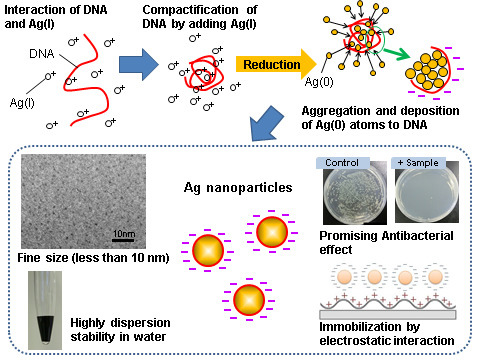
DNA/Ag Nanoparticles as Antibacterial Agents against Gram-Negative Bacteria
Abstract
:1. Introduction
2. Results
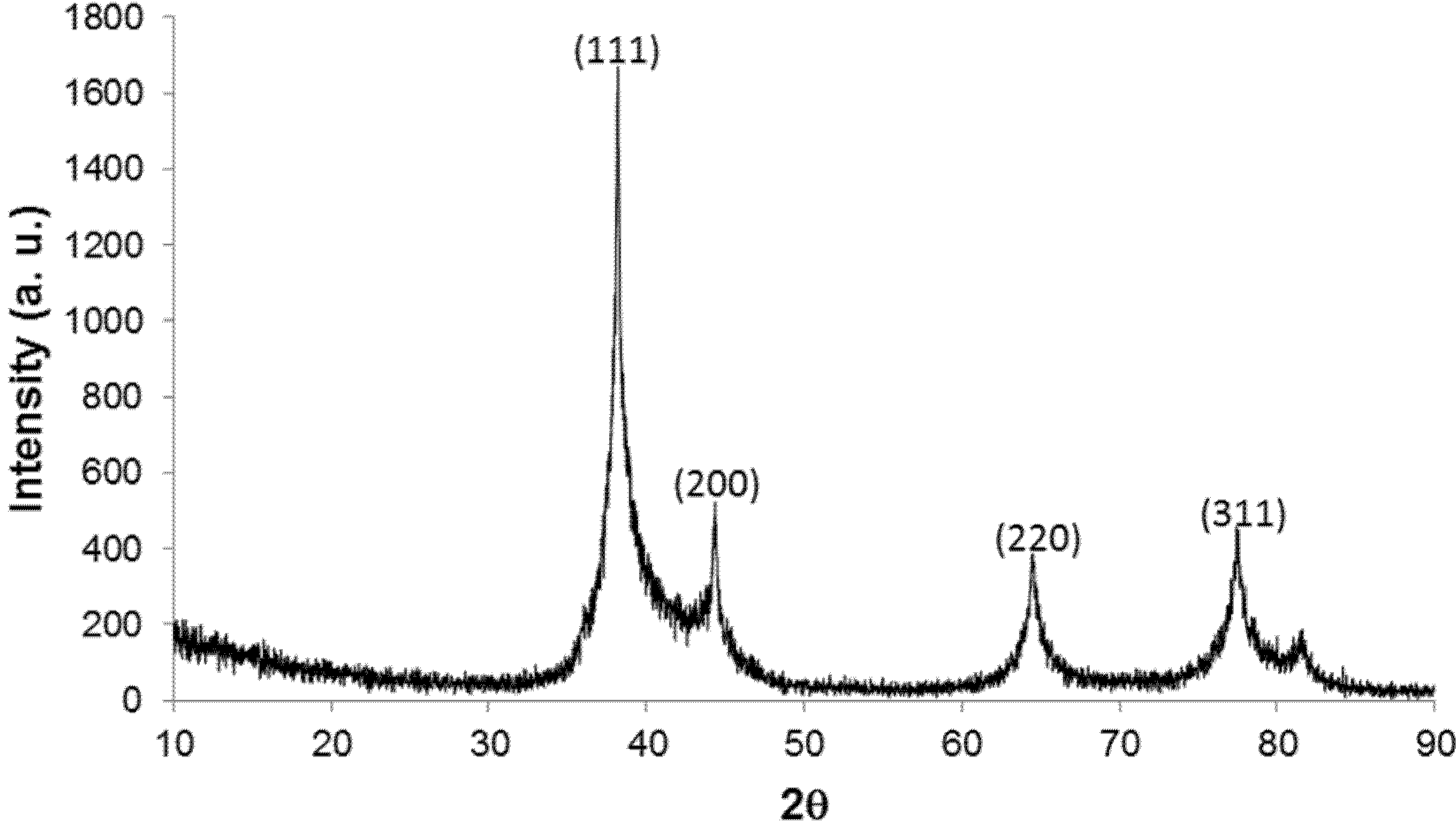
2.1. Characteristics of Size Distribution and Crystal Structure of the DNA/Ag Nanoparticles

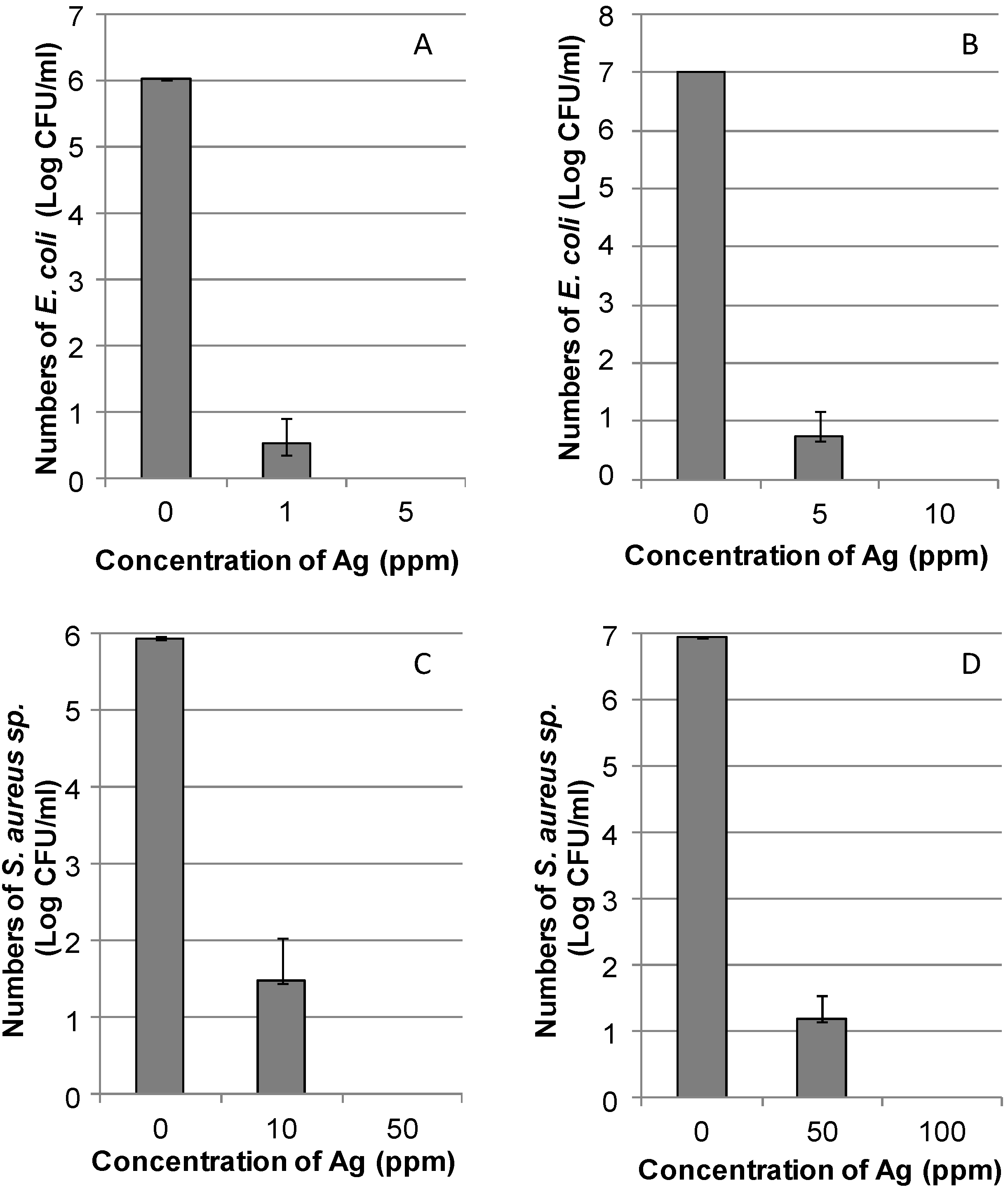
2.2. Use of the DNA/Ag Nanoparticles as Antibacterial Materials
2.2.1. Evaluation of the Antibacterial Efficiency of the DNA/Ag Nanoparticles
2.2.2. Preparation of the DNA/Ag Nanoparticle Immobilized Fabrics
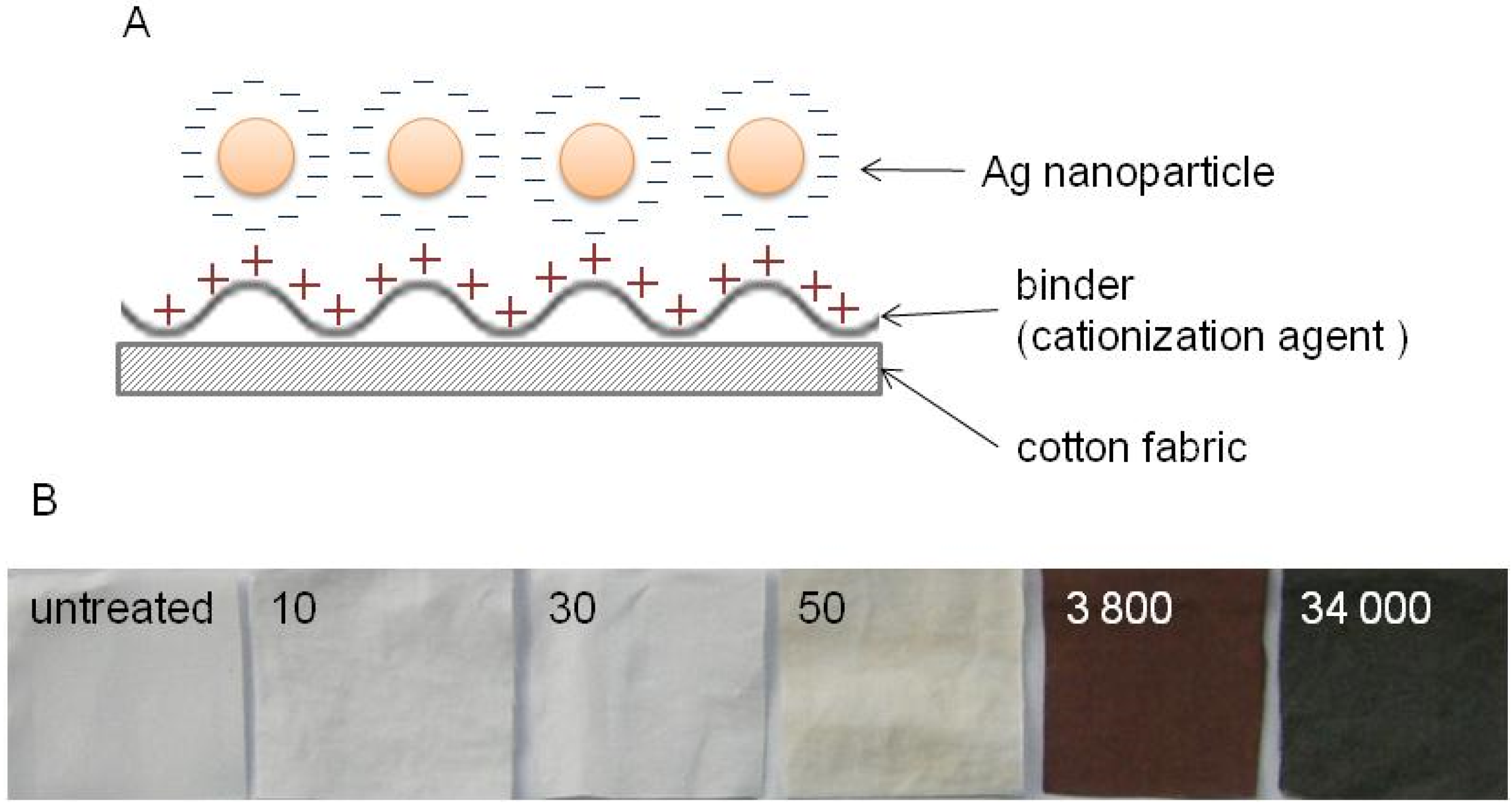
2.2.3. Ag(I) Ions Release Ratio from the DNA/Ag Nanoparticle Immobilized Fabrics
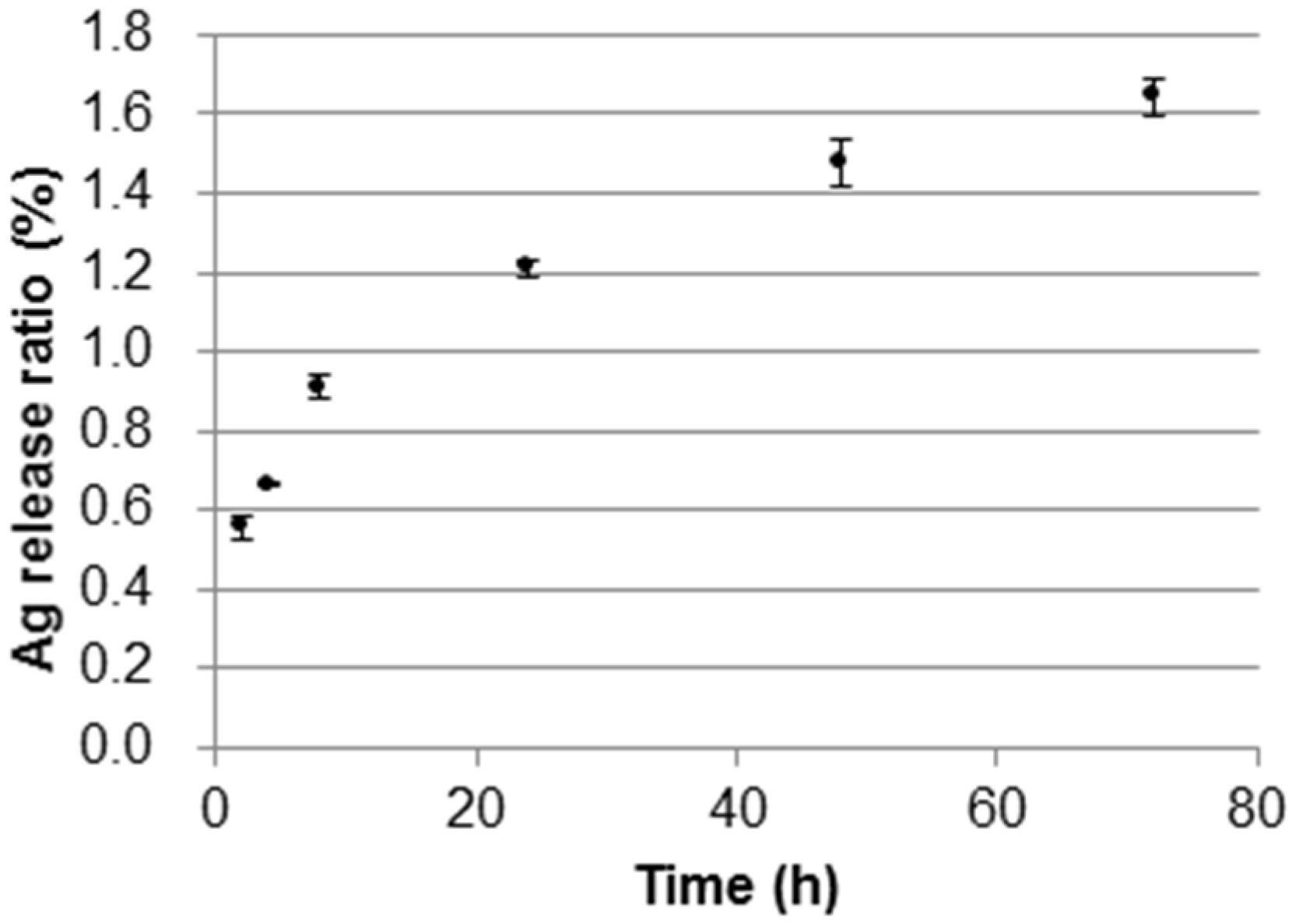
2.2.4. Evaluation of the Antibacterial Efficiency of the DNA/Ag Nanoparticle Immobilized Fabrics
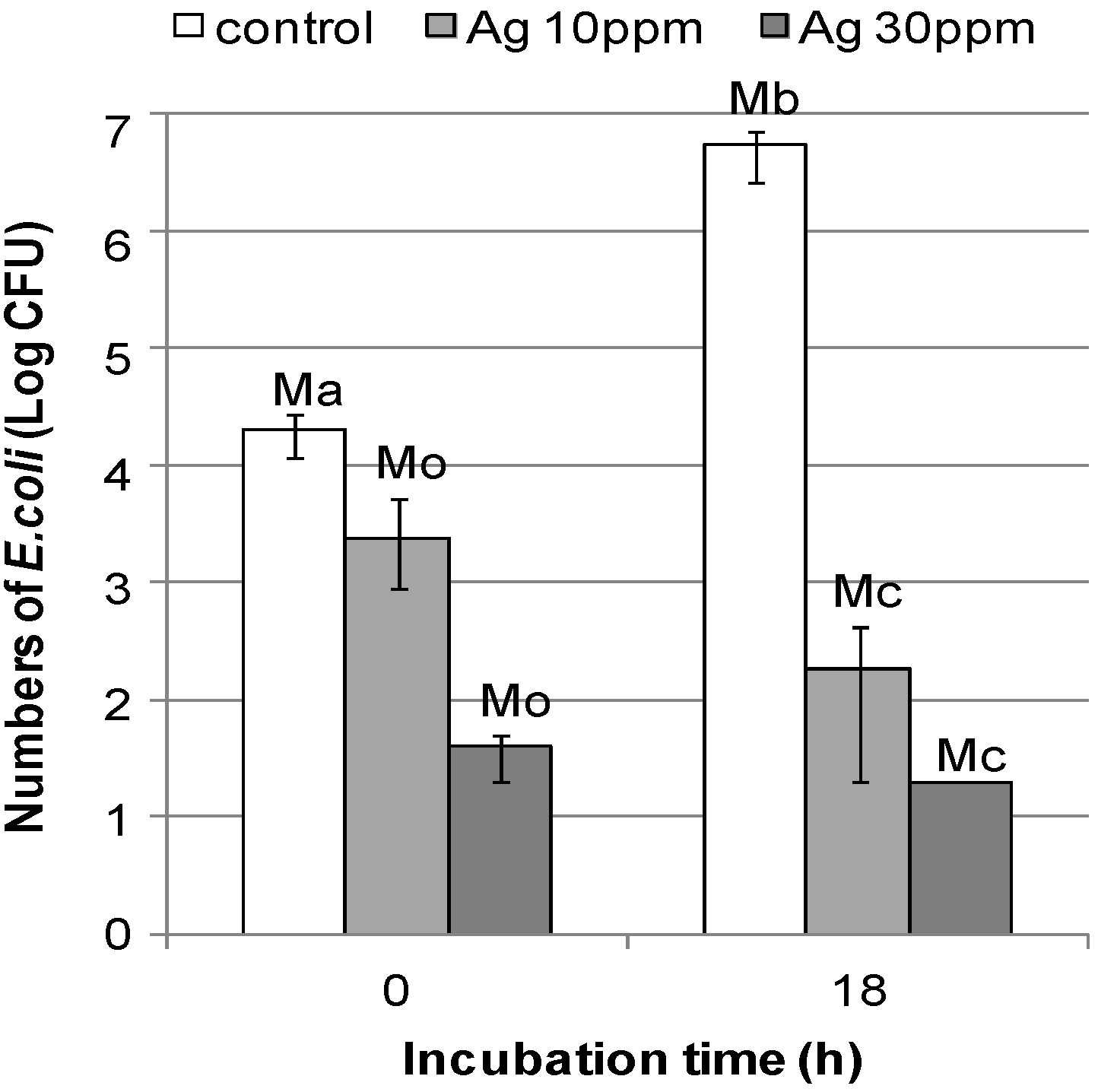
| Testing Item | Test Results (Colony Numbers) | Reference [33] | ||
|---|---|---|---|---|
| Controls | DNA/Ag Nanoparticle Immobilized Fabrics | |||
| 10 ppm | 30 ppm | |||
| Ma | 2.05 × 104 | - | - | |
| Mb | 5.40 × 106 | - | - | |
| Mo | - | 2.35 × 103 | <40 | |
| Mc | - | 1.82 × 102 | <20 | |
| BG | 2.4 | - | - | |
| BS | >3.6 | >2.7 | >2.2 Inhibitory effect | |
| BC | >2.3 | >3.0 | >0 Killing effect | |
3. Discussion
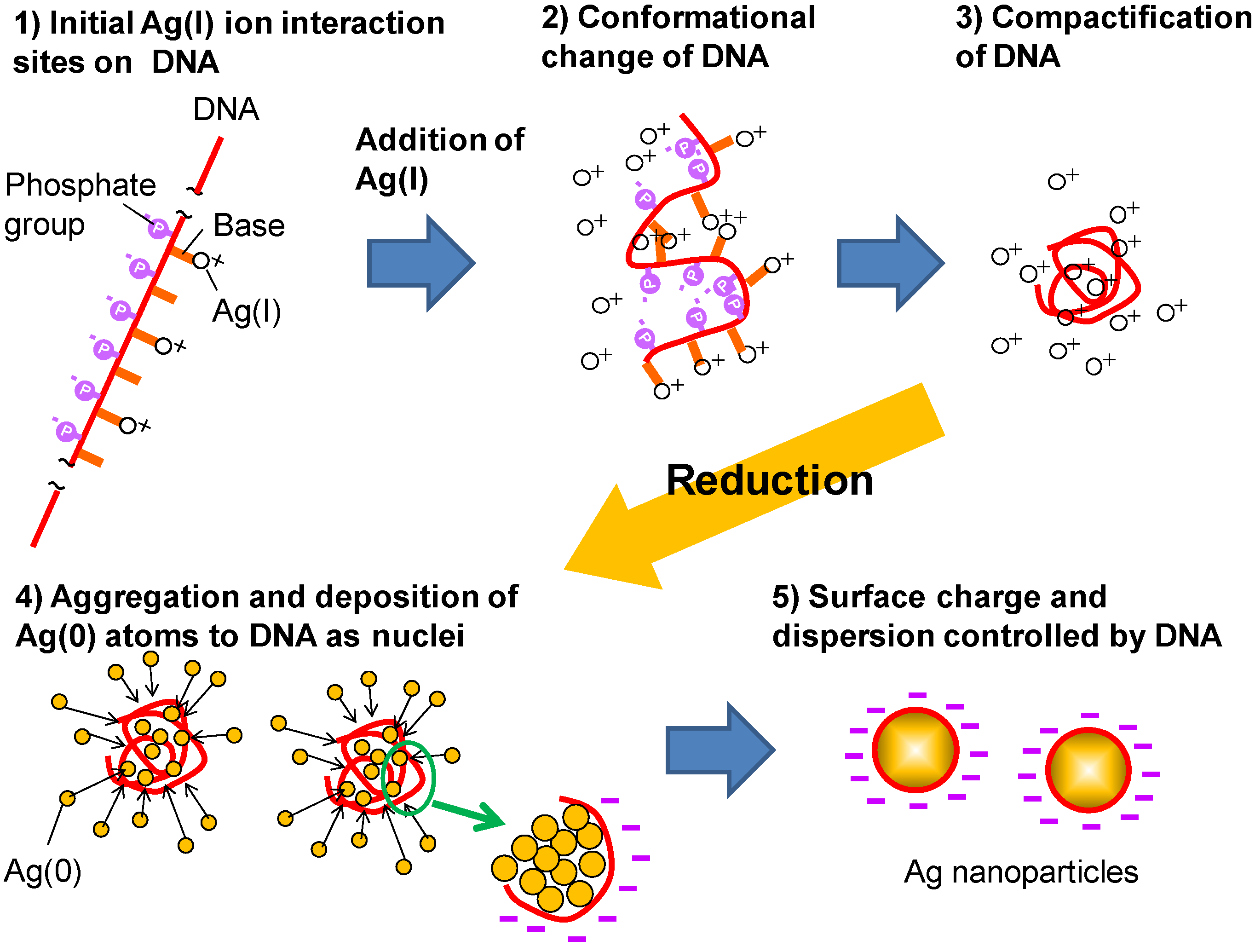
3.1. Formation Mechanism of DNA/Ag Nanoparticles
3.2. Antibacterial Effect of the DNA/Ag Nanoparticles
4. Experimental Section
4.1. Materials
4.2. Preparation and Characterization of the DNA/Ag Nanoparticles
4.3. Evaluation of the DNA/Ag Nanoparticles as Antibacterial Materials
4.3.1. Antibacterial Evaluation of the DNA/Ag Nanoparticles
4.3.2. Preparation of the DNA/Ag Nanoparticle Immobilized Fabrics
4.3.3. Ag(I) Ions Release from the DNA/Ag Nanoparticle Immobilized Fabrics
4.3.4. Antibacterial Evaluation of the DNA/Ag Nanoparticle Immobilized Fabrics
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mulvaney, P. Surface plasmon spectroscopy of nanosized metal particles. Langmuir 1996, 12, 788–800. [Google Scholar] [CrossRef]
- Toshima, N.; Yonezawa, T. Bimetallic nanoparticles—Novel materials for chemical and physical applications. New J. Chem. 1998, 22, 1179–1201. [Google Scholar] [CrossRef]
- Burda, C.; Chen, X.B.; Narayanan, R.; El-Sayed, M.A. Chemistry and properties of nanocrystals of different shapes. Chem. Rev. 2005, 105, 1025–1102. [Google Scholar] [CrossRef] [PubMed]
- Sondi, I.; Salopek-Sondi, B. Silver nanoparticles as antimicrobial agent: A case study on E. coli as a model for Gram-negative bacteria. J. Colloid Interface Sci. 2004, 275, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Kuk, E.; Yu, K.N.; Kim, J.H.; Park, S.J.; Lee, H.J.; Kim, S.H.; Park, Y.K.; Park, Y.H.; Hwang, C.Y.; et al. Antimicrobial effects of silver nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2007, 3, 95–101. [Google Scholar] [CrossRef]
- Chernousova, S.; Epple, M. Silver as antibacterial agent: Ion, nanoparticle, and metal. Angew. Chem. Int. Ed. 2012, 51, 2–20. [Google Scholar] [CrossRef]
- Rai, M.; Yadav, A.; Gade, A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol. Adv. 2009, 27, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Rai, M.K.; Deshmukh, S.D.; Ingle, A.P.; Gade, A.K. Silver nanoparticles: The powerful nanoweapon against multidrug-resistant bacteria. J. Appl. Microbiol. 2012, 112, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Galdiero, S.; Falanga, A.; Vitiello, M.; Cantisani, M.; Marra, V.; Galdiero, M. Silver nanoparticles as potential antiviral agents. Molecules 2011, 16, 8894–8918. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Peng, H.; Huang, W.; Zhou, Y.; Yan, D. Facile preparation and characterization of highly antimicrobial colloid Ag or Au nanoparticles. J. Colloid Interface Sci. 2008, 325, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Lok, C.N.; Ho, C.M.; Chen, R.; He, Q.Y.; Yu, W.Y.; Sun, H.Z.; Tam, P.K.H.; Chiu, J.F.; Che, C.M. Proteomic analysis of the mode of antibacterial action of silver nanoparticles. J. Proteome. Res. 2006, 5, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Silver, S. Bacterial silver resistance: Molecular biology and uses and misuses of silver compounds. FEMS Microbiol. Rev. 2003, 27, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.L.; Wu, J.; Chen, G.Q.; Cui, F.Z.; Kim, T.N.; Kim, J.O. A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J. Biomed. Mater. Res. 2000, 52, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Choi, O.; Deng, K.K.; Kim, N.J.; Ross, L.; Surampalli, R.Y.; Hu, Z.Q. The inhibitory effects of silver nanoparticles, silver ions, and silver chloride colloids on microbial growth. Water Res. 2008, 42, 3066–3074. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Castanon, G.A.; Nino-Martinez, N.; Martinez-Gutierrez, F.; Martinez-Mendoza, J.R.; Ruiz, F. Synthesis and antibacterial activity of silver nanoparticles with different sizes. J. Nanopart. Res. 2008, 10, 1343–1348. [Google Scholar] [CrossRef]
- Pal, S.; Tak, Y.K.; Song, J.M. Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the Gram-negative bacterium Escherichia coli. Appl. Environ. Microbiol. 2007, 73, 1712–1720. [Google Scholar] [CrossRef] [PubMed]
- Radzig, M.A.; Nadtochenko, V.A.; Koksharova, O.A.; Kiwi, J.; Lipasova, V.A.; Khmel, I.A. Antibacterial effects of silver nanoparticles on Gram-negative bacteria: Influence on the growth and biofilms formation, mechanisms of action. Colloids Surf. B 2013, 102, 300–306. [Google Scholar] [CrossRef]
- Inoue, Y.; Hoshino, M.; Takahashi, H.; Noguchi, T.; Murata, T.; Kanzaki, Y.; Hamashima, H.; Sasatsu, M. Bactericidal activity of Ag-zeolite mediated by reactive oxygen species under aerated conditions. J. Inorg. Biochem. 2002, 92, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Duran, N.; Marcato, P.D.; De Souza, G.I.H.; Alves, O.L.; Esposito, E. Antibacterial effect of silver nanoparticles produced by fungal process on textile fabrics and their effluent treatment. J. Biomed. Nanotechnol. 2007, 3, 203–208. [Google Scholar] [CrossRef]
- Tankhiwale, R.; Bajpai, S.K. Graft copolymerization onto cellulose-based filter paper and its further development as silver nanoparticles loaded antibacterial food-packaging material. Colloids Surf. B 2009, 69, 164–168. [Google Scholar] [CrossRef]
- Sharma, V.K.; Yngard, R.A.; Lin, Y. Silver nanoparticles: Green synthesis and their antimicrobial activities. Adv. Colloid Interface Sci. 2009, 145, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.Z.; Yuan, Q.; Yang, X.R. Preparation and characterization of metal-chitosan nanocomposites. Colloids Surf. B 2004, 39, 31–37. [Google Scholar] [CrossRef]
- Liu, Y.S.; Chen, S.M.; Zhong, L.; Wu, G.Z. Preparation of high-stable silver nanoparticle dispersion by using sodium alginate as a stabilizer under gamma radiation. Radiat. Phys. Chem. 2009, 78, 251–255. [Google Scholar] [CrossRef]
- Kasyutich, O.; Ilari, A.; Fiorillo, A.; Tatchev, D.; Hoell, A.; Ceci, P. Silver ion incorporation and nanoparticle formation inside the cavity of Pyrococcus furiosus ferritin: Structural and size-distribution analyses. J. Am. Chem. Soc. 2010, 132, 3621–3627. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Zhang, R.C.; Ni, Y.X.; Du, Q.A.; Wang, X.; Zhang, J.L.; Li, W. Catalytic performance of Ag nanoparticles templated by polymorphic DNA. Catal. Lett. 2010, 139, 145–150. [Google Scholar] [CrossRef]
- Petty, J.T.; Zheng, J.; Hud, N.V.; Dickson, R.M. DNA-templated Ag nanocluster formation. J. Am. Chem. Soc. 2004, 126, 5207–5212. [Google Scholar] [CrossRef] [PubMed]
- Molotsky, T.; Tamarin, T.; Ben Moshe, A.; Markovich, G.; Kotlyar, A.B. Synthesis of chiral silver clusters on a DNA template. J. Phys. Chem. C 2010, 114, 15951–15954. [Google Scholar] [CrossRef]
- Lan, G.Y.; Chen, W.Y.; Chang, H.T. Control of synthesis and optical properties of DNA templated silver nanoclusters by varying DNA length and sequence. RSC Adv. 2011, 1, 802–807. [Google Scholar] [CrossRef]
- Zhang, X.L.; Yu, M.; Liu, J.H.; Li, S.M. Fabrication and characterization of Ag nanoparticles based on plasmid DNA as templates. Mater. Lett. 2011, 65, 719–721. [Google Scholar] [CrossRef]
- Zinchenko, A.A.; Baigl, D.M.; Chen, N.; Pyshkina, O.; Endo, K.; Sergeyev, V.G.; Yoshikawa, K. Conformational behavior of giant DNA through binding with Ag+ and metallization. Biomacromolecules 2008, 9, 1981–1987. [Google Scholar] [CrossRef] [PubMed]
- Takeshima, T.; Sun, L.; Wang, Y.; Yamada, Y.; Nishi, N.; Yonezawa, T.; Fugetsu, B. Salmon milt DNA as a template for the mass production of Ag nanoparticles. Polym. J. 2014, 46, 36–41. [Google Scholar] [CrossRef]
- JIS L 1902:2008. In Testing for Antimicrobial Activity and Efficacy on Textile Products; Japanese Industrial Standards; Japanese Industrial Standards Committee: Tokyo, Japan, 2008.
- JEC301. In The Certification Standards of Antibacterial Finished Textile Products; Japan Textile Evaluation Technology Council: Osaka, Japan, 2012.
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takeshima, T.; Tada, Y.; Sakaguchi, N.; Watari, F.; Fugetsu, B. DNA/Ag Nanoparticles as Antibacterial Agents against Gram-Negative Bacteria. Nanomaterials 2015, 5, 284-297. https://doi.org/10.3390/nano5010284
Takeshima T, Tada Y, Sakaguchi N, Watari F, Fugetsu B. DNA/Ag Nanoparticles as Antibacterial Agents against Gram-Negative Bacteria. Nanomaterials. 2015; 5(1):284-297. https://doi.org/10.3390/nano5010284
Chicago/Turabian StyleTakeshima, Tomomi, Yuya Tada, Norihito Sakaguchi, Fumio Watari, and Bunshi Fugetsu. 2015. "DNA/Ag Nanoparticles as Antibacterial Agents against Gram-Negative Bacteria" Nanomaterials 5, no. 1: 284-297. https://doi.org/10.3390/nano5010284
APA StyleTakeshima, T., Tada, Y., Sakaguchi, N., Watari, F., & Fugetsu, B. (2015). DNA/Ag Nanoparticles as Antibacterial Agents against Gram-Negative Bacteria. Nanomaterials, 5(1), 284-297. https://doi.org/10.3390/nano5010284