009, 9, 1270–1277. [Google Scholar] [CrossRef] [PubMed]
- Hong, E.; Halman, J.R.; Shah, A.B.; Khisamutdinov, E.F.; Dobrovolskaia, M.A.; Afonin, K.A. Structure and Composition Define Immunorecognition of Nucleic Acid Nanoparticles. Nano Lett. 2018, 18, 4309–4321. [Google Scholar] [CrossRef] [PubMed]
- Khisamutdinov, E.F.; Jasinski, D.L.; Li, H.; Zhang, K.; Chiu, W.; Guo, P. Fabrication of RNA 3D Nanoprisms for Loading and Protection of Small RNAs and Model Drugs. PANS 2016, 28, 10079–10087. [Google Scholar] [CrossRef] [PubMed]
- Afonin, K.A.; Bindewald, E.; Yaghoubian, A.J.; Voss, N.; Jacovetty, E.; Shapiro, B.A.; Jaeger, L. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol. 2010, 5, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Halman, J.R.; Satterwhite, E.; Roark, B.; Chandler, M.; Viard, M.; Ivanina, A.; Bindewald, E.; Kasprzak, W.K.; Panigaj, M.; Bui, M.N.; et al. Functionally-interdependent shape-switching nanoparticles with controllable properties. Nucleic Acids Res. 2017, 45, 2210–2220. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G. A review of therapeutic aptamer conjugates with emphasis on new approaches. Pharmaceuticals 2013, 6, 340–357. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, S.; Nosrati, R.; Yousefi, M.; Nezami, A.; Soltani, F.; Taghdisi, S.M.; Abnous, K.; Alibolandi, M.; Ramezani, M. Aptamer-based biosensors and nanosensors for the detection of vascular endothelial growth factor (VEGF): A review. Biosens. Bioelectron. 2018, 110, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Farzin, L.; Shamsipur, M.; Sheibani, S. A review: Aptamer-based analytical strategies using the nanomaterials for environmental and human monitoring of toxic heavy metals. Talanta 2017, 174, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Dolgosheina, E.V.; Unrau, P.J. Fluorophore-binding RNA aptamers and their applications. Wiley Interdiscip. Rev. RNA 2016, 7, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Eydeler, K.; Magbanua, E.; Werner, A.; Ziegelmuller, P.; Hahn, U. Fluorophore binding aptamers as a tool for RNA visualization. Biophys. J. 2009, 96, 3703–3707. [Google Scholar] [CrossRef] [PubMed]
- Stojanovic, M.N.; Kolpashchikov, D.M. Modular aptameric sensors. JACS 2004, 126, 9266–9270. [Google Scholar] [CrossRef] [PubMed]
- Guet, D.; Burns, L.T.; Maji, S.; Boulanger, J.; Hersen, P.; Wente, S.R.; Salamero, J.; Dargemont, C. Combining Spinach-tagged RNA and gene localization to image gene expression in live yeast. Nat. Commun. 2015, 6, 8882. [Google Scholar] [CrossRef] [PubMed]
- Nilaratanakul, V.; Hauer, D.A.; Griffin, D.E. Development and characterization of Sindbis virus with encoded fluorescent RNA aptamer Spinach2 for imaging of replication and immune-mediated changes in intracellular viral RNA. J. Gen. Virol. 2017, 98, 992–1003. [Google Scholar] [CrossRef] [PubMed]
- DasGupta, S.; Shelke, S.A.; Li, N.S.; Piccirilli, J.A. Spinach RNA aptamer detects lead(II) with high selectivity. Chem. Commun. 2015, 51, 9034–9037. [Google Scholar] [CrossRef] [PubMed]
- Babendure, J.R.; Adams, S.R.; Tsien, R.Y. Aptamers switch on fluorescence of triphenylmethane dyes. JACS 2003, 125, 14716–14717. [Google Scholar] [CrossRef] [PubMed]
- Paige, J.S.; Wu, K.Y.; Jaffrey, S.R. RNA mimics of green fluorescent protein. Science 2011, 333, 642–646. [Google Scholar] [CrossRef] [PubMed]
- Filonov, G.S.; Moon, J.D.; Svensen, N.; Jaffrey, S.R. Broccoli: Rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. JACS 2014, 136, 16299–16308. [Google Scholar] [CrossRef] [PubMed]
- Sando, S.; Narita, A.; Hayami, M.; Aoyama, Y. Transcription monitoring using fused RNA with a dye-binding light-up aptamer as a tag: A blue fluorescent RNA. Chem. Commun. 2008, 3858–3860. [Google Scholar] [CrossRef] [PubMed]
- Dolgosheina, E.V.; Jeng, S.C.; Panchapakesan, S.S.; Cojocaru, R.; Chen, P.S.; Wilson, P.D.; Hawkins, N.; Wiggins, P.A.; Unrau, P.J. RNA mango aptamer-fluorophore: A bright, high-affinity complex for RNA labeling and tracking. ACS Chem. Biol. 2014, 9, 2412–2420. [Google Scholar] [CrossRef] [PubMed]
- Qiu, M.K.; Khisamutdinov, E.; Zhao, Z.Y.; Pan, C.; Choi, J.W.; Leontis, N.B.; Guo, P.X. RNA nanotechnology for computer design and in vivo computation. Philos Trans. R. Soc. A 2013, 371. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, E.; Gil, B. Cell biology. RNA computing in a living cell. Science 2008, 322, 387–388. [Google Scholar] [CrossRef] [PubMed]
- Ogihara, M.; Ray, A. Molecular computation: DNA computing on a chip. Nature 2000, 403, 143–144. [Google Scholar] [CrossRef] [PubMed]
- Benenson, Y.; Paz-Elizur, T.; Adar, R.; Keinan, E.; Livneh, Z.; Shapiro, E. Programmable and autonomous computing machine made of biomolecules. Nature 2001, 414, 430–434. [Google Scholar] [CrossRef] [PubMed]
- Abels Seth, G.; Khisamutdinov Emil, F. Nucleic Acid Computing and its Potential to Transform Silicon-Based Technology. DNA RNA Nanotechnol. 2015, 2, 13–22. [Google Scholar] [CrossRef]
- Adleman, L.M. Molecular Computation of Solutions to Combinatorial Problems. Science 1994, 266, 1021–1024. [Google Scholar] [CrossRef] [PubMed]
- Carell, T. Molecular computing: DNA as a logic operator. Nature 2011, 469, 45–46. [Google Scholar] [CrossRef] [PubMed]
- Normile, D. Molecular computing. DNA-based computer takes aim at genes. Science 2002, 295, 951. [Google Scholar] [CrossRef] [PubMed]
- Seelig, G.; Soloveichik, D.; Zhang, D.Y.; Winfree, E. Enzyme-free nucleic acid logic circuits. Science 2006, 314, 1585–1588. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; LaBean, T.H.; Relf, J.H.; Seeman, N.C. Logical computation using algorithmic self-assembly of DNA triple-crossover molecules. Nature 2000, 407, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Benenson, Y. RNA-based computation in live cells. Curr. Opin. Biotechnol. 2009, 20, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Benenson, Y. Biocomputers: From test tubes to live cells. Mol. Biosyst. 2009, 5, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Benenson, Y. Engineering RNAi circuits. Meth. Enzymol. 2011, 497, 187–205. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Wroblewska, L.; Prochazka, L.; Weiss, R.; Benenson, Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 2011, 333, 1307–1311. [Google Scholar] [CrossRef] [PubMed]
- Zadegan, R.M.; Jepsen, M.D.; Hildebrandt, L.L.; Birkedal, V.; Kjems, J. Construction of a fuzzy and Boolean logic gates based on DNA. Small 2015, 11, 1811–1817. [Google Scholar] [CrossRef] [PubMed]
- Campolongo, M.J.; Kahn, J.S.; Cheng, W.L.; Yang, D.Y.; Gupton-Campolongo, T.; Luo, D. Adaptive DNA-based materials for switching, sensing, and logic devices. J. Mater. Chem. 2011, 21, 6113–6121. [Google Scholar] [CrossRef]
- Qian, L.; Winfree, E. Scaling Up Digital Circuit Computation with DNA Strand Displacement Cascades. Science 2011, 332, 1196–1201. [Google Scholar] [CrossRef] [PubMed]
- Shlyahovsky, B.; Li, Y.; Lioubashevski, O.; Elbaz, J.; Willner, I. Logic Gates and Antisense DNA Devices Operating on a Translator Nucleic Acid Scaffold. ACS Nano 2009, 3, 1831–1843. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Li, T.; Zhang, L.; Dong, S.; Wang, E. G-quadruplex DNAzyme based molecular catalytic beacon for label-free colorimetric logic gates. Biomaterials 2011, 32, 7318–7324. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yin, P.; Green, A.A. Ribocomputing: Cellular Logic Computation Using RNA Devices. Biochemistry 2018, 57, 883–885. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Espinosa, E.; Casadesus, J.; Vogel, J. Small RNA-based feedforward loop with AND-gate logic regulates extrachromosomal DNA transfer in Salmonella. PNAS 2015, 112, E4772–E4781. [Google Scholar] [CrossRef] [PubMed]
- Schaerli, Y.; Gili, M.; Isalan, M. A split intein T7 RNA polymerase for transcriptional AND-logic. NAR 2014, 42, 12322–12328. [Google Scholar] [CrossRef] [PubMed]
- Penchovsky, R.; Breaker, R.R. Computational design and experimental validation of oligonucleotide-sensing allosteric ribozymes. Nat. Biotechnol. 2005, 23, 1424–1433. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Hickey, S.F.; Keyser, S.G.; Hammond, M.C. In Vitro and In Vivo Enzyme Activity Screening via RNA-Based Fluorescent Biosensors for S-Adenosyl-l-homocysteine (SAH). JACS 2016, 138, 7040–7047. [Google Scholar] [CrossRef] [PubMed]
- Jaffrey, S.R. RNA-Based Fluorescent Biosensors for Detecting Metabolites in vitro and in Living Cells. Adv. Pharmacol. 2018, 82, 187–203. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; White, R.J.; Xia, F.; Zuo, X.L.; Vallee-Belisle, A.; Plaxco, K.W. DNA biomolecular-electronic encoder and decoder devices constructed by multiplex biosensors. NPG Asia Mater. 2012, 4. [Google Scholar] [CrossRef]
- Xu, S.L.; Li, H.L.; Miao, Y.Q.; Liu, Y.Q.; Wang, E.K. Implementation of half adder and half subtractor with a simple and universal DNA-based platform. NPG Asia Mater. 2013, 5. [Google Scholar] [CrossRef]
- Nguyen, D.H.; DeFina, S.C.; Fink, W.H.; Dieckmann, T. Binding to an RNA aptamer changes the charge distribution and conformation of malachite green. JACS 2002, 124, 15081–15084. [Google Scholar] [CrossRef]
- Baugh, C.; Grate, D.; Wilson, C. 2.8 A crystal structure of the malachite green aptamer. J. Mol. Biol. 2000, 301, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Zadeh, J.N.; Steenberg, C.D.; Bois, J.S.; Wolfe, B.R.; Pierce, M.B.; Khan, A.R.; Dirks, R.M.; Pierce, N.A. NUPACK: Analysis and Design of Nucleic Acid Systems. J. Comput. Chem. 2011, 32, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. NAR 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Elbaz, J.; Lioubashevski, O.; Wang, F.; Remacle, F.; Levine, R.D.; Willner, I. DNA computing circuits using libraries of DNAzyme subunits. Nat. Nanotechnol. 2010, 5, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Pischel, U. Chemical approaches to molecular logic elements for addition and subtraction. Angew. Chem. 2007, 46, 4026–4040. [Google Scholar] [CrossRef] [PubMed]
- Shlyakhtenko, L.S.; Gall, A.A.; Lyubchenko, Y.L. Mica functionalization for imaging of DNA and protein-DNA complexes with atomic force microscopy. MIMB 2013, 931, 295–312. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goldsworthy, V.; LaForce, G.; Abels, S.; Khisamutdinov, E.F. Fluorogenic RNA Aptamers: A Nano-platform for Fabrication of Simple and Combinatorial Logic Gates. Nanomaterials 2018, 8, 984. https://doi.org/10.3390/nano8120984
Goldsworthy V, LaForce G, Abels S, Khisamutdinov EF. Fluorogenic RNA Aptamers: A Nano-platform for Fabrication of Simple and Combinatorial Logic Gates. Nanomaterials. 2018; 8(12):984. https://doi.org/10.3390/nano8120984
Chicago/Turabian StyleGoldsworthy, Victoria, Geneva LaForce, Seth Abels, and Emil F. Khisamutdinov. 2018. "Fluorogenic RNA Aptamers: A Nano-platform for Fabrication of Simple and Combinatorial Logic Gates" Nanomaterials 8, no. 12: 984. https://doi.org/10.3390/nano8120984
APA StyleGoldsworthy, V., LaForce, G., Abels, S., & Khisamutdinov, E. F. (2018). Fluorogenic RNA Aptamers: A Nano-platform for Fabrication of Simple and Combinatorial Logic Gates. Nanomaterials, 8(12), 984. https://doi.org/10.3390/nano8120984