Gated Nanosensor for Sulphate-Reducing Bacteria Detection
Abstract
1. Introduction
2. Materials and Methods
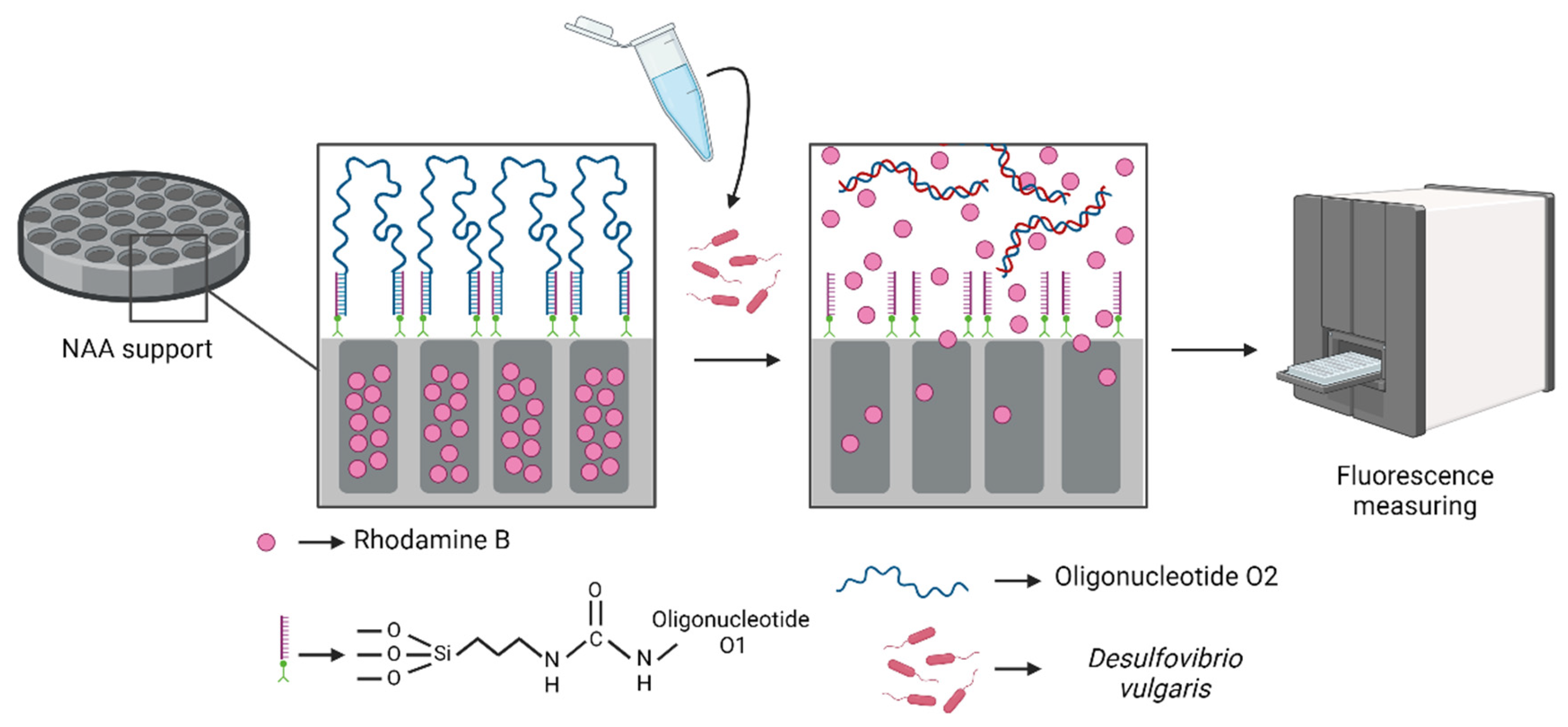
2.1. Probe Design
2.2. Chemical Reagents
2.3. Microscopy and Spectroscopy
2.4. Fabrication of Oligonucleotide-Capped Materials
2.5. Cargo Quantification
2.6. Detection Procedure
2.7. Sensitivity Analysis
2.8. Selectivity Analysis
2.9. Testing System’s Performance in Water Samples
3. Results and Discussion
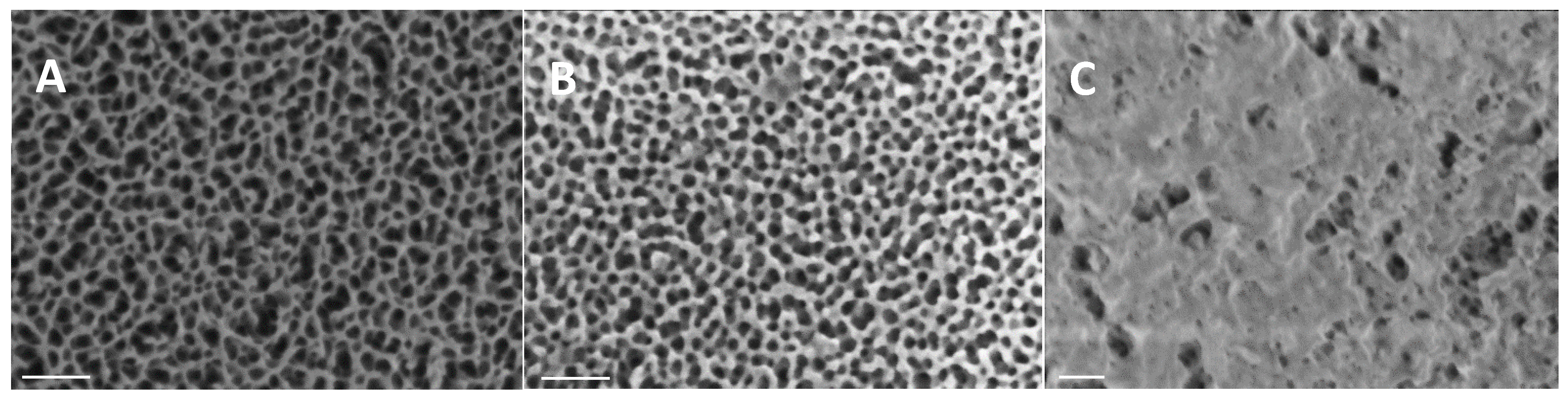
3.1. Fabrication and Characterization of Sensors
3.2. Release Assays
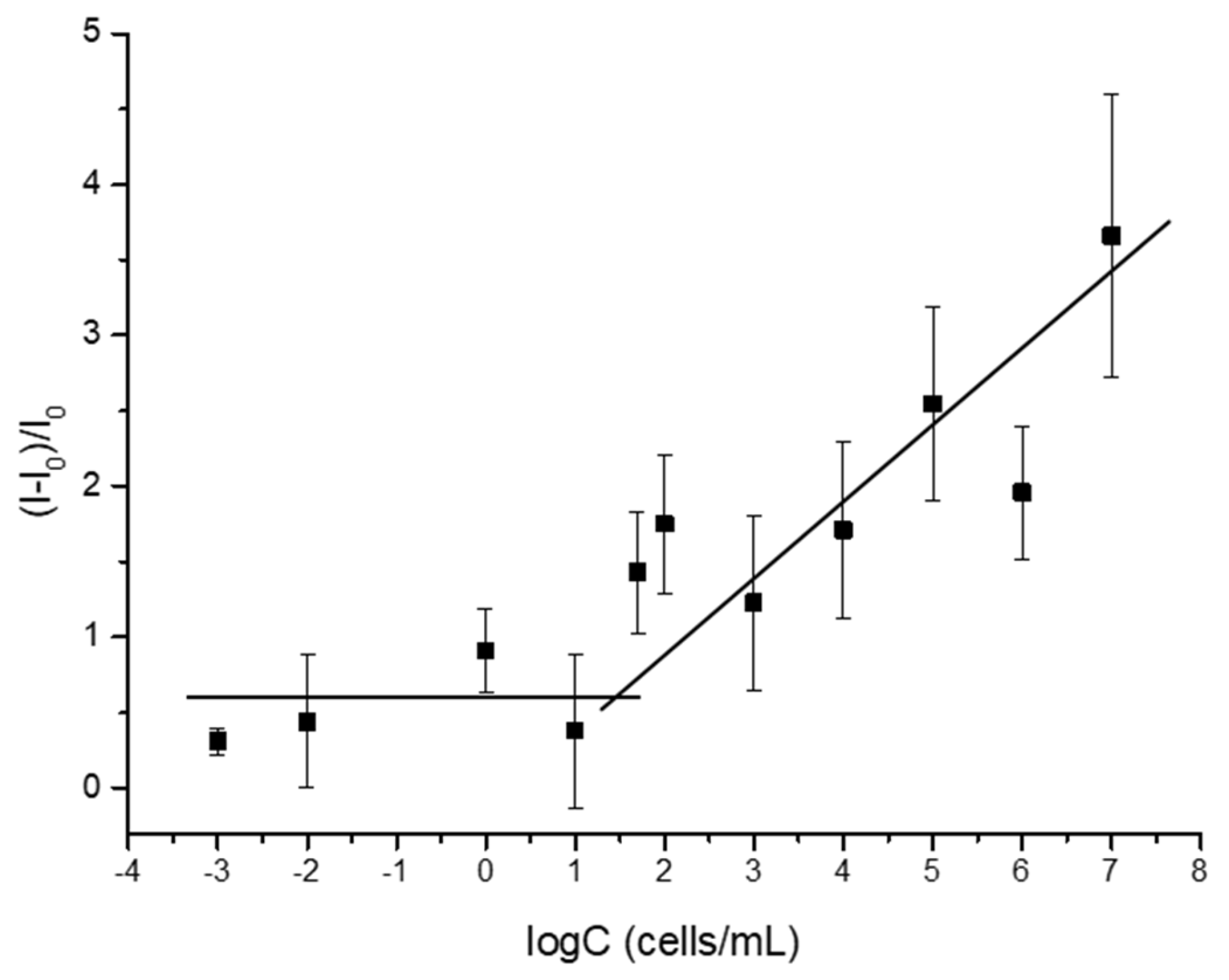
3.3. Sensitivity Assay
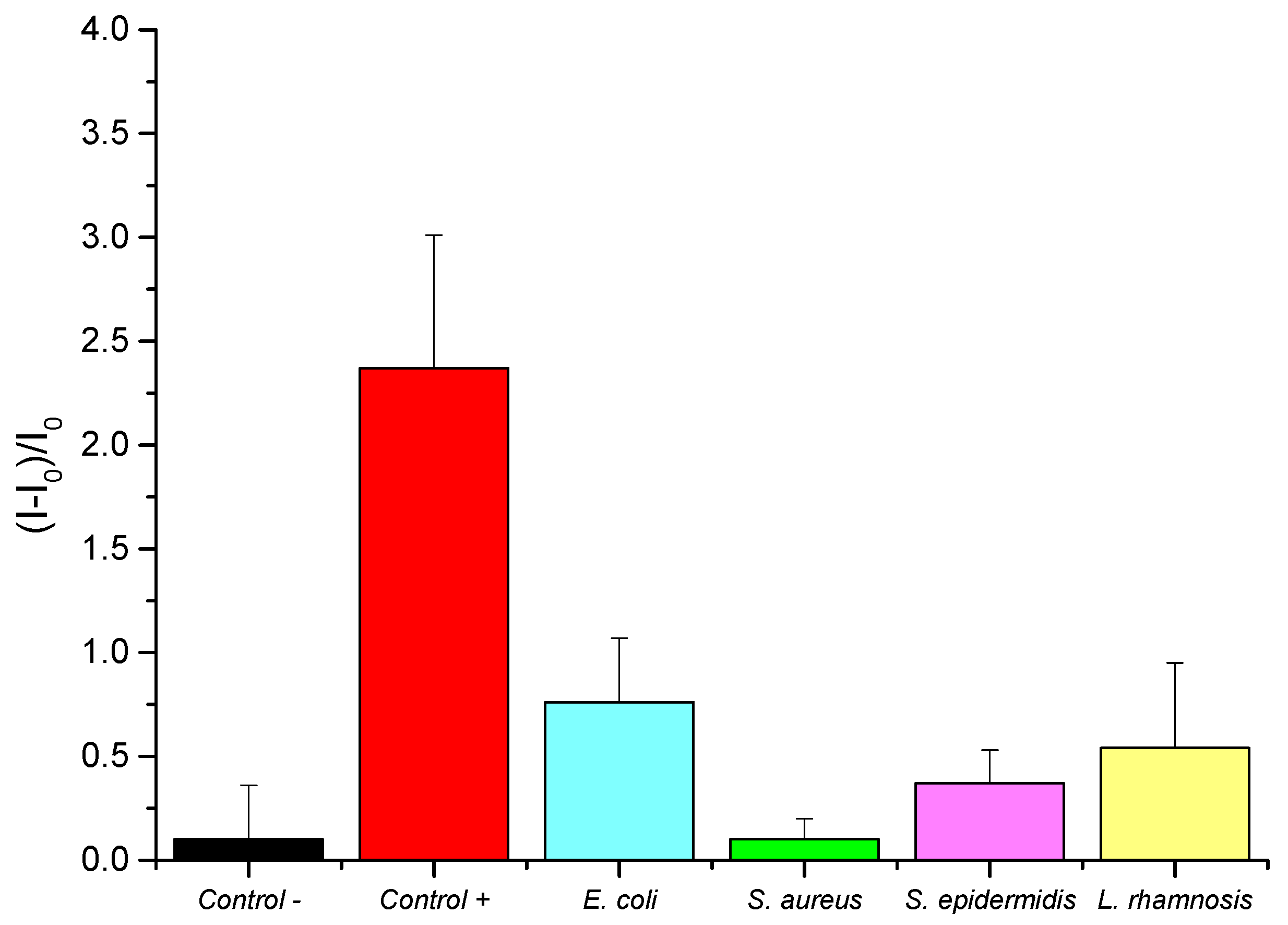
3.4. Selectivity Analysis
3.5. Performance Analysis in Inoculated Water Samples
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MIC | Microbiologically influenced corrosion |
| SRB | Sulphate-reducing bacteria |
| ATP | Adenosine triphosphate |
| RNA | Ribonucleic acid |
| DNA | Deoxyribonucleic acid |
| PCR | Polymerase chain reaction |
| NAA | Nanoporous anodic alumina |
References
- Kumar, M.A.; Kasirajan, K.T.; Parthiban, K. Production and characterization of exopolysaccharides (EPS) from biofilm forming marine bacterium. Braz. Arch. Biol. Technol. 2011, 54, 259–265. [Google Scholar] [CrossRef]
- Marschall, C.; Frenzel, P.; Cypionka, H. Influence of oxygen on sulfate reduction and growth of sulfate-reducing bacteria. Arch. Microbiol. 1993, 159, 168–173. [Google Scholar] [CrossRef]
- Eguía-López, E.; Trueba-Ruiz, A.; Río-Calonge, B.; Girón-Portilla, M.A.; Bielva-Tejera, C. Recent studies on antifouling systems to artificial structures in marine ecosystem. J. Marit. Res. 2006, 3, 73–89. [Google Scholar]
- Beech, I.B.; Sunner, J. Biocorrosion: Towards understanding interactions between biofilms and metals. Curr. Opin. Biotechnol. 2004, 15, 181–186. [Google Scholar] [CrossRef]
- Zadvorny, O.A.; Zorin, N.A.; Gogotoy, I.N. Transformation of metals and metal ions by hydrogenases from phototrophic bacteria. Arch. Microbiol. 2006, 184, 279–285. [Google Scholar] [CrossRef]
- Yuan, S.; Liang, B.; Zhao, Y.; Pehkonen, S.O. Surface chemistry and corrosion behavior of 304 stainless steel in simulated seawater containing inorganic sulphide and sulphate-reducing bacteria. Corros. Sci. 2013, 74, 353–366. [Google Scholar] [CrossRef]
- Ben-Dov, E.; Brenner, A.; Kushmaro, A. Quantification of Sulfate-reducing Bacteria in Industrial Wastewater, by Real-time Polymerase Chain Reaction (PCR) Using dsrA and apsA Genes. Microb. Ecol. 2007, 54, 439–451. [Google Scholar] [CrossRef]
- Gibson, G.R.; Cummings, J.H.; Macfarlane, G.T. Growth and activities of sulphate-reducing bacteria in gut contents of healthy subjects and patients with ulcerative colitis. FEMS Microbiol. Lett. 1991, 86, 103–111. [Google Scholar] [CrossRef]
- Klein, M.; Friedrich, M.; Roger, A.J.; Hugenholtz, P.; Fishbain, S.; Abicht, H.; Blackall, L.L.; Stahl, D.A.; Wagner, M. Multiple Lateral Transfers of Dissimilatory Sulfite Reductase Genes between Major Lineages of Sulfate-Reducing Prokaryotes. J. Bacteriol. 2001, 183, 6028–6035. [Google Scholar] [CrossRef]
- Tanner, R.S. Monitoring sulfate-reducing bacteria: Comparison of enumeration media. J. Microbiol. Methods 1989, 10, 83–90. [Google Scholar] [CrossRef]
- Daly, K.; Sharp, R.J.; McCarthy, A.J. Development of oligonucleotide probes and PCR primers for detecting phylogenetic subgroups of sulfate-reducing bacteria. Microbiology 2000, 146, 1693–1705. [Google Scholar] [CrossRef] [PubMed]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef] [PubMed]
- Beale, D.J.; Karpe, A.V.; Jadhav, S.; Muster, T.H.; Palombo, E.A. Omics-based approaches and their use in the assessment of microbial-influenced corrosion of metals. Corros. Rev. 2015, 34, 1–15. [Google Scholar] [CrossRef]
- Kondo, R.; Nedwell, D.B.; Purdy, K.J.; Queiroz-Silva, S. Detection and Enumeration of Sulphate-Reducing Bacteria in Estuarine Sediments by Competitive PCR. Geomicrobiol. J. 2004, 21, 145–157. [Google Scholar] [CrossRef]
- Purdy, K.J.; Nedwell, D.; Embley, T.M.; Takii, S. Use of 16S rRNA-targeted oligonucleotide probes to investigate the occurrence and selection of sulfate-reducing bacteria in response to nutrient addition to sediment slurry microcosms from a Japanese estuary. FEMS Microbiol. Ecol. 1997, 24, 221–234. [Google Scholar] [CrossRef]
- Salimiyan rizi, K.; Ashrafi, A. Biosensors, mechatronics, & microfluidics for early detection & monitoring of microbial corrosion: A comprehensive critical review. Results Mater. 2023, 18, 100402. [Google Scholar] [CrossRef]
- Chen, S.; Cheng, Y.F.; Voordouw, G. Three-dimensional graphene nanosheet doped with gold nanoparticles as electrochemical DNA biosensor for bacterial detection. Sens. Actuators B Chem. 2018, 262, 860–868. [Google Scholar] [CrossRef]
- Asif, M.; Ashraf, G.; Aziz, A.; Iftikhar, T.; Wang, Z.; Xiao, F.; Sun, Y. Tuning the Redox Chemistry of Copper Oxide Nanoarchitectures Integrated with rGOP via Facet Engineering: Sensing H2S toward SRB Detection. ACS Appl. Mater. Interfaces 2022, 14, 19480–19490. [Google Scholar] [CrossRef]
- Lopes, R.N.; Rodrigues, D.M.C.; Allil, R.C.S.B.; Werneck, M.M. Plastic optical fiber immunosensor for fast detection of sulfate-reducing bacteria. Measurement 2018, 125, 377–385. [Google Scholar] [CrossRef]
- Qi, P.; Zhang, D.; Zeng, Y.; Wan, Y. Biosynthesis of CdS nanoparticles: A fluorescent sensor for sulfate-reducing bacteria detection. Talanta 2016, 147, 142–146. [Google Scholar] [CrossRef]
- Zheng, L.; Ye, X.; Qi, P.; Zhang, D.; Sun, Y. Fluorometric detection of sulfate-reducing bacteria via the aggregation-induced emission of glutathione-gold(I) complexe. Microchim. Acta 2019, 186, 382. [Google Scholar] [CrossRef] [PubMed]
- Vigneshvar, S.; Sudhakumari, C.C.; Septhilkumaran, B.; Prakash, H. Recent Advances in Biosensor Technology for Potential Applications—An Overview. Front. Bioeng. Biotechnol. 2016, 4, 11. [Google Scholar] [CrossRef]
- Aznar, E.; Oroval, M.; Pascual, L.; Murguía, J.R.; Martínez-Máñez, R.; Sancenón, F. Gated materials for on-command release of guest molecules. Chem. Rev. 2016, 116, 561–718. [Google Scholar] [CrossRef]
- Pascual, L.; Baroja, I.; Aznar, E.; Sancenón, F.; Marcos, M.D.; Murguía, J.R.; Amorós, P.; Rurack, K.; Martínez-Máñez, R. Oligonucleotide-capped mesoporous silica nanoparticles as DNA-responsive dye delivery systems for genomic DNA detection. Chem. Commun. 2015, 51, 1414–1416. [Google Scholar] [CrossRef]
- García-Fernández, A.; Aznar, E.; Martínez-Máñez, R.; Sancenón, F. New Advances in In Vivo Applications of Gated Mesoporous Silica as Drug Delivery Nanocarriers. Small 2020, 16, 1902242. [Google Scholar] [CrossRef]
- Llopis-Lorente, A.; Lozano-Torres, B.; Bernardos, A.; Martínez-Máñez, R.; Sancenón, F. Mesoporous silica materials for controlled delivery based on enzymes. J. Mater. Chem. B 2017, 5, 3069–3083. [Google Scholar] [CrossRef]
- Sancenón, F.; Pascual, L.; Oroval, M.; Aznar, E.; Martínez-Máñez, R. Gated silica mesoporous materials in sensing applications. ChemistryOpen 2015, 4, 418–437. [Google Scholar] [CrossRef]
- Pla, L.; Lozano-Torres, B.; Martínez-Máñez, R.; Sancenón, F.; Ros-Lis, J.V. Overview of the Evolution of Silica-Based Chromo-Fluorogenic Nanosensors. Sensors 2019, 19, 5138. [Google Scholar] [CrossRef]
- Castillo, R.R.; Baeza, A.; Vallet-Regí, M. Recent applications of the combination of mesoporous silica nanoparticles with nucleic acids: Development of bioresponsive devices, carriers and sensors. Biomater. Sci. 2017, 5, 353–377. [Google Scholar] [CrossRef]
- Ribes, À.; Santiago-Felipe, S.; Bernardos, A.; Marcos, M.D.; Pardo, T.; Sancenón, F.; Aznar, E. Two new fluorogenic aptasensors based on capped mesoporous silica nanoparticles to detect ochratoxin A. ChemistryOpen 2017, 6, 653–659. [Google Scholar] [CrossRef]
- Climent, E.; Martínez-Máñez, R.; Sancenón, F.; Marcos, M.D.; Soto, J.; Maquieira, A.; Amorós, P. Controlled Delivery Using Oligonucleotide-Capped Mesoporous Silica Nanoparticles. Angew. Chem. Int. Ed. 2010, 49, 7281–7283. [Google Scholar] [CrossRef] [PubMed]
- Baranowska, M.; Slota, A.J.; Eravuchira, P.J.; Macias, G.; Xifré-Pérez, E.; Pallares, J.; Marsal, L.F. Protein attachment to nanoporous anodic alumina for biotechnological applications: Influence of pore size, protein size and functionalization path. Colloids Surf. B Biointerfaces 2014, 122, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Kumeria, T.; Losic, D. Nanoporous Anodic Alumina: A Versatile Platform for Optical Biosensors. Materials 2014, 7, 4297–4320. [Google Scholar] [CrossRef]
- Rajeev, G.; Prieto Simon, B.; Marsal, L.F.; Voelcker, N.H. Advances in Nanoporous Anodic Alumina-Based Biosensors to Detect Biomarkers of Clinical Significance: A Review. Adv. Healthc. Mater. 2018, 7, 1700904. [Google Scholar] [CrossRef]
- Alberti, S.; Soler-Illi, G.J.A.A.; Azzaroni, O. Gated supramolecular chemistry in hybrid mesoporous silica nanoarchitectures: Controlled delivery and molecular transport in response to chemical, physical and biological stimuli. Chem. Commun. 2015, 51, 6050–6075. [Google Scholar] [CrossRef]
- Lee, W.; Park, S.J. Porous Anodic Aluminum Oxide: Anodization and Templated Synthesis of Functional Nanostructures. Chem. Rev. 2014, 114, 7487–7556. [Google Scholar] [CrossRef]
- Ribes, A.; Santiago-Felipe, S.; Avino, A.; Candela-Noguera, V.; Eritja, R.; Sancenón, F.; Aznar, E. Design of oligonucleotide-capped mesoporous silica nanoparticles for the detection of miRNA-145 by duplex and triplex formation. Sens. Actuators B Chem. 2018, 277, 598–603. [Google Scholar] [CrossRef]
- Garrido-Cano, I.; Pla, L.; Santiago-Felipe, S.; Simon, S.; Ortega, B.; Bermejo, B.; Martínez-Máñez, R. Nanoporous anodic alumina-based sensor for miR-99a-5p detection as an effective early breast cancer diagnostic tool. ACS Sens. 2021, 6, 1022–1029. [Google Scholar] [CrossRef]
- Climent, E.; Mondragón, L.; Martínez-Máñez, R.; Sancenón, F.; Marcos, M.D.; Murguía, J.R.; Amorós, P.; Rurack, K.; Pérez-Payá, E. Selective, highly sensitive, and rapid detection of genomic DNA by using gated materials: Mycoplasma detection. Angew. Chem. Int. Ed. 2013, 52, 8938–8942. [Google Scholar] [CrossRef]
- Pla, L.; Xifré-Pérez, E.; Ribes, À.; Aznar, E.; Marcos, M.D.; Marsal, L.F.; Sancenón, F. A mycoplasma genomic DNA probe using gated nanoporous anodic alumina. ChemPlusChem 2017, 82, 337–341. [Google Scholar] [CrossRef]
- Ribes, À.; Aznar, E.; Santiago-Felipe, S.; Xifre-Perez, E.; Tormo-Mas, M.À.; Peman, J.; Martínez-Máñez, R. Selective and sensitive probe based in oligonucleotide-capped nanoporous alumina for the rapid screening of infection produced by Candida albicans. ACS Sens. 2019, 4, 1291–1298. [Google Scholar] [CrossRef] [PubMed]
- Pla, L.; Santiago-Felipe, S.; Tormo-Mas, M.Á.; Pemán, J.; Sancenón, F.; Aznar, E.; Martínez-Máñez, R. Aptamer-Capped nanoporous anodic alumina for Staphylococcus aureus detection. Sens. Actuators B Chem. 2020, 320, 128281. [Google Scholar] [CrossRef]
- Pla, L.; Santiago-Felipe, S.; Tormo-Mas, M.À.; Ruiz-Gaitán, A.; Pemán, J.; Valentín, E.; Sancenón, F.; Aznar, E.; Martínez-Máñez, R. Oligonucleotide-capped nanoporous anodic alumina biosensor as diagnostic tool for rapid and accurate detection of Candida auris in clinical samples. Emerg. Microbes Infect. 2021, 10, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Caballos, I.; Aranda, M.N.; López-Palacios, A.; Pla, L.; Santiago-Felipe, S.; Hernández-Montoto, A.; Tormo-Mas, M.À.; Pemán, J.; Gómez-Ruiz, M.D.; Calabuig, E.; et al. Aptamer-Capped Nanoporous Anodic Alumina for SARS-CoV-2 Spike Protein Detection. Adv. Mater. Technol. 2023, 8, 2201913. [Google Scholar] [CrossRef]
- López-Palacios, A.; Aranda, M.N.; Caballos, I.; Hernández-Montoto, A.; Calabuig, E.; Gómez-Ruiz, M.D.; Tormo-Mas, M.Á.; Pemán, J.; Sancenón, F.; Martínez-Máñez, R.; et al. SARS-COV-2 viral RNA detection through oligonucleotide-capped nanoporous anodic alumina supports. Sens Actuators Rep. 2025, 9, 100298. [Google Scholar] [CrossRef]
- Hernández-Montoto, A.; Aranda, M.N.; Caballos, I.; López-Palacios, A.; Tormo-Mas, M.À.; Pemán, J.; Prieto-Rodríguez, M.; Picornell, C.; Aznar, E.; Martínez-Máñez, R. Human Papilloma Virus DNA Detection in Clinical Samples Using Fluorogenic Probes Based on Oligonucleotide Gated Nanoporous Anodic Alumina Films. Adv. Healthc. Mater. 2023, 12, 2203326. [Google Scholar] [CrossRef]
- Hansen, T.A. Metabolism of sulfate-reducing prokaryotes. Antonie Van Leeuwenhoek 1994, 66, 165–185. [Google Scholar] [CrossRef]
- Heildelberg, J.F.; Seshadri, R.; Haveman, S.A.; Hemme, C.L.; Paulsen, I.T.; Kolonay, J.F.; Eisen, J.A.; Ward, N.; Methe, B.; Brinkac, L.M.; et al. The genoma sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 2004, 22, 554–559. [Google Scholar] [CrossRef]
- Marangoni, P.R.D.; Robl, D.; Berton, M.A.C.; Garcia, C.M.; Bozza, A.; Porsani, M.V.; Dalzoto, P.R.; Vicente, V.A.; Pimentel, I.C. Occurrence of sulphate reducing bacteria (SRB) associated with biocorrosion on metallic surfaces in a hydroelectric power station in Ibirama (SC)—Brazil. Braz. Arch. Biol. Technol. 2013, 56, 801–809. [Google Scholar] [CrossRef]
- Dar, S.A.; Yao, L.; Dongen, U.; Kuenen, J.G.; Muyzer, G. Analysis of diversity and activity of sulfate-reducing bacterial communities in sulfidogenic bioreactors using 16S rRNA and dsrB genes as molecular markers. Appl. Environ. Microbiol. 2007, 73, 594–604. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Neu, H.C.; Ashman, D.F.; Price, T.D. Effect of ethylenediaminetetraacetic acid-Tris(hydroxymethyl)aminomethane on release of the acid-soluble nucleotide pool and on breakdown of ribosomal ribonucleic acid in Escherichia coli. J. Bacteriol. 1967, 93, 1360–1368. [Google Scholar] [CrossRef]
- Donohue, T.J.; Cain, B.D.; Kaplan, S. Alterations in the phospholipid composition of Rhodopseudomonas sphaeroides and other bacteria induced by Tris. J. Bacteriol. 1982, 152, 595–606. [Google Scholar] [CrossRef]
- Hancock, R.E.; Wong, P.G. Compounds which increase the permeability of the Pseudomonas aeruginosa outer membrane. Antimicrob. Agents Chemother. 1984, 26, 48–52. [Google Scholar] [CrossRef]
- Wurm, D.J.; Slouka, C.; Bosilj, T.; Herwig, C.; Spadiut, O. How to trigger periplasmic release in recombinant Escherichia coli: A comparative analysis. Eng. Life Sci. 2017, 17, 215–222. [Google Scholar] [CrossRef]
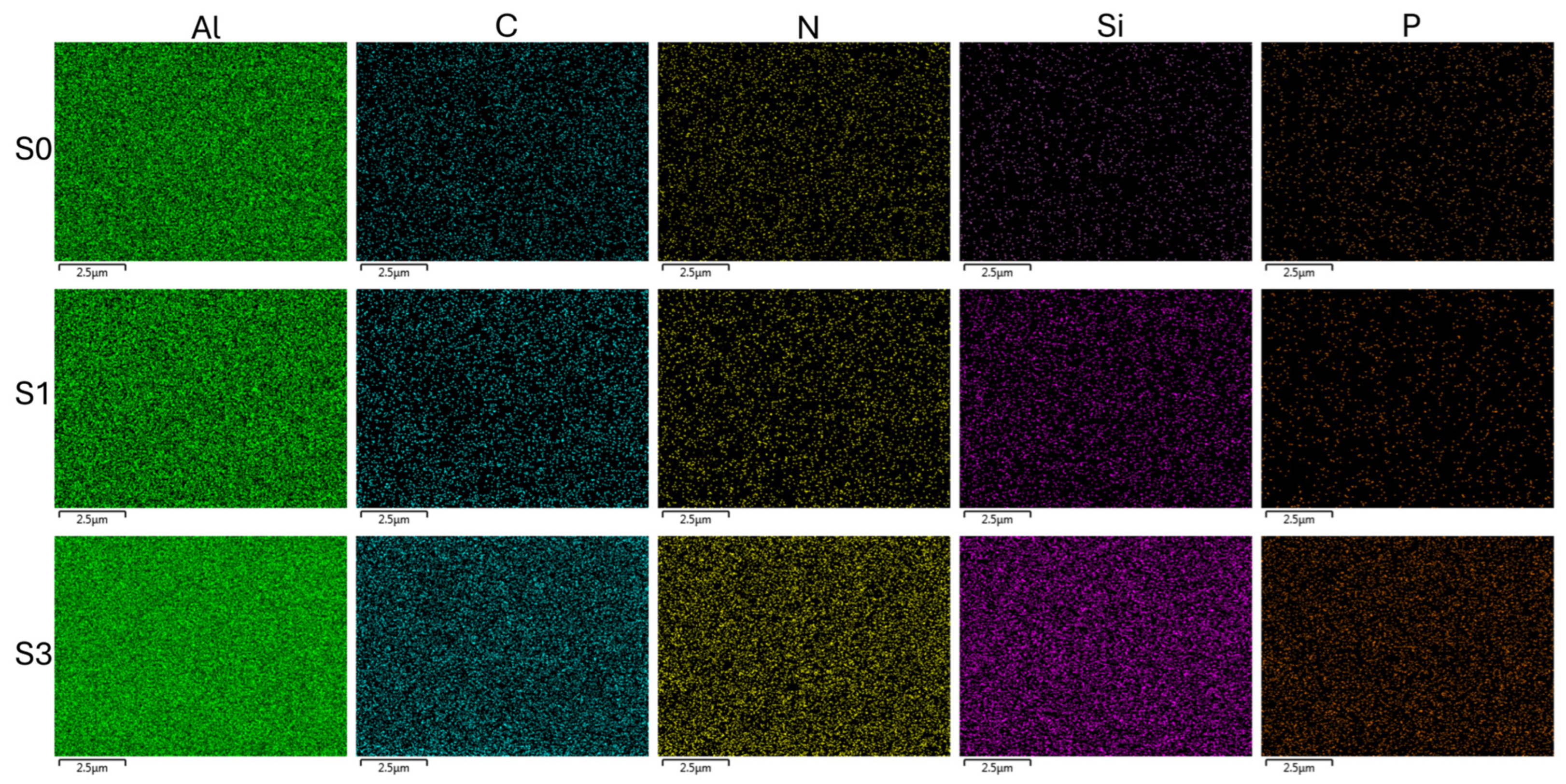


| C/Al | N/Al | Si/Al | P/Al | |
|---|---|---|---|---|
| S0 | 0.14 ± 0.01 | - | - | - |
| S1 | 0.48 ± 0.10 | 0.07 ± 0.02 | 0.10 ± 0.04 | - |
| S3 | 0.35 ± 0.01 | 0.05 ± 0.01 | 0.04 ± 0.02 | 0.04 ± 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Palacios, A.; Morellá-Aucejo, Á.; Moreno, Y.; Ponz-Carcelén, R.; Pedro-Monzonís, M.; Marcos, M.D.; Bernardos, A.; Sancenón, F.; Aznar, E.; Martínez-Máñez, R.; et al. Gated Nanosensor for Sulphate-Reducing Bacteria Detection. Nanomaterials 2025, 15, 774. https://doi.org/10.3390/nano15100774
López-Palacios A, Morellá-Aucejo Á, Moreno Y, Ponz-Carcelén R, Pedro-Monzonís M, Marcos MD, Bernardos A, Sancenón F, Aznar E, Martínez-Máñez R, et al. Gated Nanosensor for Sulphate-Reducing Bacteria Detection. Nanomaterials. 2025; 15(10):774. https://doi.org/10.3390/nano15100774
Chicago/Turabian StyleLópez-Palacios, Alba, Ángela Morellá-Aucejo, Yolanda Moreno, Román Ponz-Carcelén, María Pedro-Monzonís, M. Dolores Marcos, Andrea Bernardos, Félix Sancenón, Elena Aznar, Ramón Martínez-Máñez, and et al. 2025. "Gated Nanosensor for Sulphate-Reducing Bacteria Detection" Nanomaterials 15, no. 10: 774. https://doi.org/10.3390/nano15100774
APA StyleLópez-Palacios, A., Morellá-Aucejo, Á., Moreno, Y., Ponz-Carcelén, R., Pedro-Monzonís, M., Marcos, M. D., Bernardos, A., Sancenón, F., Aznar, E., Martínez-Máñez, R., & Hernández-Montoto, A. (2025). Gated Nanosensor for Sulphate-Reducing Bacteria Detection. Nanomaterials, 15(10), 774. https://doi.org/10.3390/nano15100774







