Graphene Oxides (GOs) with Different Lateral Dimensions and Thicknesses Affect the Molecular Response in Chironomus riparius
Abstract
1. Introduction
2. Materials and Methods
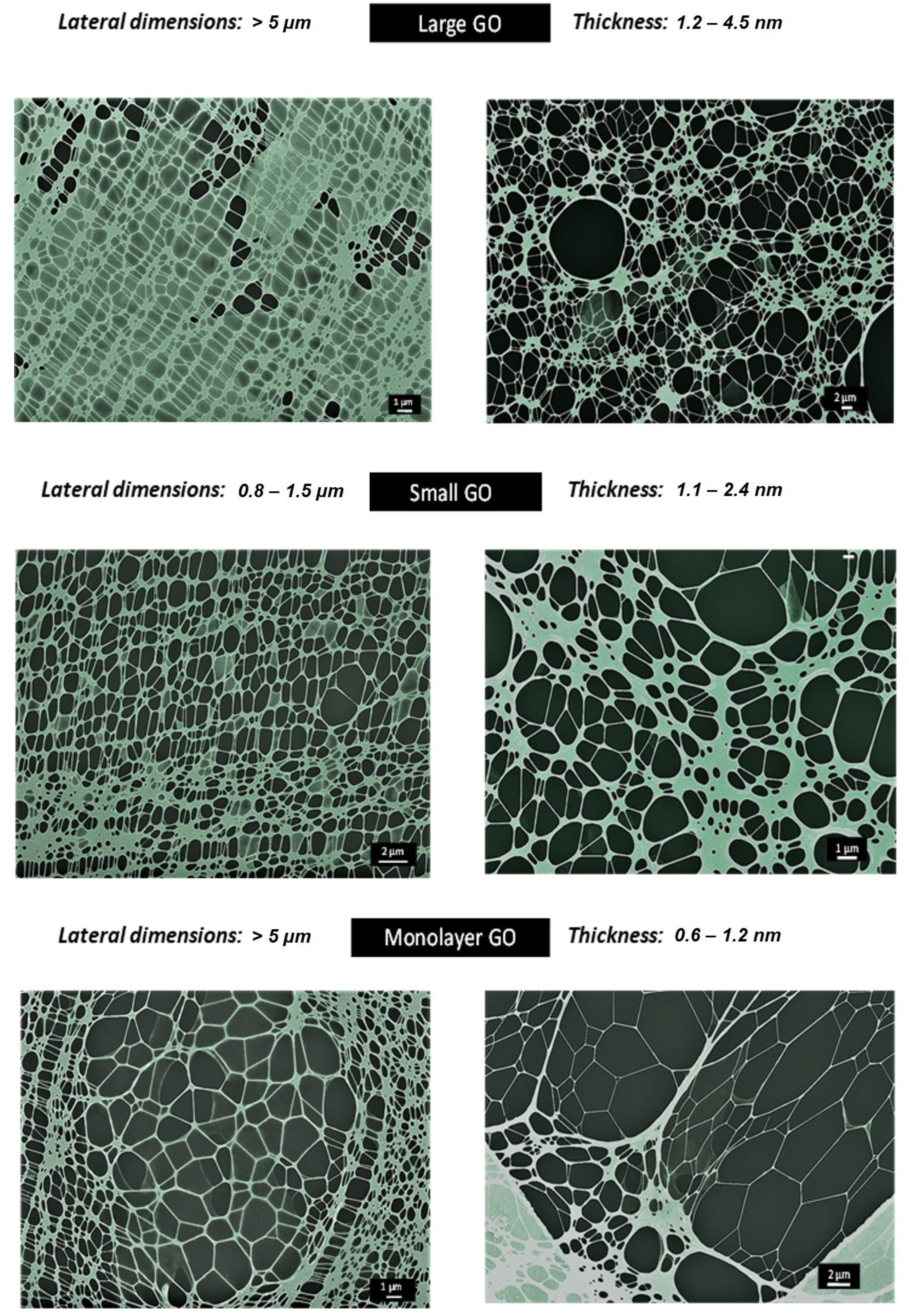
2.1. Synthesis and Characterisation of GO Materials
2.2. Animals and Treatments
2.3. RNA Extraction and Complementary DNA Synthesis
2.4. Real-Time Polymerase Chain Reaction
2.5. Statistical Analysis
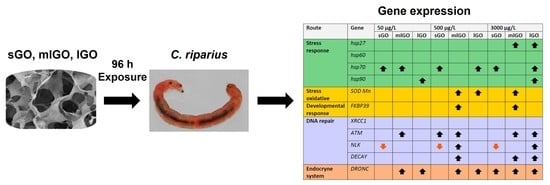
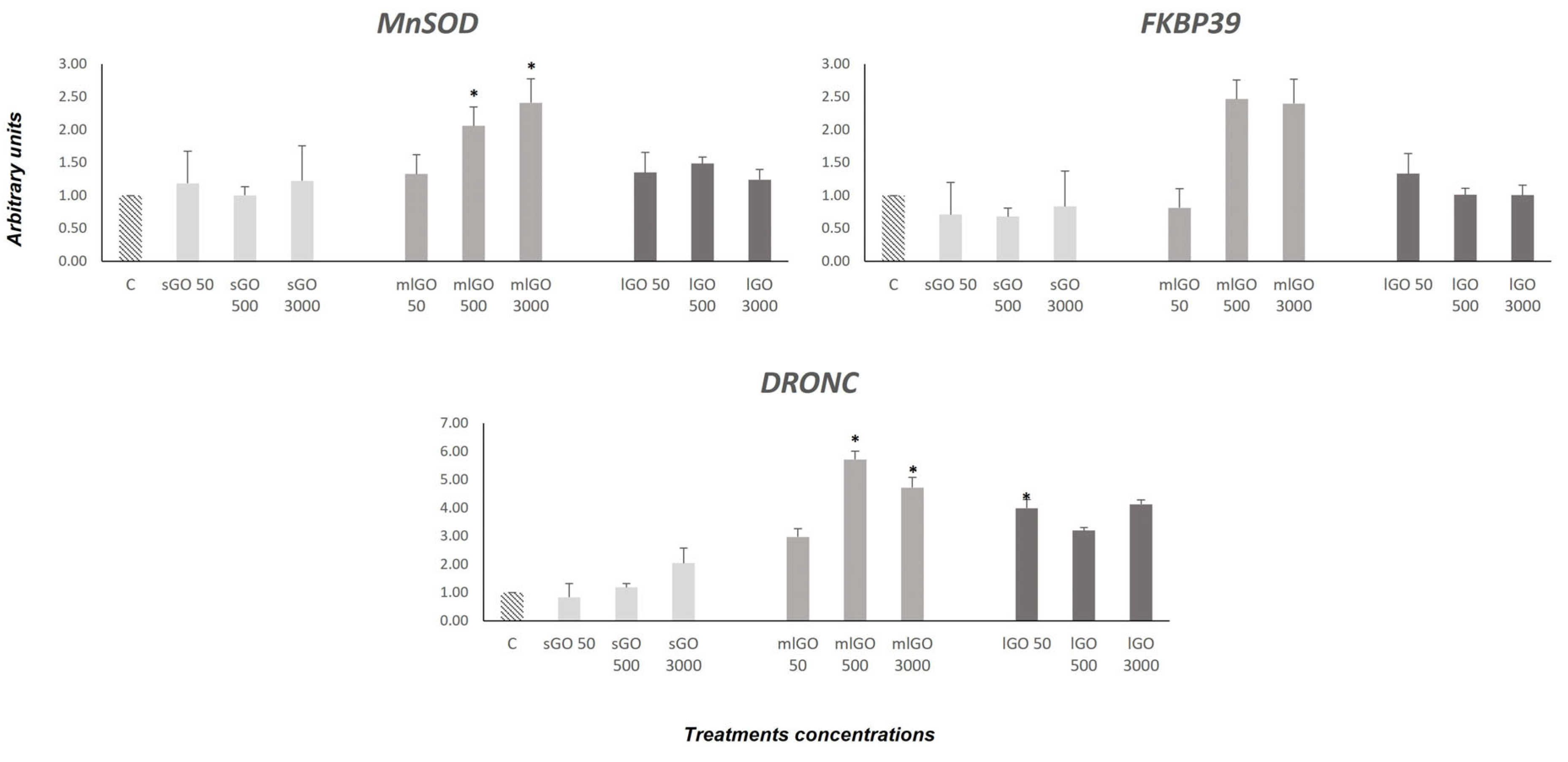
3. Results and Discussion
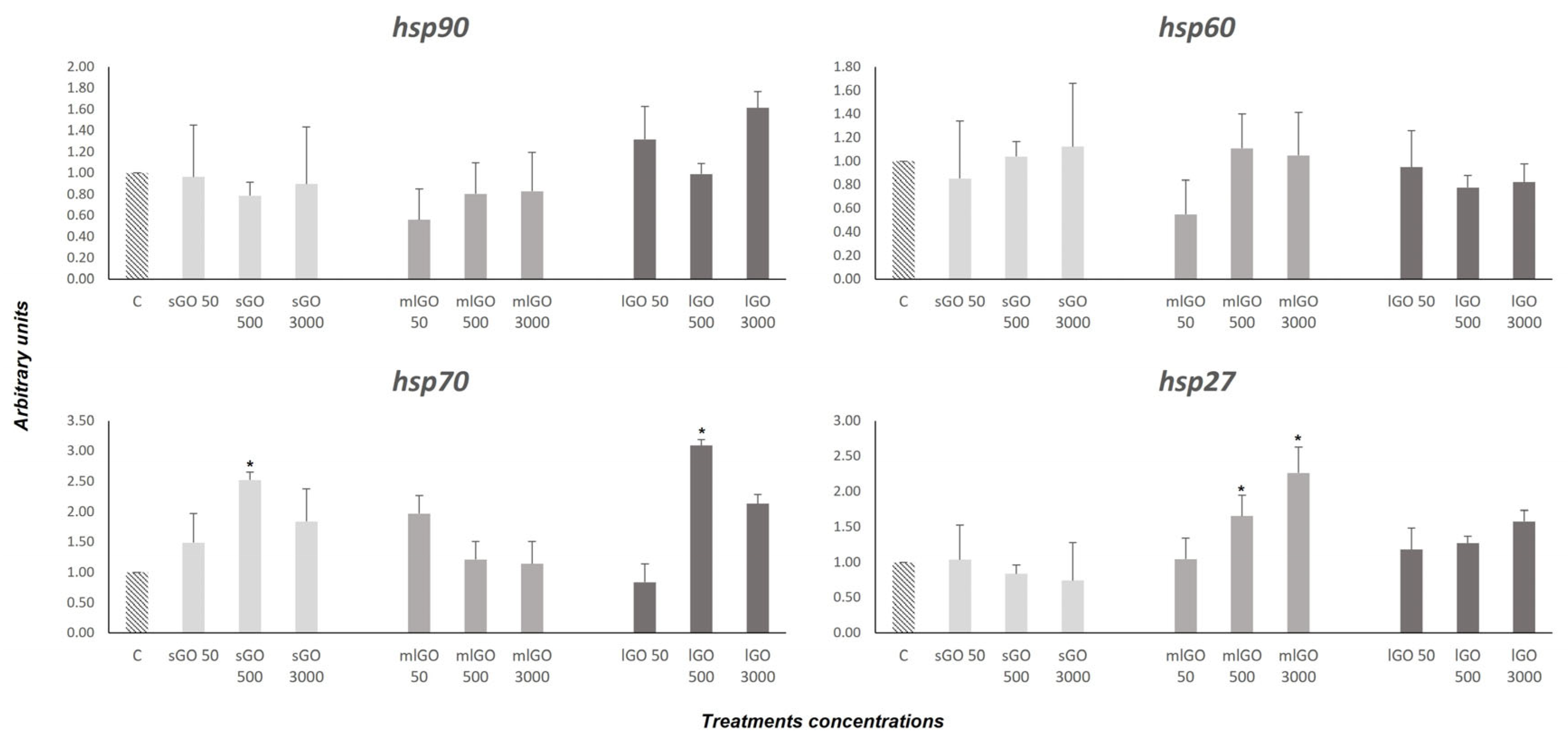
3.1. Stress Response
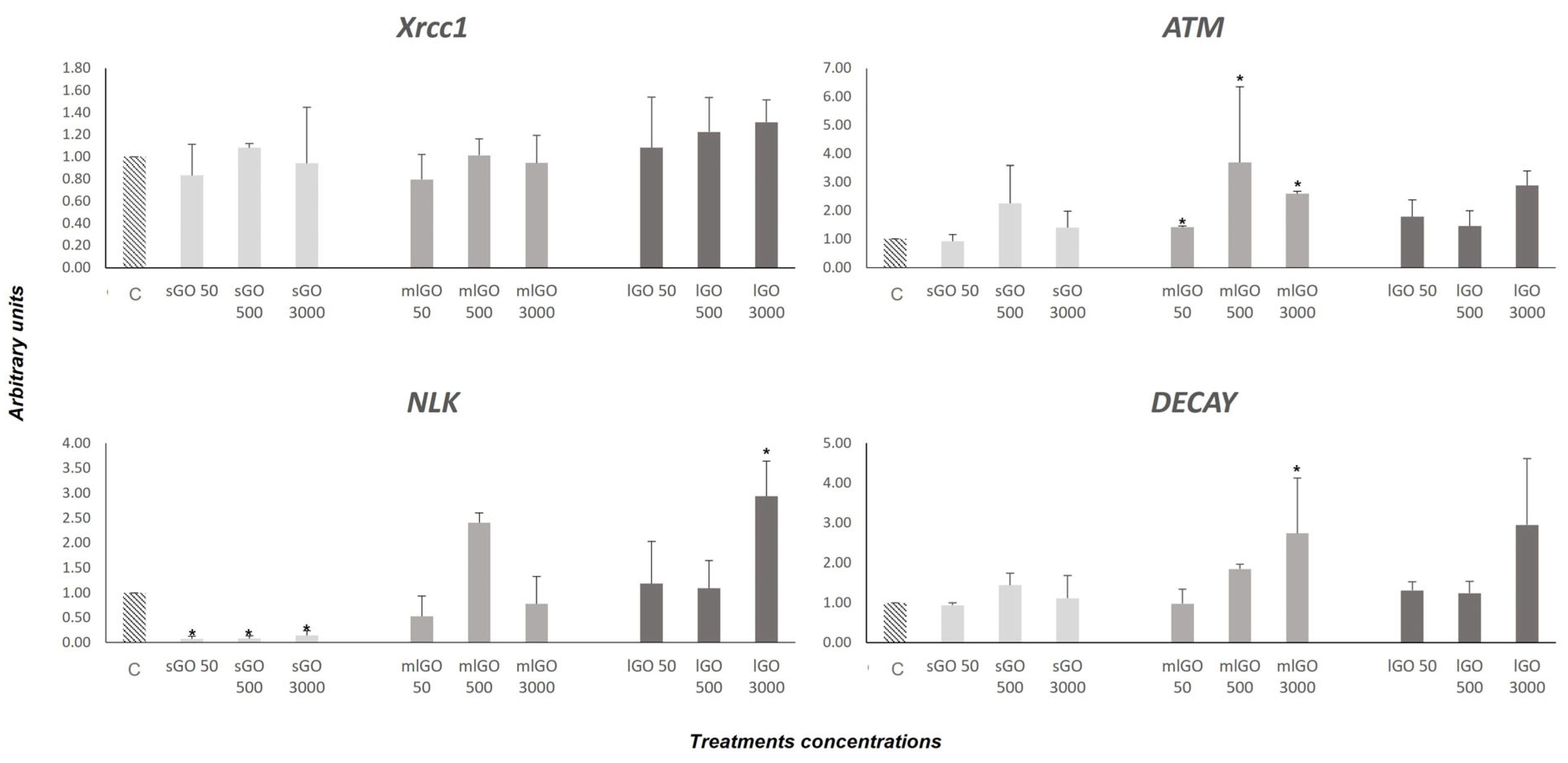
3.2. DNA Damage Response
3.3. Antioxidant, Endocrine and Developmental Responses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Esteban-Arranz, A.; Compte-Tordesillas, D.; Muñoz-Andrés, V.; Pérez-Cadenas, M.; Guerrero-Ruiz, A. Effect of Surface, Structural and Textural Properties of Graphenic Materials over Cooperative and Synergetic Adsorptions of Two Chloroaromatic Compounds from Aqueous Solution. Catal. Today 2018, 301, 104–111. [Google Scholar] [CrossRef]
- De Lázaro, I.; Vranic, S.; Marson, D.; Rodrigues, A.F.; Buggio, M.; Esteban-Arranz, A.; Mazza, M.; Posocco, P.; Kostarelos, K. Graphene Oxide as a 2D Platform for Complexation and Intracellular Delivery of SiRNA. Nanoscale 2019, 11, 13863–13877. [Google Scholar] [CrossRef] [PubMed]
- Esteban-Arranz, A.; Arranz, M.Á.; Morales, M.; Martin-Folgar, R.; Álvarez-Rodríguez, J. Thickness of Graphene Oxide-Based Materials as a Control Parameter. ChemRxiv 2021. [Google Scholar] [CrossRef]
- Arvidsson, R.; Molander, S.; Sandén, B.A. Review of Potential Environmental and Health Risks of the Nanomaterial Graphene. Hum. Ecol. Risk Assess. Int. J. 2013, 19, 873–887. [Google Scholar] [CrossRef]
- Akhavan, O.; Ghaderi, E.; Akhavan, A. Size-Dependent Genotoxicity of Graphene Nanoplatelets in Human Stem Cells. Biomaterials 2012, 33, 8017–8025. [Google Scholar] [CrossRef] [PubMed]
- Akhavan, O.; Ghaderi, E. Toxicity of Graphene and Graphene Oxide Nanowalls Against Bacteria. ACS Nano 2010, 4, 5731–5736. [Google Scholar] [CrossRef]
- Liao, K.-H.; Lin, Y.-S.; Macosko, C.W.; Haynes, C.L. Cytotoxicity of Graphene Oxide and Graphene in Human Erythrocytes and Skin Fibroblasts. ACS Appl. Mater. Interfaces 2011, 3, 2607–2615. [Google Scholar] [CrossRef]
- Liu, S.; Zeng, T.H.; Hofmann, M.; Burcombe, E.; Wei, J.; Jiang, R.; Kong, J.; Chen, Y. Antibacterial Activity of Graphite, Graphite Oxide, Graphene Oxide, and Reduced Graphene Oxide: Membrane and Oxidative Stress. ACS Nano 2011, 5, 6971–6980. [Google Scholar] [CrossRef]
- Malhotra, N.; Villaflores, O.B.; Audira, G.; Siregar, P.; Lee, J.-S.; Ger, T.-R.; Hsiao, C.-D. Toxicity Studies on Graphene-Based Nanomaterials in Aquatic Organisms: Current Understanding. Molecules 2020, 25, 3618. [Google Scholar] [CrossRef]
- Volkov, Y.; McIntyre, J.; Prina-Mello, A. Graphene Toxicity as a Double-Edged Sword of Risks and Exploitable Opportunities: A Critical Analysis of the Most Recent Trends and Developments. 2D Mater. 2017, 4, 022001. [Google Scholar] [CrossRef]
- Jia, P.-P.; Sun, T.; Junaid, M.; Yang, L.; Ma, Y.-B.; Cui, Z.-S.; Wei, D.-P.; Shi, H.-F.; Pei, D.-S. Nanotoxicity of Different Sizes of Graphene (G) and Graphene Oxide (GO) in vitro and in vivo. Environ. Pollut. 2019, 247, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-J.; Jiang, X.-F.; Junaid, M.; Ma, Y.-B.; Jia, P.-P.; Wang, H.-B.; Pei, D.-S. Graphene Oxide Nanosheets Induce DNA Damage and Activate the Base Excision Repair (BER) Signaling Pathway Both in vitro and in vivo. Chemosphere 2017, 184, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Achawi, S.; Feneon, B.; Pourchez, J.; Forest, V. Structure–Activity Relationship of Graphene-Based Materials: Impact of the Surface Chemistry, Surface Specific Area and Lateral Size on Their In Vitro Toxicity. Nanomaterials 2021, 11, 2963. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Rivers-Auty, J.; Crică, L.E.; Barr, K.; Rosano, V.; Arranz, A.E.; Loret, T.; Spiller, D.; Bussy, C.; Kostarelos, K.; et al. Dynamic Interactions and Intracellular Fate of Label-Free, Thin Graphene Oxide Sheets within Mammalian Cells: Role of Lateral Sheet Size. Nanoscale Adv. 2021, 3, 4166–4185. [Google Scholar] [CrossRef] [PubMed]
- OECD. Guidelines for the Testing of Chemicals. Acute immobilisation. Chironomus sp.; Test No. 235; Organization for Economic Co-operation and Development (OECD): Paris, France, 2011. [Google Scholar]
- OECD. Guidelines for the Testing of Chemicals. Sediment-Water Chironomid Life-Cycle Toxicity Test Using Spiked Water or Spiked Sediment; Test No. 233; Organization for Economic Co-operation and Development (OECD): Paris, France, 2010. [Google Scholar]
- Sahandi, J. Natural Food Production for Aquaculture: Cultivation and Nutrition of Chironomid Larvae (Insecta, Diptera). Adv. Environ. Sci. 2011, 3, 268–271. [Google Scholar]
- Martin-Folgar, R.; Esteban-Arranz, A.; Negri, V.; Morales, M. Toxicological Effects of Three Different Types of Highly Pure Graphene Oxide in the Midge Chironomus riparius. Sci. Total Environ. 2022, 815, 152465. [Google Scholar] [CrossRef]
- Martínez-Paz, P.; Negri, V.; Esteban-Arranz, A.; Martínez-Guitarte, J.L.J.L.; Ballesteros, P.; Morales, M. Effects at Molecular Level of Multi-Walled Carbon Nanotubes (MWCNT) in Chironomus riparius (DIPTERA) Aquatic Larvae. Aquat. Toxicol. 2019, 209, 42–48. [Google Scholar] [CrossRef]
- Waissi, G.C.; Bold, S.; Pakarinen, K.; Akkanen, J.; Leppänen, M.T.; Petersen, E.J.; Kukkonen, J.V.K. Chironomus Riparius Exposure to Fullerene-Contaminated Sediment Results in Oxidative Stress and May Impact Life Cycle Parameters. J. Hazard. Mater. 2017, 322, 301–309. [Google Scholar] [CrossRef]
- Lv, X.; Yang, Y.; Tao, Y.; Jiang, Y.; Chen, B.; Zhu, X.; Cai, Z.; Li, B. A Mechanism Study on Toxicity of Graphene Oxide to Daphnia magna: Direct Link between Bioaccumulation and Oxidative Stress. Environ. Pollut. 2018, 234, 953–959. [Google Scholar] [CrossRef]
- Kim, M.; Eom, H.-J.; Choi, I.; Hong, J.; Choi, J. Graphene Oxide-Induced Neurotoxicity on Neurotransmitters, AFD Neurons and Locomotive Behavior in Caenorhabditis elegans. Neurotoxicology 2020, 77, 30–39. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhi, L.; Wu, Q.; Yu, Y.; Sun, Q.; Wang, D. P38 MAPK-SKN-1/Nrf Signaling Cascade Is Required for Intestinal Barrier against Graphene Oxide Toxicity in Caenorhabditis elegans. Nanotoxicology 2016, 10, 1469–1479. [Google Scholar] [CrossRef] [PubMed]
- Soares, J.; Pereira, T.; Costa, K.; Maraschin, T.; Basso, N.; Bogo, M. Developmental Neurotoxic Effects of Graphene Oxide Exposure in Zebrafish Larvae (Danio rerio). Colloids Surf. B Biointerfaces 2017, 157, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Jasim, D.A.; Lozano, N.; Kostarelos, K. Synthesis of Few-Layered, High-Purity Graphene Oxide Sheets from Different Graphite Sources for Biology. 2D Mater. 2016, 3, 014006. [Google Scholar] [CrossRef]
- Rodrigues, A.F.; Newman, L.; Lozano, N.; Mukherjee, S.P.; Fadeel, B.; Bussy, C.; Kostarelos, K. A Blueprint for the Synthesis and Characterisation of Thin Graphene Oxide with Controlled Lateral Dimensions for Biomedicine. 2D Mater. 2018, 5, 035020. [Google Scholar] [CrossRef]
- Morales, M.; Martínez-Paz, P.; Ozáez, I.; Martínez-Guitarte, J.L.; Morcillo, G. DNA Damage and Transcriptional Changes Induced by Tributyltin (TBT) after Short in Vivo Exposures of Chironomus riparius (Diptera) Larvae. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2013, 158, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Yin, J.; Liang, Y.; Yuan, S.; Wang, F.; Song, M.; Wang, H. Oxidative Stress and Immunotoxicity Induced by Graphene Oxide in Zebrafish. Aquat. Toxicol. 2016, 174, 54–60. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, Q.; Zou, W.; Hu, X. Molecular Mechanisms of Developmental Toxicity Induced by Graphene Oxide at Predicted Environmental Concentrations. Environ. Sci. Technol. 2017, 51, 7861–7871. [Google Scholar] [CrossRef]
- Martinez-Paz, P.; Morales, M.; Martin, R.; Martinez-Guitarte, J.L.; Morcillo, G. Characterization of the Small Heat Shock Protein Hsp27 Gene in Chironomus riparius (Diptera) and Its Expression Profile in Response to Temperature Changes and Xenobiotic Exposures. Cell Stress Chaperones 2014, 19, 529–540. [Google Scholar] [CrossRef]
- Morales, M.; de la Fuente, M.; Martín-Folgar, R. BPA and Its Analogues (BPS and BPF) Modify the Expression of Genes Involved in the Endocrine Pathway and Apoptosis and a Multi Drug Resistance Gene of the Aquatic Midge Chironomus riparius (Diptera). Environ. Pollut. 2020, 265, 114806. [Google Scholar] [CrossRef]
- Morales, M.; Planello, R.; Martinez-Paz, P.; Herrero, O.; Cortes, E.; Martinez-Guitarte, J.L.; Morcillo, G. Characterization of Hsp70 Gene in Chironomus riparius: Expression in Response to Endocrine Disrupting Pollutants as a Marker of Ecotoxicological Stress. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2011, 153, 150–158. [Google Scholar] [CrossRef]
- Martínez-Paz, P.; Morales, M.; Martínez-Guitarte, J.L.; Morcillo, G. Characterization of a Cytochrome P450 Gene (CYP4G) and Modulation under Different Exposures to Xenobiotics (Tributyltin, Nonylphenol, Bisphenol A) in Chironomus riparius Aquatic Larvae. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2012, 155, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Carrasco-Navarro, V.; Muñiz-González, A.-B.; Sorvari, J.; Martínez-Guitarte, J.-L. Altered Gene Expression in Chironomus riparius (Insecta) in Response to Tire Rubber and Polystyrene Microplastics. Environ. Pollut. 2021, 285, 117462. [Google Scholar] [CrossRef] [PubMed]
- Martín-Folgar, R.; Martínez-Guitarte, J.-L. Cadmium Alters the Expression of Small Heat Shock Protein Genes in the Aquatic Midge Chironomus riparius. Chemosphere 2017, 169, 485–492. [Google Scholar] [CrossRef]
- Aquilino, M.; Sánchez-Argüello, P.; Martínez-Guitarte, J.-L. Genotoxic Effects of Vinclozolin on the Aquatic Insect Chironomus riparius (Diptera, Chironomidae). Environ. Pollut. 2018, 232, 563–570. [Google Scholar] [CrossRef]
- Martín-Folgar, R.; Martínez-Guitarte, J.-L. Effects of Single and Mixture Exposure of Cadmium and Copper in Apoptosis and Immune Related Genes at Transcriptional Level on the Midge Chironomus riparius Meigen (Diptera, Chironomidae). Sci. Total Environ. 2019, 677, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, J.G.; Kristensen, T.N.; Loeschcke, V. The Evolutionary and Ecological Role of Heat Shock Proteins. Ecol. Lett. 2003, 6, 1025–1037. [Google Scholar] [CrossRef]
- Bakthisaran, R.; Tangirala, R.; Rao, C.M. Small Heat Shock Proteins: Role in Cellular Functions and Pathology. Biochim. Biophys. Acta Proteins Proteom. 2015, 1854, 291–319. [Google Scholar] [CrossRef]
- Morrow, G.; Hightower, L.E.; Tanguay, R.M. Small Heat Shock Proteins: Big Folding Machines. Cell Stress Chaperones 2015, 20, 207–212. [Google Scholar] [CrossRef]
- Muñiz-González, A.-B.; Silva, C.J.M.; Patricio Silva, A.L.; Campos, D.; Pestana, J.L.T.; Martínez-Guitarte, J.-L. Suborganismal Responses of the Aquatic Midge Chironomus riparius to Polyethylene Microplastics. Sci. Total Environ. 2021, 783, 146981. [Google Scholar] [CrossRef]
- Park, K.; Kwak, I.-S. Characterization of Heat Shock Protein 40 and 90 in Chironomus riparius Larvae: Effects of Di(2-Ethylhexyl) Phthalate Exposure on Gene Expressions and Mouthpart Deformities. Chemosphere 2008, 74, 89–95. [Google Scholar] [CrossRef]
- Joly, A.L.; Wettstein, G.; Mignot, G.; Ghiringhelli, F.; Garrido, C. Dual Role of Heat Shock Proteins as Regulators of Apoptosis and Innate Immunity. J. Innate Immun. 2010, 2, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, M.; Zhou, J.; Zhang, X. HSP27, 70 and 90, Anti-Apoptotic Proteins, in Clinical Cancer Therapy (Review). Int. J. Oncol. 2014, 45, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Bruey, J.-M.; Ducasse, C.; Bonniaud, P.; Ravagnan, L.; Susin, S.A.; Diaz-Latoud, C.; Gurbuxani, S.; Arrigo, A.-P.; Kroemer, G.; Solary, E.; et al. Hsp27 Negatively Regulates Cell Death by Interacting with Cytochrome C. Nat. Cell Biol. 2000, 2, 645. [Google Scholar] [CrossRef] [PubMed]
- Dziewięcka, M.; Karpeta-Kaczmarek, J.; Augustyniak, M.; Majchrzycki, Ł.; Augustyniak-Jabłokow, M.A. Evaluation of in Vivo Graphene Oxide Toxicity for Acheta Domesticus in Relation to Nanomaterial Purity and Time Passed from the Exposure. J. Hazard. Mater. 2016, 305, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Majchrzycki, Ł.; Augustyniak-Jabłokow, M.A.; Strzelczyk, R.; Maćkowiak, M. Magnetic Centres in Functionalized Graphene. Acta Phys. Pol. A 2015, 127, 540–542. [Google Scholar] [CrossRef]
- Dorstyn, L.; Read, S.H.; Quinn, L.M.; Richardson, H.; Kumar, S. DECAY, a Novel Drosophila caspase Related to Mammalian Caspase-3 and Caspase-7. J. Biol. Chem. 1999, 274, 30778–30783. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Choi, Y.C.; Kim, R.; Lee, S.K. Multiwall Carbon Nanotube-Induced Apoptosis and Antioxidant Gene Expression in the Gills, Liver, and Intestine of Oryzias latipes. Biomed. Res. Int. 2015, 2015, 485343. [Google Scholar] [CrossRef]
- Cha, Y.J.; Lee, J.; Choi, S.S. Apoptosis-Mediated in Vivo Toxicity of Hydroxylated Fullerene Nanoparticles in Soil Nematode Caenorhabditis elegans. Chemosphere 2012, 87, 49–54. [Google Scholar] [CrossRef]
- Pelin, M.; Fusco, L.; Martín, C.; Sosa, S.; Frontiñán-Rubio, J.; González-Domínguez, J.M.; Durán-Prado, M.; Vázquez, E.; Prato, M.; Tubaro, A. Graphene and Graphene Oxide Induce ROS Production in Human HaCaT Skin Keratinocytes: The Role of Xanthine Oxidase and NADH Dehydrogenase. Nanoscale 2018, 10, 11820–11830. [Google Scholar] [CrossRef]
- Lammel, T.; Navas, J.M. Graphene Nanoplatelets Spontaneously Translocate into the Cytosol and Physically Interact with Cellular Organelles in the Fish Cell Line PLHC-1. Aquat. Toxicol. 2014, 150, 55–65. [Google Scholar] [CrossRef]
- Srikanth, K.; Sundar, L.S.; Pereira, E.; Duarte, A.C. Graphene Oxide Induces Cytotoxicity and Oxidative Stress in Bluegill Sunfish Cells. J. Appl. Toxicol. 2018, 38, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Souza, J.P.; Venturini, F.P.; Santos, F.; Zucolotto, V. Chronic Toxicity in Ceriodaphnia dubia Induced by Graphene Oxide. Chemosphere 2018, 190, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hu, X.; Sun, J.; Zhou, Q. Specific Nanotoxicity of Graphene Oxide during Zebrafish Embryogenesis. Nanotoxicology 2015, 10, 42–52. [Google Scholar] [CrossRef]
- Brem, R. XRCC1 Is Required for DNA Single-Strand Break Repair in Human Cells. Nucleic Acids Res. 2005, 33, 2512–2520. [Google Scholar] [CrossRef]
- Hanssen-Bauer, A.; Solvang-Garten, K.; Akbari, M.; Otterlei, M. X-Ray Repair Cross Complementing Protein 1 in Base Excision Repair. Int. J. Mol. Sci. 2012, 13, 17210. [Google Scholar] [CrossRef]
- Guleria, A.; Chandna, S. ATM Kinase: Much More than a DNA Damage Responsive Protein. DNA Repair. 2016, 39, 1–20. [Google Scholar] [CrossRef] [PubMed]
- IIJIMA, K.; OHARA, M.; SEKI, R.; TAUCHI, H. Dancing on Damaged Chromatin: Functions of ATM and the RAD50/MRE11/NBS1 Complex in Cellular Responses to DNA Damage. J. Radiat. Res. 2008, 49, 451–464. [Google Scholar] [CrossRef]
- Barzilai, A. ATM Deficiency and Oxidative Stress: A New Dimension of Defective Response to DNA Damage. DNA Repair. 2002, 1, 3–25. [Google Scholar] [CrossRef]
- Wang, Y.; Branicky, R.; Noë, A.; Hekimi, S. Superoxide Dismutases: Dual Roles in Controlling ROS Damage and Regulating ROS Signaling. J. Cell Biol. 2018, 217, 1915–1928. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Z.; Robinson, G.E.; Palli, S.R. Identification and Characterization of a Juvenile Hormone Response Element and Its Binding Proteins. J. Biol. Chem. 2007, 282, 37605–37617. [Google Scholar] [CrossRef]
- Orłowski, M.; Popławska, K.; Pieprzyk, J.; Szczygieł-Sommer, A.; Więch, A.; Zarębski, M.; Tarczewska, A.; Dobrucki, J.; Ożyhar, A. Molecular Determinants of Drosophila Immunophilin FKBP39 Nuclear Localization. Biol. Chem. 2018, 399, 467–484. [Google Scholar] [CrossRef] [PubMed]
- Juhász, G.; Puskás, L.G.; Komonyi, O.; Érdi, B.; Maróy, P.; Neufeld, T.P.; Sass, M. Gene Expression Profiling Identifies FKBP39 as an Inhibitor of Autophagy in Larval Drosophila Fat Body. Cell Death Differ. 2007, 14, 1181–1190. [Google Scholar] [CrossRef] [PubMed]
- Theopold, U.; Dal Zotto, L.; Hultmark, D. FKBP39, a Drosophila Member of a Family of Proteins That Bind the Immunosuppressive Drug FK506. Gene 1995, 156, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Khan, C.; Muliyil, S.; Ayyub, C.; Rao, B.J. DNA Damage Signalling in D. melanogaster Requires Non-Apoptotic Function of Initiator Caspase Dronc. J. Cell Sci. 2017, 130, 2984–2995. [Google Scholar] [CrossRef] [PubMed]
| Gene | Primers | Process | |
|---|---|---|---|
| rpL13F | ACCAGCTAGAAAGCACCGTC | [32] | |
| rpL13R | ATGGGCATCTGACGATTGGG | ||
| GAPDHF | GGTATTTCATTGAATGATCACTTTG | [33] | |
| GAPDHR | TAATCCTTGGATTGCATGTACTTG | ||
| hsp90F | AGGCTGAAGCTGACAAGAATG | Stress response | [34] |
| hsp90R | TCATGCGATAAATGCGAGCAG | ||
| hsp70F | ACTTGAACCAGTTGAGCGT | Stress response | [27] |
| hsp70R | TTGCCACAGAAGAAATCTTG | ||
| hsp60F | TGCTGTCCTTAAAGTCGGTGG | Stress response | [35] |
| hsp60R | TCCACCACC-CAACGATTC | ||
| hsp27F | TCAACACACAGGACCG | Stress response | [30] |
| hsp27R | ATCCTTTATTGGTGATTAATTATG | ||
| XRCC1 F | GACGATTTGCATTGGATAGT | DNA repair. SSB/DSB | [36] |
| XRCC1 R | ATCAACATATCGCCATCAG | ||
| ATMF | ACATTTGGCGTAGATCAGGCA | DNA repair. SSB | [36] |
| ATM | ACGAGATGCATCAAATCATGC | ||
| NLKF | CATCTCACCAGATCGTCTCT | DNA repair. SSB | [36] |
| NLKR | GAATTTATTTGATTATGCGGC | ||
| DECAY F | AAAGTGTTCCGATTATGGC | DNA repair. Apoptosis | [36] |
| DECAY R | TTCACACCAGTTAAAATCCAC | ||
| SODMnF | AAGTCGCTGCTGTTGGAGTT | Oxidative stress | [34] |
| SODMnR | TGGAACTAAGCCGGTTGTGG | ||
| FKBP39F | AGGCTGGGATATCGGACTCAT | Development | [34] |
| FKBP39R | GTAAGCAAATGCAGGCGGG | ||
| DRONCF | GAAATGTCACAGATTTCAGTGCC | Death regulator Nedd2-like caspase | [37] |
| DRONCR | GTGAATATCGTAAGCATGTTCTGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin-Folgar, R.; Esteban-Arranz, A.; Negri, V.; Morales, M. Graphene Oxides (GOs) with Different Lateral Dimensions and Thicknesses Affect the Molecular Response in Chironomus riparius. Nanomaterials 2023, 13, 967. https://doi.org/10.3390/nano13060967
Martin-Folgar R, Esteban-Arranz A, Negri V, Morales M. Graphene Oxides (GOs) with Different Lateral Dimensions and Thicknesses Affect the Molecular Response in Chironomus riparius. Nanomaterials. 2023; 13(6):967. https://doi.org/10.3390/nano13060967
Chicago/Turabian StyleMartin-Folgar, Raquel, Adrián Esteban-Arranz, Viviana Negri, and Mónica Morales. 2023. "Graphene Oxides (GOs) with Different Lateral Dimensions and Thicknesses Affect the Molecular Response in Chironomus riparius" Nanomaterials 13, no. 6: 967. https://doi.org/10.3390/nano13060967
APA StyleMartin-Folgar, R., Esteban-Arranz, A., Negri, V., & Morales, M. (2023). Graphene Oxides (GOs) with Different Lateral Dimensions and Thicknesses Affect the Molecular Response in Chironomus riparius. Nanomaterials, 13(6), 967. https://doi.org/10.3390/nano13060967