Reduction in Toxicity of Polystyrene Nanoplastics Combined with Phenanthrene through Binding of Jellyfish Mucin with Nanoplastics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Jellyfish Mu Extraction
2.2. Physicochemical Properties of PS-NP, PS-NP + Phe, and Mu Mixtures
2.3. Test Organism and Exposure Conditions
2.4. Quantitative Analysis of the Binding Capacity of Jellyfish Mu and PS-NP
2.5. Analysis of Substances Penetrated and Absorbed by the Zebrafish Embryos
2.6. Gene Analysis
2.6.1. mRNA Sequence Analysis
2.6.2. Data Analysis
2.6.3. Quantitative Analysis of Gene Expression
2.7. Statistical Analysis
3. Experimental Results and Discussion
3.1. Physicochemical Properties of PS-NP, Phe, and Mu Mixtures
3.2. Evaluation of the Binding Capacity of Jellyfish Mu and PS-NP
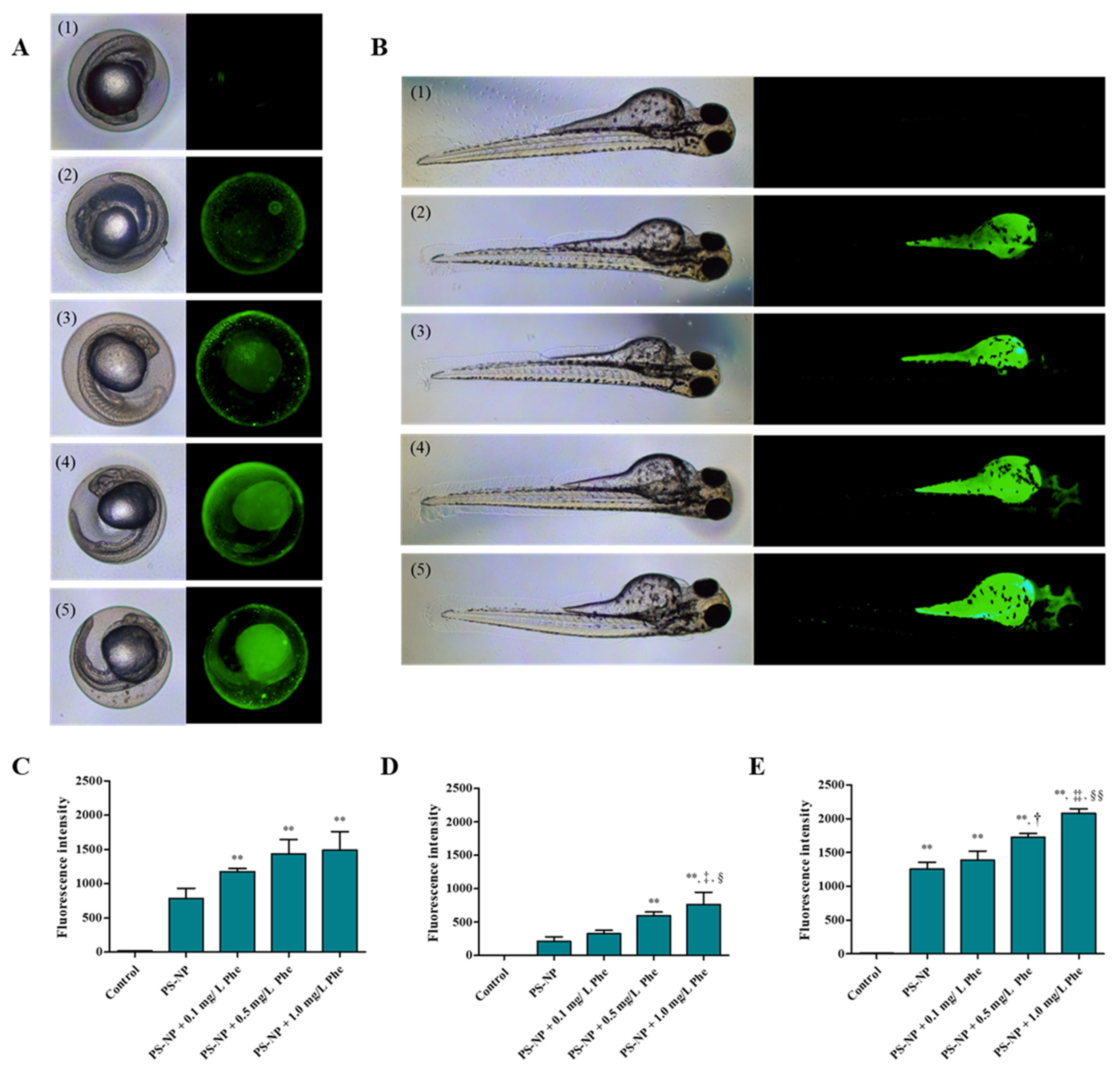
3.3. Analysis of Substances Penetrated and Absorbed by the Zebrafish Embryos
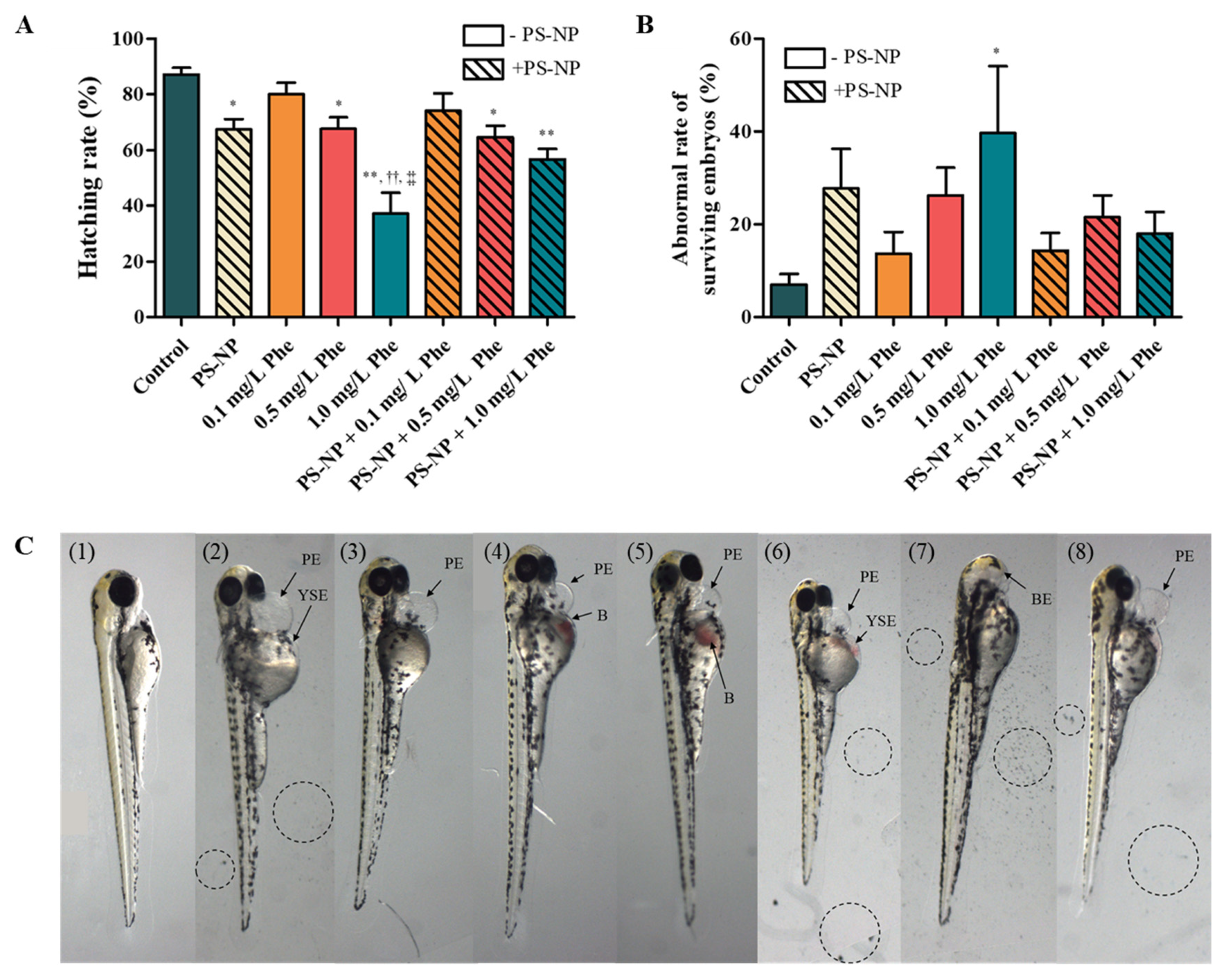
3.3.1. Effects of PS-NP, Phe and PS-NP + Phe without Mu
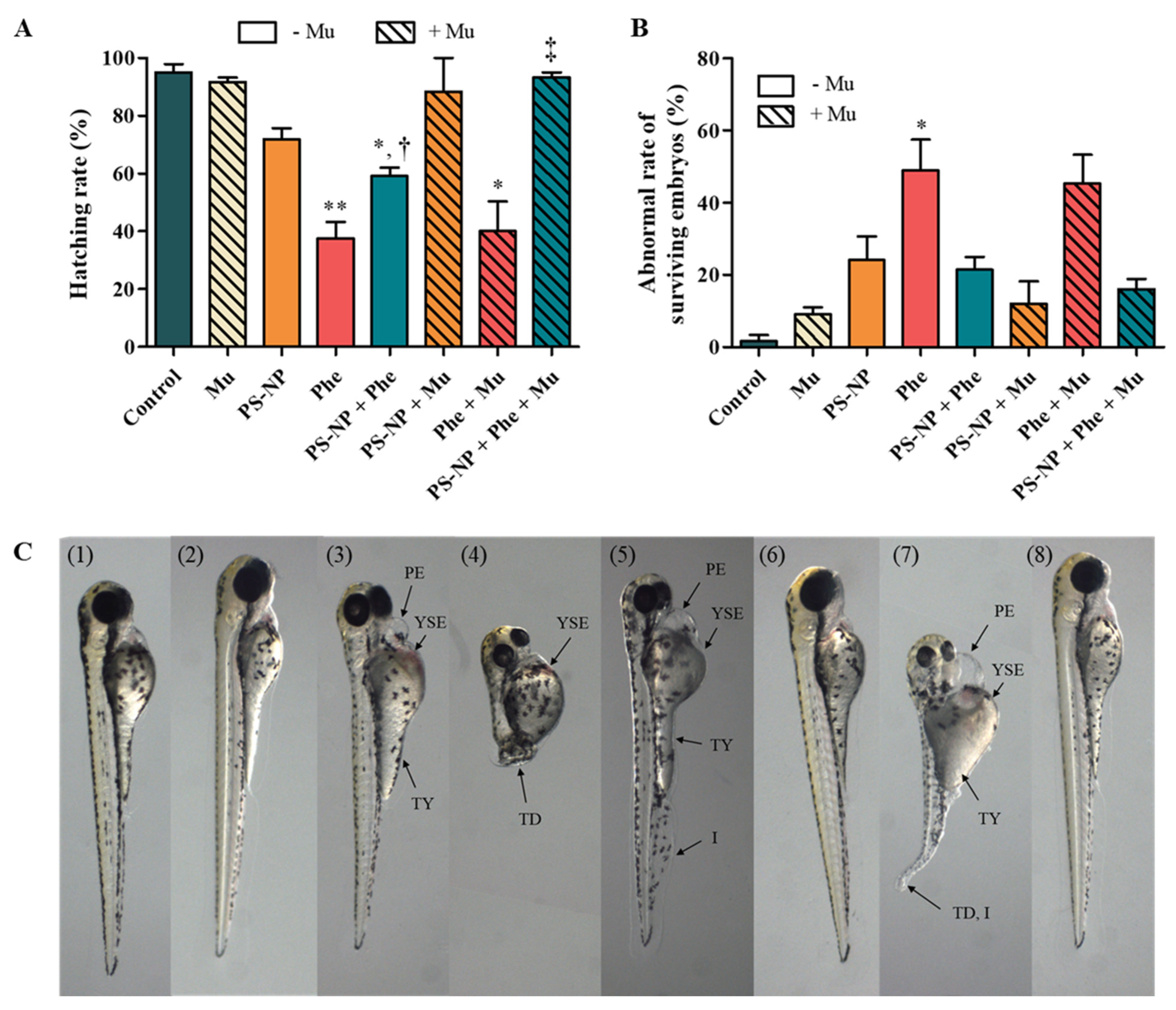
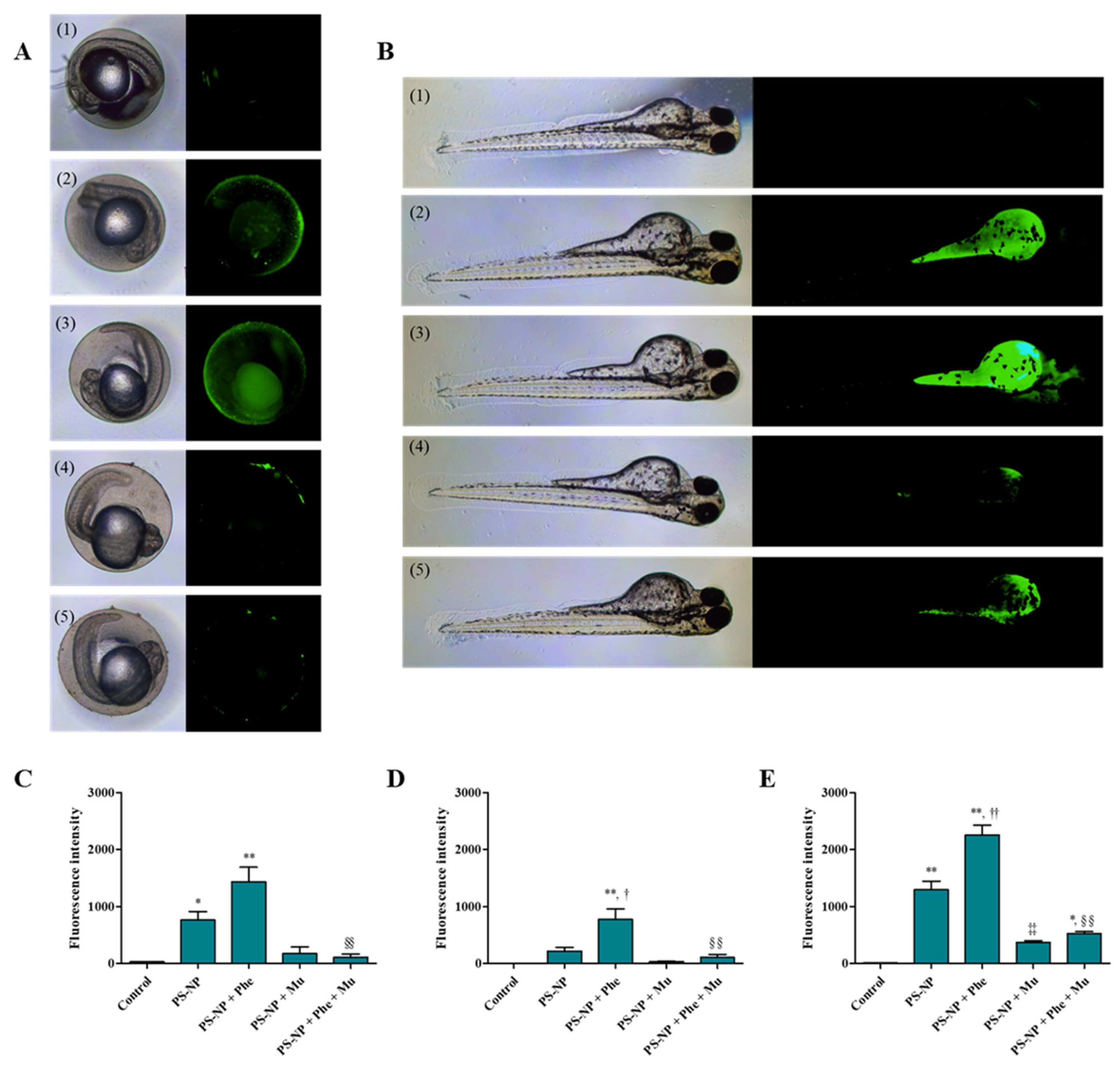
3.3.2. Effect of Mu on the Absorption and Toxicity of PS-NP, Phe and PS-NP + Phe
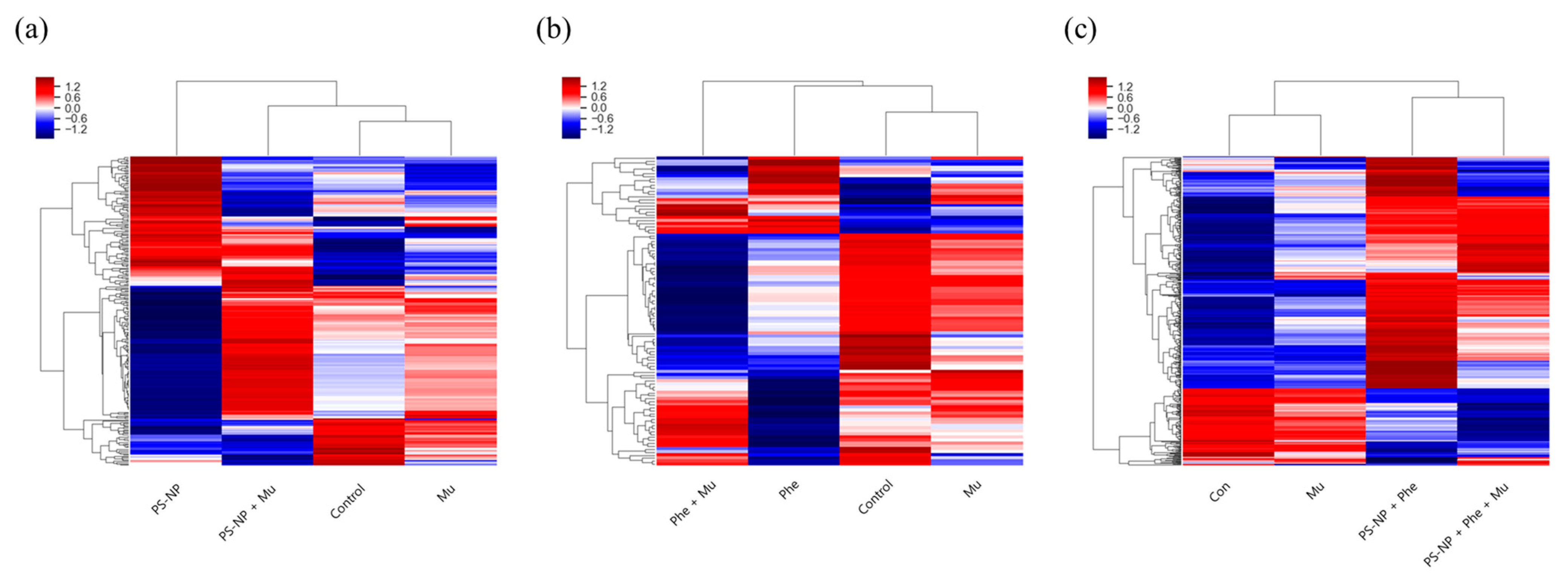
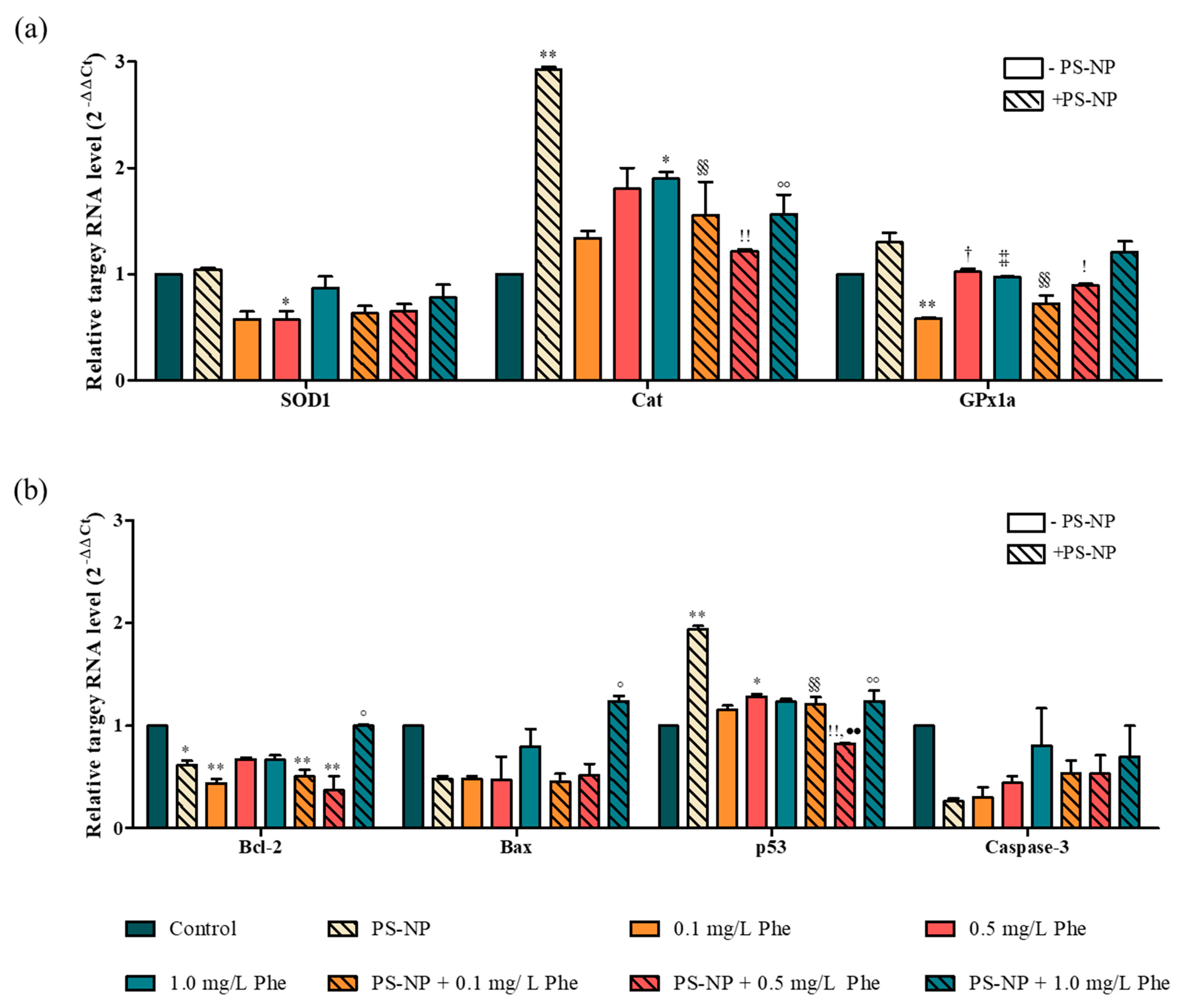
3.4. Gene Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kik, K.; Bukowska, B.; Sicińska, P. Polystyrene Nanoparticles: Sources, Occurrence in the Environment, Distribution in Tissues, Accumulation and Toxicity to Various Organisms. Environ. Pollut. 2020, 262, 114297. [Google Scholar] [CrossRef] [PubMed]
- Pitt, J.A.; Kozal, J.S.; Jayasundara, N.; Massarsky, A.; Trevisan, R.; Geitner, N.; Wiesner, M.; Levin, E.D.; di Giulio, R.T. Uptake, Tissue Distribution, and Toxicity of Polystyrene Nanoparticles in Developing Zebrafish (Danio Rerio). Aquat. Toxicol. 2018, 194, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Andrady, A.L. Microplastics in the Marine Environment. Mar. Pollut. Bull. 2011, 62, 1596–1605. [Google Scholar] [CrossRef] [PubMed]
- Sudhakar, M.; Trishul, A.; Doble, M.; Suresh Kumar, K.; Syed Jahan, S.; Inbakandan, D.; Viduthalai, R.R.; Umadevi, V.R.; Sriyutha Murthy, P.; Venkatesan, R. Biofouling and Biodegradation of Polyolefins in Ocean Waters. Polym. Degrad. Stab. 2007, 92, 1743–1752. [Google Scholar] [CrossRef]
- Watters, D.L.; Yoklavich, M.M.; Love, M.S.; Schroeder, D.M. Assessing Marine Debris in Deep Seafloor Habitats off California. Mar. Pollut. Bull. 2010, 60, 131–138. [Google Scholar] [CrossRef]
- O’Brine, T.; Thompson, R.C. Degradation of Plastic Carrier Bags in the Marine Environment. Mar. Pollut. Bull. 2010, 60, 2279–2283. [Google Scholar] [CrossRef]
- Cedervall, T.; Hansson, L.A.; Mattsson, K. Nano-Plastics in the Aquatic Environment. Environ. Sci. Processes Impacts 2015, 17, 1712–1721. [Google Scholar] [CrossRef]
- da Costa, J.P.; Santos, P.S.M.; Duarte, A.C.; Rocha-Santos, T. (Nano)Plastics in the Environment—Sources, Fates and Effects. Sci. Total Environ. 2016, 566, 15–26. [Google Scholar] [CrossRef]
- Ng, E.-L.; Huerta Lwanga, E.; Eldridge, S.M.; Johnston, P.; Hu, H.W.; Geissen, V.; Chen, D. An Overview of Microplastic and Nanoplastic Pollution in Agroecosystems. Sci. Total Environ. 2018, 627, 1377–1388. [Google Scholar] [CrossRef]
- Lei, L.; Liu, M.; Song, Y.; Lu, S.; Hu, J.; Cao, C.; Xie, B.; Shi, H.; He, D. Polystyrene (Nano)Microplastics Cause Size-Dependent Neurotoxicity, Oxidative Damage and Other Adverse Effects in Caenorhabditis Elegans. Environ. Sci. Nano 2018, 5, 2009–2020. [Google Scholar] [CrossRef]
- Schirinzi, G.F.; Pérez-Pomeda, I.; Sanchís, J.; Rossini, C.; Farré, M.; Barceló, D. Cytotoxic Effects of Commonly Used Nanomaterials and Microplastics on Cerebral and Epithelial Human Cells. Environ. Res. 2017, 159, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Eom, H.J.; Nam, S.E.; Rhee, J.S. Polystyrene Microplastics Induce Mortality through Acute Cell Stress and Inhibition of Cholinergic Activity in a Brine Shrimp. Mol. Cell. Toxicol. 2020, 16, 233–243. [Google Scholar] [CrossRef]
- Kim, J.; Rhee, J.S. Biochemical and Physiological Responses of the Water Flea Moina Macrocopa to Microplastics: A Multigenerational Study. Mol. Cell. Toxicol. 2021, 17, 523–532. [Google Scholar] [CrossRef]
- Alimba, C.G.; Faggio, C. Microplastics in the Marine Environment: Current Trends in Environmental Pollution and Mechanisms of Toxicological Profile. Environ. Toxicol. Pharmacol. 2019, 68, 61–74. [Google Scholar] [CrossRef]
- Matos, B.; Martins, M.; Samamed, A.C.; Sousa, D.; Ferreira, I.; Diniz, M.S. Toxicity Evaluation of Quantum Dots (ZnS and CdS) Singly and Combined in Zebrafish (Danio Rerio). Int. J. Environ. Res. Public Health 2019, 17, 232. [Google Scholar] [CrossRef] [Green Version]
- Sendra, M.; Pereiro, P.; Yeste, M.P.; Mercado, L.; Figueras, A.; Novoa, B. Size Matters: Zebrafish (Danio Rerio) as a Model to Study Toxicity of Nanoplastics from Cells to the Whole Organism. Environ. Pollut. 2021, 268, 115769. [Google Scholar] [CrossRef]
- González-fernández, C.; Díaz Baños, F.G.; Esteban, M.Á.; Cuesta, A. Functionalized Nanoplastics (NPs) Increase the Toxicity of Metals in Fish Cell Lines. Int. J. Mol. Sci. 2021, 22, 7141. [Google Scholar] [CrossRef]
- Anguissola, S.; Garry, D.; Salvati, A.; O’Brien, P.J.; Dawson, K.A. High Content Analysis Provides Mechanistic Insights on the Pathways of Toxicity Induced by Amine-Modified Polystyrene Nanoparticles. PLoS ONE 2014, 9, e108025. [Google Scholar] [CrossRef]
- Inkielewicz-Stepniak, I.; Tajber, L.; Behan, G.; Zhang, H.; Radomski, M.W.; Medina, C.; Santos-Martinez, M.J. The Role of Mucin in the Toxicological Impact of Polystyrene Nanoparticles. Materials 2018, 11, 724. [Google Scholar] [CrossRef] [Green Version]
- Besseling, E.; Wegner, A.; Foekema, E.M.; van den Heuvel-Greve, M.J.; Koelmans, A.A. Effects of Microplastic on Fitness and PCB Bioaccumulation by the Lugworm Arenicola Marina (L.). Environ. Sci. Technol. 2013, 47, 593–600. [Google Scholar] [CrossRef]
- Nakashima, E.; Isobe, A.; Kako, S.; Itai, T.; Takahashi, S. Quantification of Toxic Metals Derived from Macroplastic Litter on Ookushi Beach, Japan. Environ. Sci. Technol. 2012, 46, 10099–10105. [Google Scholar] [CrossRef] [PubMed]
- Rios, L.M.; Moore, C.; Jones, P.R. Persistent Organic Pollutants Carried by Synthetic Polymers in the Ocean Environment. Mar. Pollut. Bull. 2007, 54, 1230–1237. [Google Scholar] [CrossRef] [PubMed]
- Verla, A.W.; Enyoh, C.E.; Verla, E.N.; Nwarnorh, K.O. Microplastic–Toxic Chemical Interaction: A Review Study on Quantified Levels, Mechanism and Implication. SN Appl. Sci. 2019, 1, 1–30. [Google Scholar] [CrossRef] [Green Version]
- Honda, M.; Suzuki, N. Toxicities of Polycyclic Aromatic Hydrocarbons for Aquatic Animals. Int. J. Environ. Res. Public Health 2020, 17, 1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alves, R.N.; Mariz, C.F., Jr.; de Paulo, D.V.; Carvalho, P.S.M. Toxicity of Effluents from Gasoline Stations Oil-Water Separators to Early Life Stages of Zebrafish Danio Rerio. Chemosphere 2017, 178, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Devi, N.L.; Yadav, I.C.; Shihua, Q.; Dan, Y.; Zhang, G.; Raha, P. Environmental Carcinogenic Polycyclic Aromatic Hydrocarbons in Soil from Himalayas, India: Implications for Spatial Distribution, Sources Apportionment and Risk Assessment. Chemosphere 2016, 144, 493–502. [Google Scholar] [CrossRef]
- Hannam, M.L.; Bamber, S.D.; Moody, A.J.; Galloway, T.S.; Jones, M.B. Immunotoxicity and Oxidative Stress in the Arctic Scallop Chlamys Islandica: Effects of Acute Oil Exposure. Ecotoxicol. Environ. Saf. 2010, 73, 1440–1448. [Google Scholar] [CrossRef]
- Jacob, J. The Significance of Polycyclic Aromatic Hydrocarbons as Environmental Carcinogens. 35 Years Research on PAH—A Retrospective. Polycycl. Aromat. Compd. 2008, 28, 242–272. [Google Scholar] [CrossRef]
- MacDonald, G.Z.; Hogan, N.S.; Köllner, B.; Thorpe, K.L.; Phalen, L.J.; Wagner, B.D.; van den Heuvel, M.R. Immunotoxic Effects of Oil Sands-Derived Naphthenic Acids to Rainbow Trout. Aquat. Toxicol. 2013, 126, 95–103. [Google Scholar] [CrossRef]
- Rengarajan, T.; Rajendran, P.; Nandakumar, N.; Lokeshkumar, B.; Rajendran, P.; Nishigaki, I. Exposure to Polycyclic Aromatic Hydrocarbons with Special Focus on Cancer. Asian Pac. J. Trop. Biomed. 2015, 5, 182–189. [Google Scholar] [CrossRef] [Green Version]
- Cherr, G.N.; Fairbairn, E.; Whitehead, A. Impacts of Petroleum-Derived Pollutants on Fish Development. Annu. Rev. Anim. Biosci. 2016, 5, 185–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knulst, J.C.; Westling, H.O.; Brorström-Lundén, E. Airborne Organic Micropollutant Concentrations in Mosses and Humus as Indicators for Local versus Long-Range Sources. Environ. Monit. Assess. 1995, 36, 75–91. [Google Scholar] [CrossRef] [PubMed]
- Boyle, K.; Örmeci, B. Microplastics and Nanoplastics in the Freshwater and Terrestrial Environment: A Review. Water 2020, 12, 2633. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, M.-E.; Chung, J.-W. Comparison of Heavy Metal Adsorption by Manganese Oxide-Coated Activated Carbon According to Manufacture Method. J. Korean Soc. Environ. Eng. 2014, 36, 7–12. [Google Scholar] [CrossRef]
- Kim, S.S.; Lee, J.A.; Yeo, M.K. Reduction in Toxicity of Nano-Ag-Polyvinyl-Pyrrolidone Using Hydra Proteins and Peptides during Zebrafish Embryogenesis. Nanomaterials 2019, 9, 1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.A.; Yeo, M.K.; Kim, S.S. Hydra Protein Reduces the Toxicity of Ag–PVP Nanoparticles in a 3D A549 Cell Line. Mol. Cell. Toxicol. 2020, 16, 73–81. [Google Scholar] [CrossRef]
- Ha, J.; Kim, E.; Lee, B.G.; Yeo, M.K. Capture and Toxicity Assessment of Ag Citrate Nanoparticles Using Jellyfish Extract. Mol. Cell. Toxicol. 2020, 16, 431–439. [Google Scholar] [CrossRef]
- Kim, E.; Yeo, M.-K.; Lee, B.G.; Geum, W. Binding of Silver Nanowaste Using Jellyfish Immune Reaction Extract and an Assessment of Aquatic Toxicity. Mol. Cell. Toxicol. 2021. [Google Scholar] [CrossRef]
- Masuda, A.; Baba, T.; Dohmae, N.; Yamamura, M.; Wada, H.; Ushida, K. Mucin (Qniumucin), a Glycoprotein from Jellyfish, and Determination of Its Main Chain Structure. J. Nat. Prod. 2007, 70, 1089–1092. [Google Scholar] [CrossRef]
- Ohta, N.; Sato, M.; Ushida, K.; Kokubo, M.; Baba, T.; Taniguchi, K.; Urai, M.; Kihira, K.; Mochida, J. Jellyfish Mucin May Have Potential Disease-Modifying Effects on Osteoarthritis. BMC Biotechnol. 2009, 9, 98. [Google Scholar] [CrossRef] [Green Version]
- Bakshani, C.R.; Morales-Garcia, A.L.; Althaus, M.; Wilcox, M.D.; Pearson, J.P.; Bythell, J.C.; Burgess, J.G. Evolutionary Conservation of the Antimicrobial Function of Mucus: A First Defence against Infection. NPJ Biofilms Microbiomes 2018, 4, 14. [Google Scholar] [CrossRef] [Green Version]
- Ushida, K.; Oohata, A.; Kawamura, G.; Hori, Y. Mucin (Qniumucin) Extracted from Jellyfish Can Be Applied to Ophthalmologic Researches and/or Diagnoses as a Substituting Material of Human Mucin: A Study on Contact Lenses. Investig. Ophthalmol. Vis. Sci. 2016, 57, 6186. [Google Scholar]
- ISO 7346-3:1996; UNE-EN ISO 7346-3 Water Quality, Determination of the Acute Lethal Toxicity of Substances to a Freshwater Fish [Brachydanio Rerio Hamilton-Bchanan (Teleostei, Cyprinidae)]. Part 3: Flow-through Method. ISO: Geneva, Switzerland, 1998.
- Karami, A.; Romano, N.; Galloway, T.; Hamzah, H. Virgin Microplastics Cause Toxicity and Modulate the Impacts of Phenanthrene on Biomarker Responses in African Catfish (Clarias Gariepinus). Environ. Res. 2016, 151, 58–70. [Google Scholar] [CrossRef] [PubMed]
- Teuten, E.L.; Rowland, S.J.; Galloway, T.S.; Thompson, R.C. Potential for Plastics to Transport Hydrophobic Contaminants. Environ. Sci. Technol. 2007, 41, 7759–7764. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish(Brachydanio Rerio), 4th ed.; University of Oregon Press: Eugene, OR, USA, 1995. [Google Scholar]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of Embryonic Development of the Zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering Splice Junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Pereira, A.C.; Gomes, T.; Ferreira Machado, M.R.; Rocha, T.L. The Zebrafish Embryotoxicity Test (ZET) for Nanotoxicity Assessment: From Morphological to Molecular Approach. Environ. Pollut. 2019, 252, 1841–1853. [Google Scholar] [CrossRef]
- Chao, S.J.; Huang, C.P.; Chen, P.C.; Chang, S.H.; Huang, C. Uptake of BDE-209 on Zebrafish Embryos as Affected by SiO2 Nanoparticles. Chemosphere 2018, 205, 570–578. [Google Scholar] [CrossRef]
- Cheng, J.; Flahaut, E.; Cheng, S.H. Effect of Carbon Nanotubes on Developing Zebrafish (Danio Rerio) Embryos. Environ. Toxicol. Chem. Int. J. 2007, 26, 708–716. [Google Scholar] [CrossRef]
- Walkey, C.D.; Chan, W.C.W. Understanding and Controlling the Interaction of Nanomaterials with Proteins in a Physiological Environment. Chem. Soc. Rev. 2012, 41, 2780–2799. [Google Scholar] [CrossRef]
- Kumar, A.; Dixit, C.K. Methods for Characterization of Nanoparticles. In Advances in Nanomedicine for the Delivery of Therapeutic Nucleic Acids; Woodhead Publishing: Cambridge, UK, 2017; pp. 43–58. [Google Scholar]
- Zhang, Y.; Goss, G.G. Potentiation of Polycyclic Aromatic Hydrocarbon Uptake in Zebrafish Embryos by Nanoplastics. Environ. Sci. Nano 2020, 7, 1730–1741. [Google Scholar] [CrossRef]
- Trevisan, R.; Voy, C.; Chen, S.; di Giulio, R.T. Nanoplastics Decrease the Toxicity of a Complex PAH Mixture but Impair Mitochondrial Energy Production in Developing Zebrafish. Environ. Sci. Technol. 2019, 53, 8405–8415. [Google Scholar] [CrossRef] [PubMed]
- Coffin, S.; Magnuson, J.T.; Vliet, S.M.F.; Volz, D.C.; Schlenk, D. Effects of Short-Term Exposure to Environmentally-Relevant Concentrations of Benzo(a)Pyrene-Sorbed Polystyrene to White Seabass (Atractoscion Nobilis)☆. Environ. Pollut. 2020, 263, 114617. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Yang, G.; Lu, L.; Zheng, Y.; Zhang, Q.; Zhang, X.; Tian, H.; Wang, W.; Ru, S. Low Level of Polystyrene Microplastics Decreases Early Developmental Toxicity of Phenanthrene on Marine Medaka (Oryzias Melastigma). J. Hazard. Mater. 2020, 385, 121586. [Google Scholar] [CrossRef]
- Yu, F.; Yang, C.; Zhu, Z.; Bai, X.; Ma, J. Adsorption Behavior of Organic Pollutants and Metals on Micro/Nanoplastics in the Aquatic Environment. Sci. Total Environ. 2019, 694, 133643. [Google Scholar] [CrossRef] [PubMed]
- Jonker, M.T.O.; Koelmans, A.A. Sorption of Polycyclic Aromatic Hydrocarbons and Polychlorinated Biphenyls to Soot and Soot-like Materials in the Aqueous Environment: Mechanistic Considerations. Environ. Sci. Technol. 2002, 36, 3725–3734. [Google Scholar] [CrossRef] [PubMed]
- Penner, N.A.; Nesterenko, P.N.; Hyin, M.M.; Tsyurupa, M.P.; Davankov, V.A. Investigation of the Properties of Hypercrosslinked Polystyrene as a Stationary Phase for High-Performance Liquid Chromatography. Chromatographia 1999, 50, 611–620. [Google Scholar] [CrossRef]
- Liu, L.; Fokkink, R.; Koelmans, A.A. Sorption of Polycyclic Aromatic Hydrocarbons to Polystyrene Nanoplastic. Environ. Toxicol. Chem. 2016, 35, 1650–1655. [Google Scholar] [CrossRef]
- Burgess, R.M.; Ryba, S.A.; Cantwell, M.G.; Gundersen, J.L.; Tien, R.; Perron, M.M. Interaction of Planar and Nonplanar Organic Contaminants with Coal Fly Ash: Effects of Polar and Nonpolar Solvent Solutions. Environ. Toxicol. Chem. Int. J. 2006, 25, 2028–2037. [Google Scholar] [CrossRef]
- Velzeboer, I.; Kwadijk, C.J.A.F.; Koelmans, A.A. Strong Sorption of PCBs to Nanoplastics, Microplastics, Carbon Nanotubes, and Fullerenes. Environ. Sci. Technol. 2014, 48, 4869–4876. [Google Scholar] [CrossRef]
- Rochman, C.M.; Manzano, C.; Hentschel, B.T.; Simonich, S.L.M.; Hoh, E. Polystyrene Plastic: A Source and Sink for Polycyclic Aromatic Hydrocarbons in the Marine Environment. Environ. Sci. Technol. 2013, 47, 13976–13984. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Shim, W.J.; Kwon, J.-H. Sorption Capacity of Plastic Debris for Hydrophobic Organic Chemicals. Sci. Total Environ. 2014, 470, 1545–1552. [Google Scholar] [CrossRef] [PubMed]
- Incardona, J.P.; Collier, T.K.; Scholz, N.L. Defects in Cardiac Function Precede Morphological Abnormalities in Fish Embryos Exposed to Polycyclic Aromatic Hydrocarbons. Toxicol. Appl. Pharmacol. 2004, 196, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Brandts, I.; Teles, M.; Tvarijonaviciute, A.; Pereira, M.L.; Martins, M.A.; Tort, L.; Oliveira, M. Effects of Polymethylmethacrylate Nanoplastics on Dicentrarchus Labrax. Genomics 2018, 110, 435–441. [Google Scholar] [CrossRef]
- Rawson, D.M.; Zhang, T.; Kalicharan, D.; Jongebloed, W.L. Field Emission Scanning Electron Microscopy and Transmission Electron Microscopy Studies of the Chorion, Plasma Membrane and Syncytial Layers of the Gastrula-Stage Embryo of the Zebrafish Brachydanio Rerio: A Consideration of the Structural and Functional Relationships with Respect to Cryoprotectant Penetration. Aquac. Res. 2000, 31, 325–336. [Google Scholar] [CrossRef]
- Shang, L.; Nienhaus, K.; Nienhaus, G.U. Engineered Nanoparticles Interacting with Cells: Size Matters. J. Nanobiotechnol. 2014, 12, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saikia, J.; Yazdimamaghani, M.; Hadipour Moghaddam, S.P.; Ghandehari, H. Differential Protein Adsorption and Cellular Uptake of Silica Nanoparticles Based on Size and Porosity. ACS Appl. Mater. Interfaces 2016, 8, 34820–34832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvati, A.; Pitek, A.S.; Monopoli, M.P.; Prapainop, K.; Bombelli, F.B.; Hristov, D.R.; Kelly, P.M.; Åberg, C.; Mahon, E.; Dawson, K.A. Transferrin-Functionalized Nanoparticles Lose Their Targeting Capabilities When a Biomolecule Corona Adsorbs on the Surface. Nat. Nanotechnol. 2013, 8, 137–143. [Google Scholar] [CrossRef] [Green Version]
- van Pomeren, M.; Brun, N.R.; Peijnenburg, W.J.G.M.; Vijver, M.G. Exploring Uptake and Biodistribution of Polystyrene (Nano)Particles in Zebrafish Embryos at Different Developmental Stages. Aquat. Toxicol. 2017, 190, 40–45. [Google Scholar] [CrossRef]
- Priyam, A.; Singh, P.P.; Afonso, L.O.; Schultz, A.G. Exposure to Biogenic Phosphorus Nano-Agromaterials Promotes Early Hatching and Causes No Acute Toxicity in Zebrafish Embryos. Environ. Sci. Nano 2022, 9, 1364–1380. [Google Scholar] [CrossRef]
- Langheinrich, U.; Hennen, E.; Stott, G.; Vacun, G. Zebrafish as a Model Organism for the Identification and Characterization of Drugs and Genes Affecting P53 Signaling. Curr. Biol. 2002, 12, 2023–2028. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Z.; Zheng, X.; Rich, K.M. Down-Regulation of Bcl-2 and Bcl-XL Expression with Bispecific Antisense Treatment in Glioblastoma Cell Lines Induce Cell Death. J. Neurochem. 2003, 84, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Sohn, J.H.; Chae, S.W.; Choi, K.C.; Shin, H.S. Caspase 3 and Ki-67 Immunoreactivity and Its Correlation with Frequency of Apoptosis in Gastric Adenomas and Carcinomas. Korean J. Pathol. 2001, 35, 286–290. [Google Scholar]
| Gene | Sequence (5′→3′) | References | |
|---|---|---|---|
| Forward | Reverse | ||
| β-actin | CGGGTGGAGTTTGACACTT | CTCCCTGATGCTGGGTCGTC | [50] |
| SOD1 | GGCCAACCGATAGTGTTAGA | CCAGCGTTGCCAGTTTTTAG | [51] |
| CAT | AGGGCAACTGGGATCTTACA | TTTATGGGACCAGACCTTGG | [50] |
| GPx1a | ACCTGTCCGCGAAACTATTG | TGACTGTTGTGCCTCAAAGC | [50] |
| Bcl-2 | TCACTCGTTCAGACCCTCAT | ACGCTTTCCACGCACAT | [50] |
| Bax | GGCTATTTCAACCAGGGTCC | TGCGAATCACCAATGCTGT | [50] |
| p53 | GGGCAATCAGCGAGCAAA | ACTGACCTTCCTGAGTCTCCA | [50] |
| Caspase-3 | CCGCTGCCCATCACTA | ATCCTTTCACGACCATCT | [50] |
| Time (h) | PS-NP Concentration (mg/L) |
|---|---|
| 0 | 2.00 ± 0.00 |
| 0.5 | 0.14 ± 0.04 ** |
| 1 | 0.19 ± 0.01 ** |
| 2 | 0.21 ± 0.03 ** |
| 4 | 0.17 ± 0.03 ** |
| 8 | 0.14 ± 0.01 ** |
| Control | PS-NP | 0.1 mg/L Phe | 0.5 mg/L Phe | 1.0 mg/L Phe | PS-NP + 0.1 mg/L Phe | PS-NP + 0.5 mg/L Phe | PS-NP + 1.0 mg/L Phe | |
|---|---|---|---|---|---|---|---|---|
| Inflammation | 2.50 ± 3.12 | 13.11 ± 15.16 | 3.44 ± 8.05 | 10.44 ± 7.83 | 32.67 ± 30.00 **, ††, ‡ | 0.00 ± 0.00 | 0.00 ± 0.00 | 7.67 ± 4.03 § |
| Tail deformity | 1.33 ± 3.67 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.89 ± 2.67 | 0.00 ± 0.00 | 1.00 ± 2.45 | 1.50 ± 3.67 | 1.33 ± 3.26 |
| Pericardial edema | 3.92 ± 4.67 | 5.56 ± 7.18 | 3.44 ± 4.48 | 15.22 ± 7.07 * | 9.33 ± 11.88 | 9.5 ± 10.43 | 4.33 ± 7.42 | 8.83 ± 11.03 |
| Yolk-sac edema | 3.83 ± 3.71 | 4.67 ± 5.05 | 6.22 ± 9.22 | 10.22 ± 9.92 | 12.00 ± 10.92 | 6.83 ± 7.33 | 8.50 ± 10.77 | 8.83 ± 11.03 |
| Thicker yolk | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.78 ± 2.33 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Slight yolk | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.78 ± 2.33 | 2.44 ± 7.33 | 1.00 ± 2.45 | 0.00 ± 0.00 | 1.33 ± 3.27 |
| Blood vessel | 0.00 ± 0.00 | 6.89 ± 8.43 | 0.89 ± 2.67 | 2.89 ± 6.25 | 12.22 ± 33.08 | 2.33 ± 3.67 | 4.00 ± 4.38 | 1.33 ± 3.27 |
| Blood | 2.42 ± 4.72 | 7.89 ± 7.46 | 6.00 ± 12.73 | 10.44 ± 9.41 | 9.00 ± 9.46 | 6.83 ± 6.77 | 7.67 ± 6.71 | 6.00 ± 7.18 |
| Spinal curvature | 1.83 ± 3.59 | 6.78 ± 10.08 | 2.67 ± 4.39 | 4.56 ± 4.61 | 2.89 ± 4.37 | 1.67 ± 4.08 | 2.83 ± 4.40 | 0.00 ± 0.00 |
| Control | Mu | PS-NP | Phe | PS-NP + Phe | PS-NP + Mu | Phe + Mu | PS-NP + Phe + Mu | |
|---|---|---|---|---|---|---|---|---|
| Inflammation | 0.00 ± 0.00 | 2.00 ± 3.46 | 9.83 ± 14.22 | 34.25 ± 25.85 * | 7.67 ± 3.20 † | 12.00 ± 10.82 | 35.33 ± 22.19 | 7.33 ± 6.35 |
| Tail deformity | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 2.58 ± 6.33 | 0.89 ± 2.67 | 0.00 ± 0.00 | 11.33 ± 10.26 *, ‡ | 0.00 ± 0.00 |
| Pericardial edema | 1.67 ± 2.89 | 1.67 ± 2.89 | 4.17 ± 6.62 | 11.42 ± 11.10 | 5.89 ± 9.78 | 2.67 ± 4.62 | 22.00 ± 6.25 | 3.67 ± 6.35 |
| Yolk-sac edema | 0.00 ± 0.00 | 2.00 ± 3.46 | 4.83 ± 5.84 | 21.58 ± 20.65 | 8.56 ± 9.72 | 0.00 ± 0.00 | 24.67 ± 4.51 | 3.67 ± 6.35 |
| Thicker yolk | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.42 ± 1.44 | 0.00 ± 0.00 | 0.00 ± 0.00 | 2.67 ± 4.62 | 11.33 ± 10.26 **, ‡‡ | 3.33 ± 2.89 |
| Slight yolk | 0.00 ± 0.00 | 2.00 ± 3.46 | 0.00 ± 0.00 | 6.25 ± 9.63 | 0.89 ± 2.67 | 0.00 ± 0.00 | 4.67 ± 8.08 | 0.00 ± 0.00 |
| Blood vessel | 0.00 ± 0.00 | 2.00 ± 3.46 | 6.58 ± 7.22 | 12.50 ± 29.89 | 5.00 ± 7.14 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Blood | 0.00 ± 0.00 | 0.00 ± 0.00 | 6.92 ± 7.23 | 10.42 ± 10.05 | 5.67 ± 6.10 | 0.00 ± 0.00 | 24.67 ± 4.51 ** | 3.67 ± 6.35 |
| Spinal curvature | 0.00 ± 0.00 | 0.00 ± 0.00 | 5.08 ± 9.13 | 3.08 ± 4.62 | 1.67 ± 5.00 | 1.67 ± 2.89 | 0.00 ± 0.00 | 3.33 ± 2.89 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geum, S.W.; Yeo, M.-K. Reduction in Toxicity of Polystyrene Nanoplastics Combined with Phenanthrene through Binding of Jellyfish Mucin with Nanoplastics. Nanomaterials 2022, 12, 1427. https://doi.org/10.3390/nano12091427
Geum SW, Yeo M-K. Reduction in Toxicity of Polystyrene Nanoplastics Combined with Phenanthrene through Binding of Jellyfish Mucin with Nanoplastics. Nanomaterials. 2022; 12(9):1427. https://doi.org/10.3390/nano12091427
Chicago/Turabian StyleGeum, Sun Woo, and Min-Kyeong Yeo. 2022. "Reduction in Toxicity of Polystyrene Nanoplastics Combined with Phenanthrene through Binding of Jellyfish Mucin with Nanoplastics" Nanomaterials 12, no. 9: 1427. https://doi.org/10.3390/nano12091427
APA StyleGeum, S. W., & Yeo, M.-K. (2022). Reduction in Toxicity of Polystyrene Nanoplastics Combined with Phenanthrene through Binding of Jellyfish Mucin with Nanoplastics. Nanomaterials, 12(9), 1427. https://doi.org/10.3390/nano12091427






