Tetrahedral DNA Framework-Programmed Electrochemical Biosenors with Gold Nanoparticles for Ultrasensitive Cell-Free DNA Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Instruments
2.2. Synthesis and Characterization of TDFs
2.3. Synthesis and Characterization of HCR Structures
2.4. Development of E-cfDNA Sensor
2.5. cfDNA Detection by E-cfDNA Sensor
3. Results and Discussion
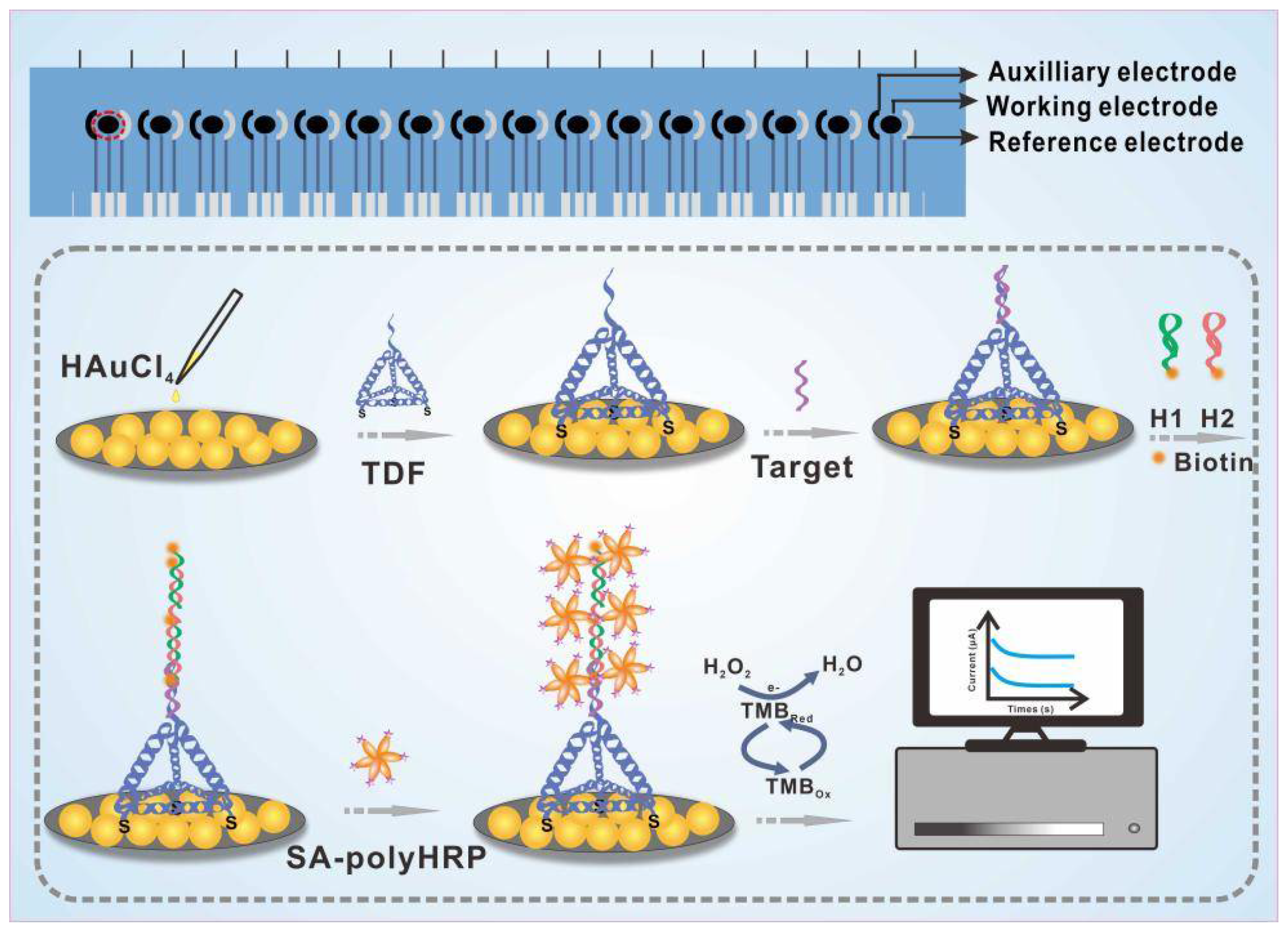
3.1. Principle of the E-cfDNA Sensor
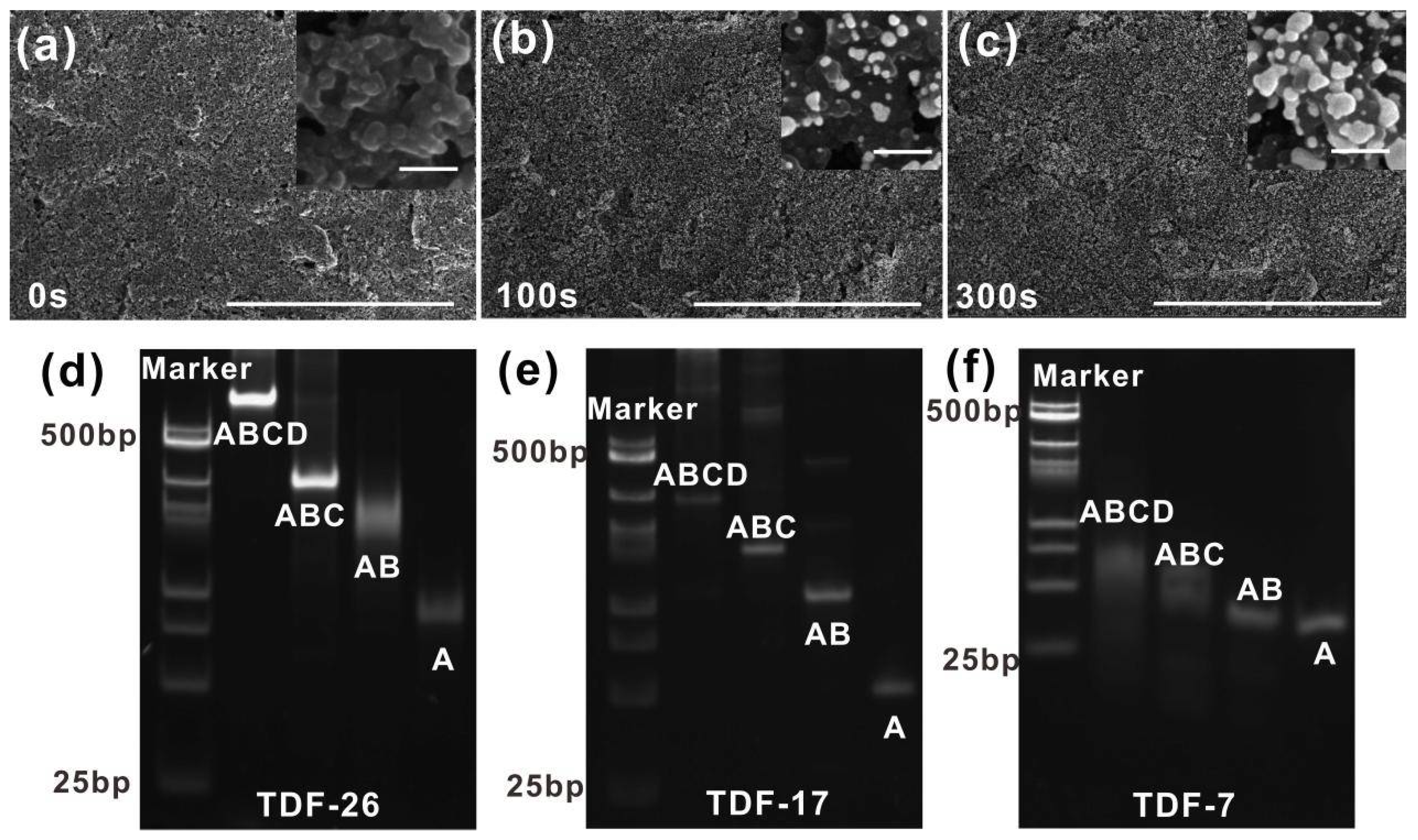
3.2. Characterization and Optimization of Treated Electrode
3.3. Comparison of Capture Performance among Different Probes
3.4. Performance Verification of the E-cfDNA Sensor Mediated by TDF Regulation
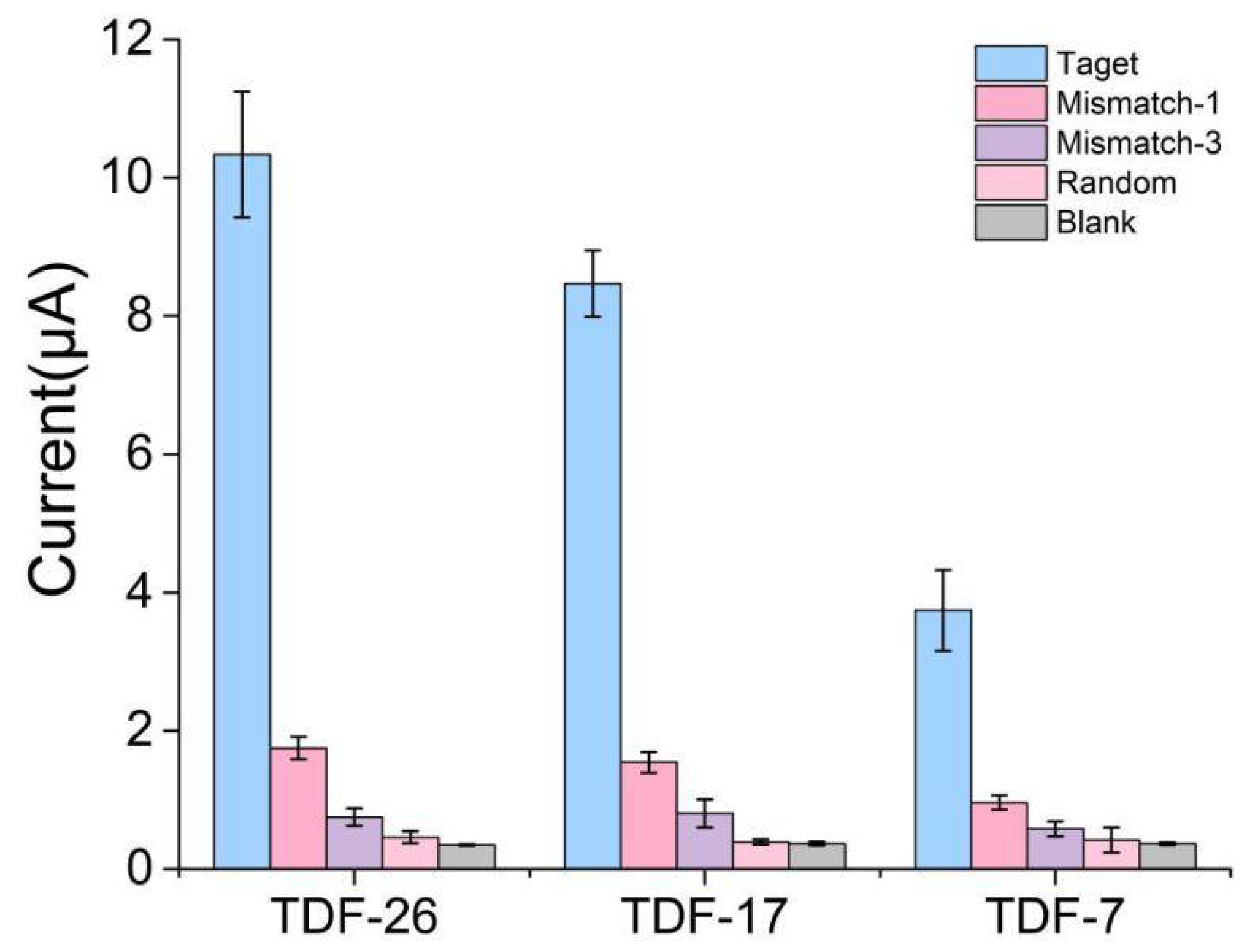
3.5. Selective Ability of the E-cfDNA Sensor
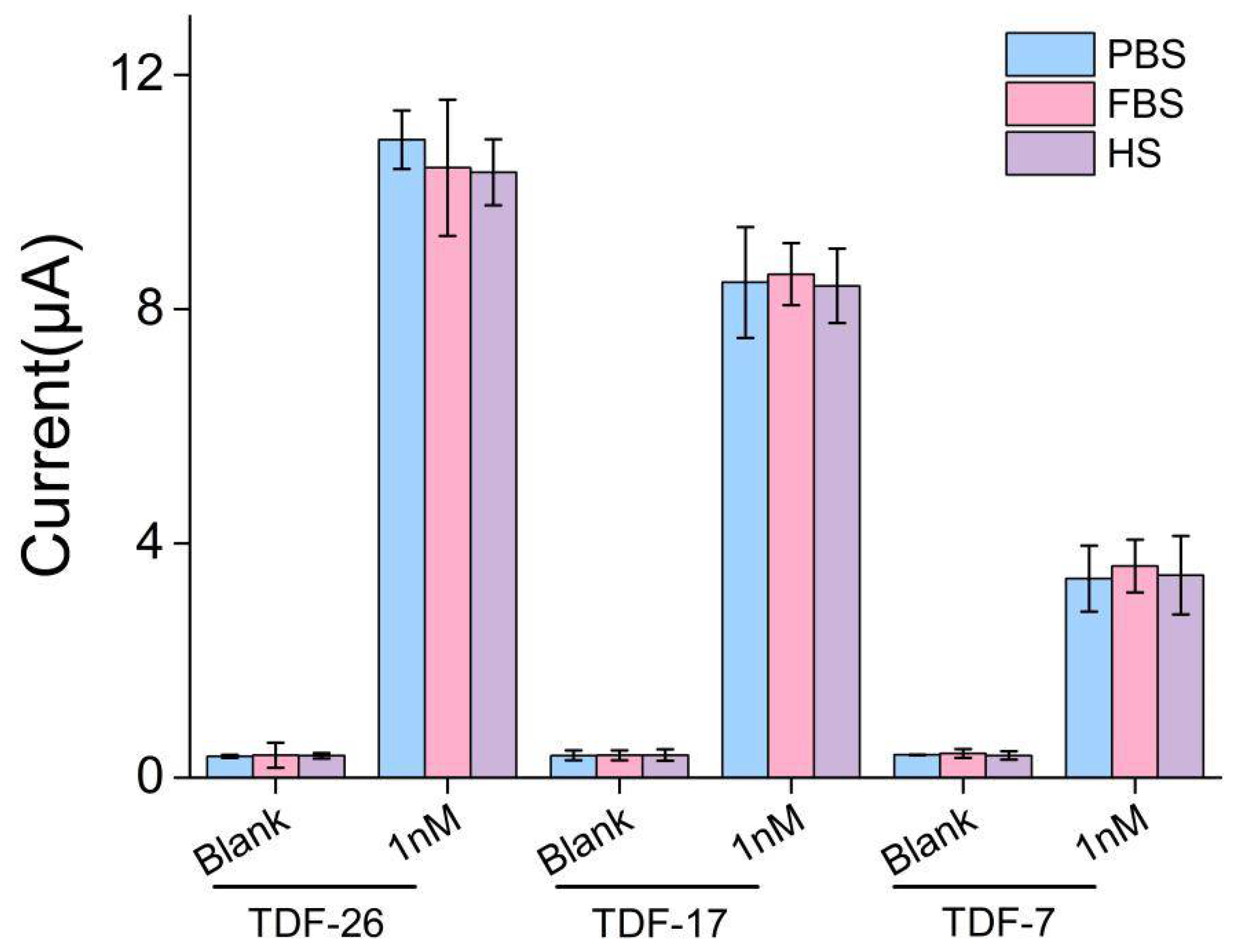
3.6. Application to the Clinical Utility
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Polski, A.; Xu, L.Y.; Prabakar, R.K.; Kim, J.W.; Shah, R.; Jubran, R.; Kuhn, P.; Cobrinik, D.; Hicks, J.; Berry, J.L. Cell-Free DNA Tumor Fraction in the Aqueous Humor Is Associated With Therapeutic Response in Retinoblastoma Patients. Transl. Vis. Sci. Technol. 2020, 9, 30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Gerratana, L.; Zhang, Y.; Flaum, L.; Shah, A.; Davis, A.; Behdad, A.; Gradishar, W.; Platanias, L.; Cristofanilli, M. ESR1 mutation in cell free DNA (cfDNA) is associated with significantly increased circulating tumor cell (CTC)-clusters and progress in stage III/IV breast cancer after systemic treatments. Cancer Res. 2019, 79 (Suppl. S4), P4-01-04. [Google Scholar] [CrossRef]
- Sato, K.A.; Nishizuka, S.S.; Iwaya, T.; Kume, K.; Otsuka, K.; Wakabayashi, G. Tumor-unique mutation detection in cell-free DNA to monitor colorectal tumor burden using a cancer-associated gene sequencing panel. Cancer Res. 2015, 75 (Suppl. S15), 5234. [Google Scholar] [CrossRef]
- Kustanovich, A.; Schwartz, R.; Peretz, T.; Grinshpun, A. Life and death of circulating cell-free DNA. Cancer Biol. Ther. 2019, 20, 1057–1067. [Google Scholar] [CrossRef] [Green Version]
- Miller, D.E.; Hawley, S.; Miller, D.E.; Hawley, S. Isolation, Sequencing and Analysis of Human Cell-Free DNA (Cfdna). J. Investig. Med. 2014, 62, 732–733. [Google Scholar]
- Assi, R.E.; Albitar, M.; Ma, W.L.; Patel, K.; Takahashi, K.; Yilmaz, M.; Short, N.J.; Jabbour, E.; Garcia-Manero, G.; Kantarjian, H.M.; et al. Comparison of somatic mutations profiles from next-generation sequencing (NGS) of cell-free DNA (cfDNA) versus bone marrow (BM) in acute myeloid leukemia (AML). J. Clin. Oncol. 2018, 36 (Suppl. S15), 7051. [Google Scholar] [CrossRef]
- Tang, W.W.; Fu, K.; Sun, H.D.; Rong, D.W.; Wang, H.J.; Cao, H.Y. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol. Cancer 2018, 17, 137. [Google Scholar] [CrossRef]
- Choi, J.H.; Lim, J.; Shin, M.; Paek, S.H.; Choi, J.W. CRISPR-Cas12a-Based Nucleic Acid Amplification-Free DNA Biosensor via Au Nanoparticle-Assisted Metal-Enhanced Fluorescence and Colorimetric Analysis. Nano Lett. 2021, 21, 693–699. [Google Scholar] [CrossRef]
- Ma, S.H.; Zhang, Y.P.; Ren, Q.Q.; Wang, X.F.; Zhu, J.H.; Yin, F.; Li, Z.G.; Zhang, M. Tetrahedral DNA nanostructure based biosensor for high-performance detection of circulating tumor DNA using all-carbon nanotube transistor. Biosens. Bioelectron. 2022, 197, 113785. [Google Scholar] [CrossRef]
- Abi, A.; Mohammadpour, Z.; Zuo, X.L.; Safavi, A. Nucleic acid-based electrochemical nanobiosensors. Biosens. Bioelectron. 2018, 102, 479–489. [Google Scholar] [CrossRef]
- Bo, X.J.; Zhou, M.; Guo, L.P. Electrochemical sensors and biosensors based on less aggregated graphene. Biosens. Bioelectron. 2017, 89, 167–186. [Google Scholar] [CrossRef]
- Sharifi, M.; Avadi, M.R.; Attar, F.; Dashtestani, F.; Ghorchian, H.; Rezayat, S.M.; Saboury, A.A.; Falahati, M. Cancer diagnosis using nanomaterials based electrochemical nanobiosensors. Biosens. Bioelectron. 2019, 126, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Torre, R.; Costa-Rama, E.; Nouws, H.P.A.; Delerue-Matos, C. Screen-Printed Electrode-Based Sensors for Food Spoilage Control: Bacteria and Biogenic Amines Detection. Biosensors 2020, 10, 139. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Liu, W.H.; Chen, S.X.; Deng, W.P.; Dou, Y.Z.; Zhao, Z.H.; Li, J.Y.; Li, Z.H.; Yin, H.; Ding, X.T.; et al. A Carbon-Based DNA Framework Nano-Bio Interface for Biosensing with High Sensitivity and a High Signal-to-Noise Ratio. ACS Sens. 2020, 5, 3979–3987. [Google Scholar] [CrossRef] [PubMed]
- Kanyong, P.; Rawlinson, S.; Davis, J. Simultaneous electrochemical determination of dopamine and 5-hydroxyindoleacetic acid in urine using a screen-printed graphite electrode modified with gold nanoparticles. Anal. Bioanal. Chem. 2016. [Google Scholar] [CrossRef]
- Gutierrez-Sanchez, C.; Pita, M.; Vaz-Dominguez, C.; Shleev, S.; De Lacey, A.L. Gold Nanoparticles as Electronic Bridges for Laccase-Based Biocathodes. J. Am. Chem. Soc. 2012, 134, 17212–17220. [Google Scholar] [CrossRef]
- Wang, J.X.; Drelich, A.J.; Hopkins, C.M.; Mecozzi, S.; Li, L.J.; Kwon, G.; Hong, S. Gold nanoparticles in virus detection: Recent advances and potential considerations for SARS-CoV-2 testing development. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2022, 14, e1754. [Google Scholar] [CrossRef]
- Yang, F.; Li, Q.; Wang, L.H.; Zhang, G.J.; Fan, C.H. Framework-Nucleic-Acid-Enabled Biosensor Development. ACS Sens. 2018, 3, 903–919. [Google Scholar] [CrossRef]
- Chand, R.; Neethirajan, S. Microfluidic platform integrated with graphene-gold nano-composite aptasensor for one-step detection of norovirus. Biosens. Bioelectron. 2017, 98, 47–53. [Google Scholar] [CrossRef]
- Senel, M.; Dervisevic, M.; Kokkokoglu, F. Electrochemical DNA biosensors for label-free breast cancer gene marker detection. Anal. Bioanal. Chem. 2019, 411, 2925–2935. [Google Scholar] [CrossRef]
- Zeng, D.D.; Zhang, H.; Zhu, D.; Li, J.; San, L.L.; Wang, Z.H.; Wang, C.G.; Wang, Y.S.; Wang, L.H.; Zuo, X.L.; et al. A novel ultrasensitive electrochemical DNA sensor based on double tetrahedral nanostructures. Biosens. Bioelectron. 2015, 71, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Pei, H.; Lu, N.; Wen, Y.L.; Song, S.P.; Liu, Y.; Yan, H.; Fan, C.H. A DNA Nanostructure-based Biomolecular Probe Carrier Platform for Electrochemical Biosensing. Adv. Mater. 2010, 22, 4754–4758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Squires, T.M.; Messinger, R.J.; Manalis, S.R. Making it stick: Convection, reaction and diffusion in surface-based biosensors. Nat. Biotechnol. 2008, 26, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Zhu, D.; Zhang, Y.N.; Wang, L.H.; Fan, C.H. DNA nanotechnology-enabled biosensors. Biosens. Bioelectron. 2016, 76, 68–79. [Google Scholar] [CrossRef]
- Mitchell, N.; Schlapak, R.; Kastner, M.; Armitage, D.; Chrzanowski, W.; Riener, J.; Hinterdorfer, P.; Ebner, A.; Howorka, S. A DNA Nanostructure for the Functional Assembly of Chemical Groups with Tunable Stoichiometry and Defined Nanoscale Geometry. Angew. Chem. Int. Ed. 2009, 48, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.H.; Wang, J.J.; Zhou, G.B.; Wang, J.B.; Wu, N.; Lu, J.X.; Gao, J.M.; Chen, X.Q.; Shi, J.Y.; Zuo, X.L.; et al. Programmable Engineering of a Biosensing Interface with Tetrahedral DNA Nanostructures for Ultrasensitive DNA Detection. Angew. Chem. Int. Ed. 2015, 54, 2151–2155. [Google Scholar] [CrossRef]
- Feng, D.Z.; Su, J.; He, G.F.; Xu, Y.; Wang, C.G.; Zheng, M.M.; Qian, Q.L.; Mi, X.Q. Electrochemical DNA Sensor for Sensitive BRCA1 Detection Based on DNA Tetrahedral-Structured Probe and Poly-Adenine Mediated Gold Nanoparticles. Biosensors 2020, 10, 78. [Google Scholar] [CrossRef]
- Wang, H.F.; Ma, R.N.; Sun, F.; Jia, L.P.; Zhang, W.; Shang, L.; Xue, Q.W.; Jia, W.L.; Wang, H.S. A versatile label-free electrochemical biosensor for circulating tumor DNA based on dual enzyme assisted multiple amplification strategy. Biosens. Bioelectron. 2018, 122, 224–230. [Google Scholar] [CrossRef]
- Luo, Z.W.; Xu, Y.; Huang, Z.J.; Chen, J.M.; Wang, X.Q.; Li, D.; Li, Y.X.; Duan, Y.X. A rapid, adaptative DNA biosensor based on molecular beacon-concatenated dual signal amplification strategies for ultrasensitive detection of p53 gene and cancer cells. Talanta 2020, 210, 120638. [Google Scholar] [CrossRef]
- Chen, S.C.; Chen, K.T.; Jou, A.F.J. Polydopamine-gold composite-based electrochemical biosensor using dual-amplification strategy for detecting pancreatic cancer-associated microRNA. Biosens. Bioelectron. 2021, 173, 112815. [Google Scholar] [CrossRef]
- Chen, Y.; Xiang, J.Y.; Liu, B.; Chen, Z.B.; Zuo, X. Gold nanoparticle-engineered electrochemical aptamer biosensor for ultrasensitive detection of thrombin. Anal. Methods 2020, 12, 3729–3733. [Google Scholar] [CrossRef] [PubMed]
- Bi, S.; Yue, S.Z.; Zhang, S.S. Hybridization chain reaction: A versatile molecular tool for biosensing, bioimaging, and biomedicine. Chem. Soc. Rev. 2017, 46, 4281–4298. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Gao, Y.; Wang, S.Q.; Qin, Y.; Xu, L.; Jin, D.; Yang, F.; Zhang, G.J. In situ hybridization chain reaction mediated ultrasensitive enzyme-free and conjugation-free electrochemcial genosensor for BRCA-1 gene in complex matrices. Biosens. Bioelectron. 2016, 80, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.L.; Lin, M.H.; Wang, P.; Pei, H.; Yan, J.; Sho, J.Y.; Huang, Q.; He, D.N.; Fan, C.H.; Zuo, X.L. Hybridization Chain Reaction Amplification of MicroRNA Detection with a Tetrahedral DNA Nanostructure-Based Electrochemical Biosensor. Anal. Chem. 2014, 86, 2124–2130. [Google Scholar] [CrossRef]
- Wen, Y.L.; Liu, G.; Pei, H.; Li, L.Y.; Xu, Q.; Liang, W.; Li, Y.; Xu, L.; Ren, S.Z.; Fan, C.H. DNA nanostructure-based ultrasensitive electrochemical microRNA biosensor. Methods 2013, 64, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Singh, N.; Tomar, V.; Chandra, R. A review on electrochemical detection of serotonin based on surface modified electrodes. Biosens. Bioelectron. 2018, 107, 76–93. [Google Scholar] [CrossRef]
- Govindasamy, M.; Mani, V.; Chen, S.M.; Chen, T.W.; Sundramoorthy, A.K. Methyl parathion detection in vegetables and fruits using silver@graphene nanoribbons nanocomposite modified screen printed electrode. Sci. Rep. 2017, 7, 46471. [Google Scholar] [CrossRef]
- Li, F.Q.; Mao, X.H.; Li, F.; Li, M.; Shen, J.L.; Ge, Z.L.; Fan, C.H.; Zuo, X.L. Ultrafast DNA Sensors with DNA Framework-Bridged Hybridization Reactions. J. Am. Chem. Soc. 2020, 142, 9975–9981. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, X.X.; Zhang, C.X.; Xu, Y.; Su, J.; Lu, X.L.; Shi, J.Y.; Wang, L.H.; Landry, M.P.; Zhu, Y.; et al. A DNA tetrahedral structure-mediated ultrasensitive fluorescent microarray platform for nucleic acid test. Sens. Actuators B-Chem. 2020, 321, 128538. [Google Scholar] [CrossRef]
- Lv, H.Y.; Chen, A.Y.; Cheng, W.Q.; Kong, L.S.; Zhao, M.; Ding, S.J.; Ju, H.X.; Cheng, W. Efficient DNA Walker Guided with Well-Regulated Interfacial Tracks for Ultrasensitive Electrochemiluminescence Biosensing. Anal. Chem. 2020, 92, 15624–15631. [Google Scholar] [CrossRef]
- Liu, P.F.; Zhao, K.R.; Liu, Z.J.; Wang, L.; Ye, S.Y.; Liang, G.X. Cas12a-based electrochemiluminescence biosensor for target amplification-free DNA detection. Biosens. Bioelectron. 2021, 176, 112954. [Google Scholar] [CrossRef] [PubMed]


| Name | Sequence (5′-3′) |
|---|---|
| Mismatch-1 | TGGTAACAGTGTGAGGTTTAACGGAACAAATGGAAGAAAATC |
| Mismatch-3 | TGGTAACAGTGTGAGGTTTAACGGAACAAGATGATGAAAATC |
| Random | TGGTAACAGTGTGAGGTTTAACGTCGATGCCTGATCTTGGTA |
| Target-BRCA-1 | TGGTAACAGTGTGAGGTTTAACGGAACAAAAGGAAGAAAATC |
| Techniques | Name | Linear Range | LOD | Refs. |
|---|---|---|---|---|
| Fluorescence Biosensors | CRISPR-Cas12a-based cfDNA biosensing system | 1 fM to 100 pM | 0.34 fM | [8] |
| DNA tetrahedral-based fluorescent microarray platform | 100 aM to 1 pM | 10 aM | [39] | |
| Electrochemical Biosensors | Label-free electrochemical biosensor | 0.01 fM to1 pM | 2.4 aM | [28] |
| HCR and DNA nanostructure-based electrochemical biosensor | 1 fM to 100 pM | 100 aM | [34] | |
| Electrochemiluminescence Biosensors | DNA walk-based electrochemiluminescence biosensing | 1 fM to 100 pM | 0.18 fM | [40] |
| Cas12a-based electrochemiluminescence biosensor | 1 pM to 10 nM | 0.48 pM | [41] | |
| E-cfDNA sensor | 1 aM to 1 pM | 1 aM | This work | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Wang, W.; Xu, Y.; Zhao, X.; Li, S.; Qian, Q.; Mi, X. Tetrahedral DNA Framework-Programmed Electrochemical Biosenors with Gold Nanoparticles for Ultrasensitive Cell-Free DNA Detection. Nanomaterials 2022, 12, 666. https://doi.org/10.3390/nano12040666
Wang C, Wang W, Xu Y, Zhao X, Li S, Qian Q, Mi X. Tetrahedral DNA Framework-Programmed Electrochemical Biosenors with Gold Nanoparticles for Ultrasensitive Cell-Free DNA Detection. Nanomaterials. 2022; 12(4):666. https://doi.org/10.3390/nano12040666
Chicago/Turabian StyleWang, Chenguang, Wei Wang, Yi Xu, Xiaoshuang Zhao, Shuainan Li, Qiuling Qian, and Xianqiang Mi. 2022. "Tetrahedral DNA Framework-Programmed Electrochemical Biosenors with Gold Nanoparticles for Ultrasensitive Cell-Free DNA Detection" Nanomaterials 12, no. 4: 666. https://doi.org/10.3390/nano12040666
APA StyleWang, C., Wang, W., Xu, Y., Zhao, X., Li, S., Qian, Q., & Mi, X. (2022). Tetrahedral DNA Framework-Programmed Electrochemical Biosenors with Gold Nanoparticles for Ultrasensitive Cell-Free DNA Detection. Nanomaterials, 12(4), 666. https://doi.org/10.3390/nano12040666






