Bacteria-Assisted Transport of Nanomaterials to Improve Drug Delivery in Cancer Therapy
Abstract
:1. Introduction
2. Bacteria
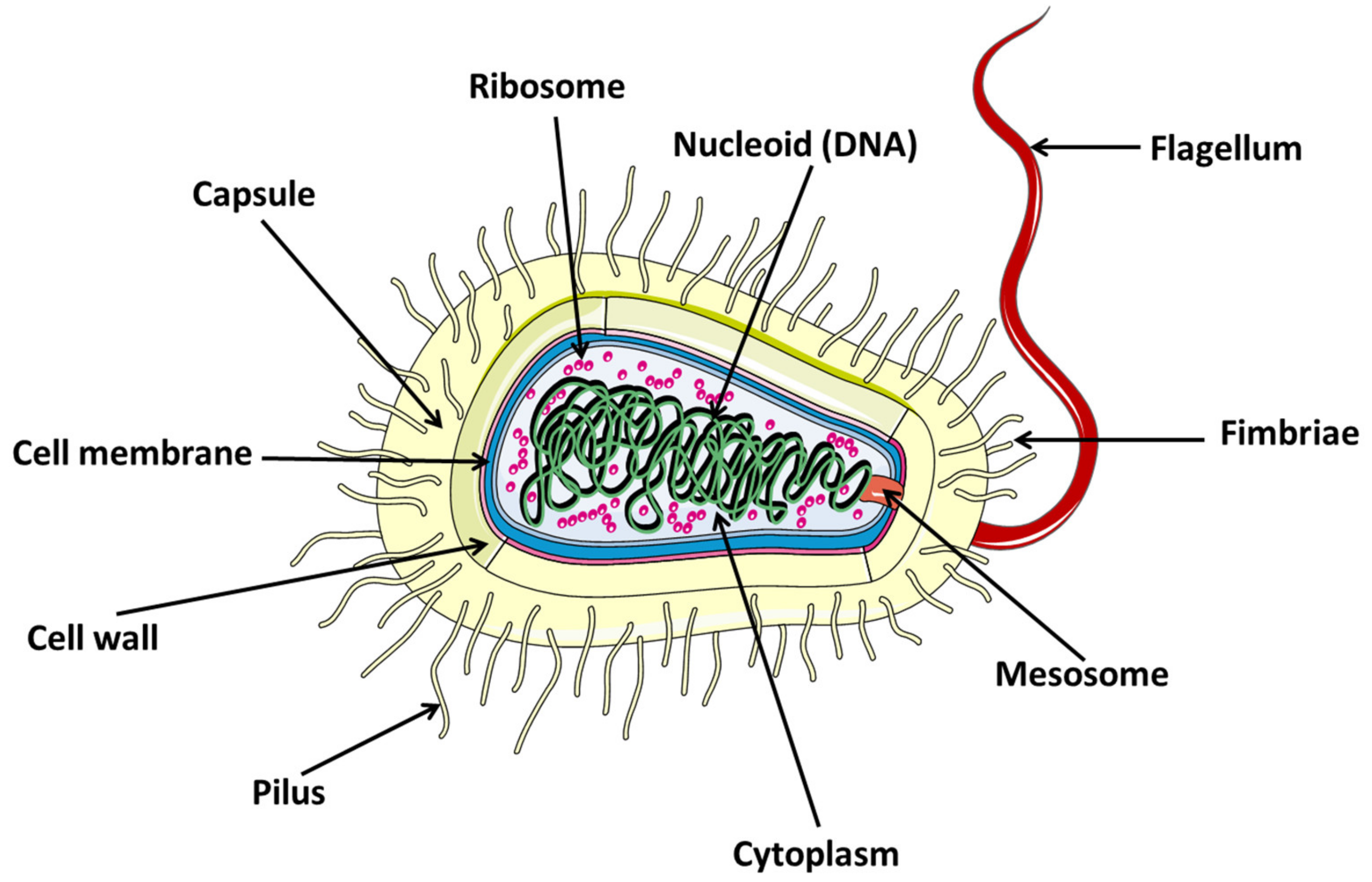
2.1. Definition
2.2. Bacteria Types
2.2.1. Bacteria Based on Basic Shape
- Cylindrical: These bacteria grow by increasing the length of the body cylinder. During their cell division, they are able to synthesize new cell poles for a short time, requiring thorough controls during cell division. In this group are Bacillus subtilis, Corynebacterium diphtheria, Helicobacter pylori, Salmonella, Escherichia coli, or Caulobacter crescentus (curved rods) [37].
- Coccal: This group grows through their division septa and must divide to grow. Their cell wall synthesis machinery is located in their division septa. In addition, because they depend on these septa and are spheroidal, they do not need to form chains, as in each generation their planes usually alternate. Thus, Neisseria gonorrhoeae divide into two alternating planes, Deinococcus radiodurans or S. aureus form bundles or clusters, and Staphylococcus aureus divide into three alternating planes [37].
- Ovococcal: Bacteria in this form usually grow through their dividing septum by modifying the extent of their length. They require changes in their mode of growth and need to place new dividing septa at the midpoint of the cell. If cell separation is not effective, these bacteria form cell chains. This group may include the bacterium Streptococcus pneumonia [37].
2.2.2. Bacteria Based on Metabolism
- Autotrophs: These bacteria have a very complex metabolism. They are capable of assimilating inorganic matter and transforming it into organic matter to produce the biomolecules necessary for their development. They are limited to using an inorganic source of carbon, such as CO2. These bacteria have no need to invade other organisms, nor do they need to break down dead organic matter to obtain the nutrients they need to survive [38].Depending on the metabolic system used by these bacteria to take inorganic compounds and transform them into organic compounds, they are divided into: Photoautotrophs (For the process of transformation of inorganic matter into organic matter, they use sunlight as a source of energy) and Chemoautotrophs (These bacteria need chemical energy to carry out their metabolic processes) [38]. Photoautotrophic bacteria can in turn be classified as oxygenic (they need photosynthesis to capture solar energy and convert it into chemical energy) and anoxygenic (these bacteria are anaerobic as they do not need oxygen for the respiration process) [39].Ultimately, these bacteria are important for ensuring the survival of other living things because they capture inorganic compounds that are toxic to other microorganisms. In addition, compounds released by these autotrophic bacteria can be assimilated by some heterotrophic bacteria.
- Heterotrophs: These bacteria use organic matter as a carbon source. This organic matter is transformed into energy and nutrients. Therefore, these organic materials are usually rich in energy, such as lipids, carbohydrates, and proteins. They need organic matter that has previously been synthetized by an autotrophic organism or other heterotrophic organisms. Other elements than carbon can be taken up as inorganic matter. Ultimately, some of these bacteria can cause infectious diseases in humans [40].
2.2.3. Bacteria Based on Cell Wall
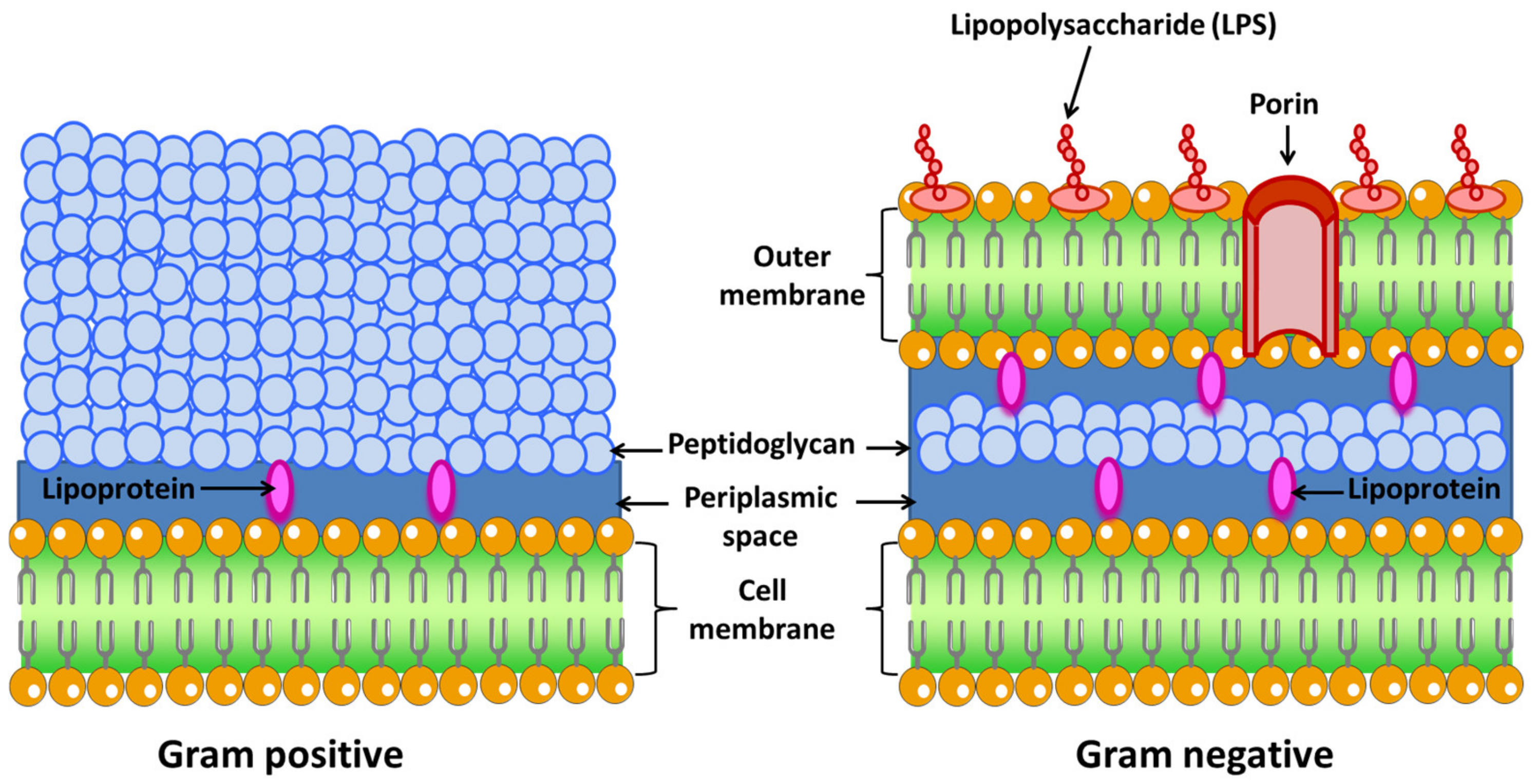
- Gram-negative: These bacteria possess a cell envelope that is composed of three main layers: the inner or cytoplasmic membrane, the peptidoglycan cell wall, and the outer membrane. The two membrane layers delimit a cellular compartment called the periplasm where a set of proteins are found [42]. From the outside to the inside, the outer membrane is the first layer. This membrane is characteristic of Gram-negative bacteria, while Gram-positive bacteria lack this organelle [43]. It is a lipid bilayer where phospholipids are found exclusively in the inner part of the membrane. The outer side is composed of glycolipids, mostly lipopolysaccharide (LPS). The human innate immune system is sensitized to LPS, an endotoxin that is recognized as antigen. It is a sure indicator of infection, since is responsible for the endotoxic shock associated with sepsis when caused by Gram-negative organisms [44].The proteins present in this membrane are usually classified into β-barrel proteins and lipoproteins. For example, in E. coli, the outer membrane contains few enzymes, but they are essential for ensuring their survival. These enzymes are a protease (OmpT) [45], an LPS-modifying enzyme (PagP) [46], and a phospholipase (PldA) [47]. The active site of these enzymes is oriented towards the outside of the cell (OmpT). The function of this membrane is to be a protective barrier. In fact, certain Gram-negative bacteria are more resistant to antibiotics than Gram-positive bacteria, i.e., Pseudomonas. In addition, LPS is central to the barrier function of this outer membrane, as it enables the maintenance and organization of this membrane. LPS is the most important surface antigen on these bacteria and therefore plays an important role in activating the immune system [48]. LPS is also responsible for Gram-negative bacteria-driven shock as it has an endotoxic action. Other functions include mediating adherence to cells and host tissues, inhibition of antibodies, and molecular mimicry [49].This membrane is bound to an underlying peptidoglycan by Braun’s lipoprotein (Lpp) [50]. Lipids attached to the amino terminus of this lipoprotein are embedded in the outer membrane. For example, Lpp is the most abundant protein in E. coli, with more than 500,000 molecules per cell [51]. This peptidoglycan is responsible for the rigidity of the bacterial cell wall and determines the morphology of the cell.Between the outer and inner membrane there is a watery cellular compartment called the periplasm, which is densely packed with proteins [52]. This compartment allows Gram-negative bacteria to capture potentially harmful enzymes, such as alkaline phosphatase or RNAase. It also contains periplasmic binding proteins important in chemotaxis, amino acid, and sugar transport, and chaperone-like molecules important in cell envelope biogenesis [53].As mentioned above, bacteria do not have intracellular organelles, so the membrane-associated functions of these organelles are performed in the inner membrane. Additionally, the proteins responsible for energy production, protein secretion, transport, and lipid biosynthesis are located in the inner membrane, although their location is different compared to eukaryotic cells [54]. This membrane is a lipid bilayer of phospholipids. For example, in E. coli phospholipids are found such as phosphatidyl glycerol, serine, and cardiolipin and phosphotidyl ethanolamine [55]. Within this group are Escherichia coli, Salmonella, Hemophilus influenzae, Neisseria, and Bordetella pertussis, among others.
- Gram-positive: The cell wall of Gram-positive bacteria is different from Gram-negative (Figure 2). First, they have no outer membrane. Lacking this membrane, the peptidoglycan layer is thicker than in Gram-negatives so that they can withstand the pressure exerted on the plasma membrane. They tend to live in harsh environments. Some of these bacteria are found in the gut. Anionic polymers called teichoic acids exist in the peptidoglycan layer, which are made up of repeated glycerol phosphates, ribitol phosphates, or glucosyl phosphates. These polymers make up 60% of the entire mass of the cell envelope of Gram-positive bacteria, making them responsible for the structure and function of the cell wall [56]. As there is no outer membrane to contain the extracellular proteins, these proteins have elements that cause them to be retained in the membrane or very close to it. Some of them are anchored to membrane-embedded lipids or have helices that pass through the membrane or bind or covalently associate with peptidoglycan [57]. This group includes Streptococcus, Staphylococcus aureus, Clostridium botulinum, and Bacillus anthracis, among others.
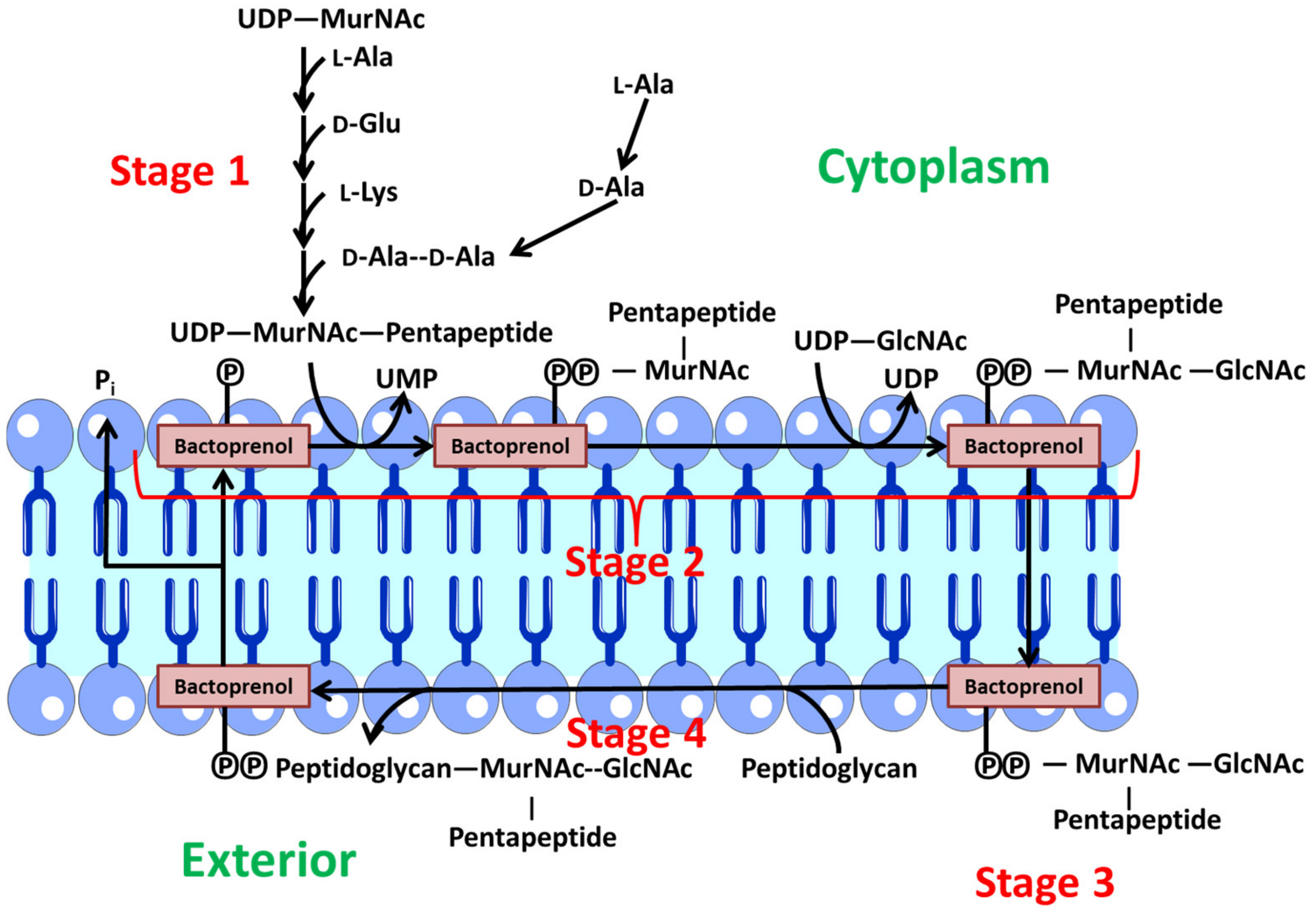
2.3. Peptidoglycans Biosynthesis
- Stage 1: Synthesis of precursors in the cytoplasm. First, the monosaccharides N-acetylmuramic acid and acetylglucosamine, which form the peptidoglycan backbone, are activated by binding to uridine diphosphate. Then, a sequential and orderly addition of the various amino acids to N-acetylmuramic acid takes place. At this point, a pentapeptide is formed. Finally, the dipeptide d-alanyl-d-alanine binds. This dipeptide is synthesized in two steps. A first stage is through a racemase that converts l-ala to d-ala and a second stage where a peptide bond is formed between two d-ala [62].
- Stage 2: In this stage, these precursors are transferred to a lipid transporter (undecaprenyl-phosphate or bactoprenol (Lip-P)) in the cytoplasmic membrane, where the disaccharide units are created with the pentapeptide. At this point, a Β (1→4) bond is generated between MurNAc and GlcNAc. Therefore, Lip-P-P-MurNAc(pentapeptide)-GlcNAc is obtained. This polypeptide is anchored to the inner part of the membrane facing the cytoplasm via bactoprenol [62].
- Stage 3: Polymerization of various disaccharides. The bactoprenol is flipped from the inner to the outer layer, so that the precursor resulting from phase 2 is oriented towards the aqueous environment outside the membrane. At this point, polymerization of several disaccharide units takes place via a transglycosidation reaction. The Lip-P-P-MurNAc(pentapeptide)-GlcNAc disaccharide unit binds to the free end of another pre-existing chain, which is also bound to another Lip-P-P molecule. At this point, one of the Lip-P-P is released in its pyrophosphorylated form. An alkaline phosphatase acts on this molecule, which is responsible for eliminating the terminal phosphate, regenerating Lip-P again, which is then free to begin another cycle [62].
- Stage 4: The polymer generated in the previous stage is a linear chain of uncross-linked peptidoglycan bound to the membrane lipid transporter. This nascent polymer reacts with another pre-existing peptidoglycan acceptor via a transpeptidation reaction. The peptide bond generated between d-Ala (position 4) and d-Ala (position 5) of the nascent peptidoglycan is replaced by another peptide bond between the carboxylic group of the d-Ala (position 4) of the nascent peptidoglycan and the free amine group of the diamino acid (position 3) of the acceptor peptidoglycan. The energy for this reaction is provided by the hydrolysis of the peptide bond formed between the two terminal d-Ala, leading to the release of a d-Ala (position 5) in each transpeptidation reaction [62].
3. Bacteria in Cancer Therapy
3.1. Tumor Physiology
- Abnormal vasculature: To meet the needs mentioned above, tumors develop their own functional vascular supply. For this, tumor cells secrete a series of pro-angiogenic factors that recruit endothelial cells for the formation of new blood vessels called angiogenesis [64]. The tumor vascular network formed is chaotic and irregular compared to the vascular supply of the normal tissue from which it begins to develop. This imbalance creates a neo-vasculature characterized by abrupt and leaky vessels that exhibit disordered branching and interconnection patterns. Ultimately, this vasculature is disordered and lacks a hierarchy of blood vessels compared to healthy tissues, where there is a regular and organized branching order [65]. These abnormalities cause a heterogeneity of tumor blood flow that interfere with the correct and homogeneous distribution of a drug within the tumor [66]. In addition, leaky blood vessels make it easier for macromolecules to reach tumor cells from the bloodstream, but also cause high interstitial pressures in tumors resulting in inhibition of drug accumulation in the tumor [67]. By this, an adverse microenvironment for cell growth is created within the tumor, which leads to the apparition of resistant cells to conventional cancer therapies as certain types of chemotherapy and radiation [68].
- High intratumoral pressure: Within tumors, tumor vessels do not supply blood efficiently due to high interstitial pressure favoring extravasation [69]. During proliferation, mechanical compression of the vessels together with high vascular permeability leads to increased interstitial fluid within the tumor. The interstitial hypertension exists due to the absence of lymphatic vessels preventing proper drainage of this extracellular fluid. In addition, this hypertension can inhibit drug diffusion and further compress the blood vessels by diverting blood from the center to the periphery of the tumor.
- High cell proliferation: Cell proliferation present different gradients due to the heterogeneity of the blood supply within the tumor microenvironment. This gradient means that cells close to the vessels increase rapidly while cells located in inner regions are deprived of nutrients. For this reason, the cell density is higher near the vessels compared to those far from the vessels. This increased cell density can also hinder drug penetration. On the other hand, hypoxic zones occur in the inner regions of the tumor that lack nutrients and oxygen supply, leading to the development of necrotic and senescent cells [70].
3.2. Hypoxia as Chemoattractant for Bacteria
4. Bacteria as Nanocarrier
4.1. Motion Capacity of Bacteria
4.2. Distribution of Drugs inside Tumors
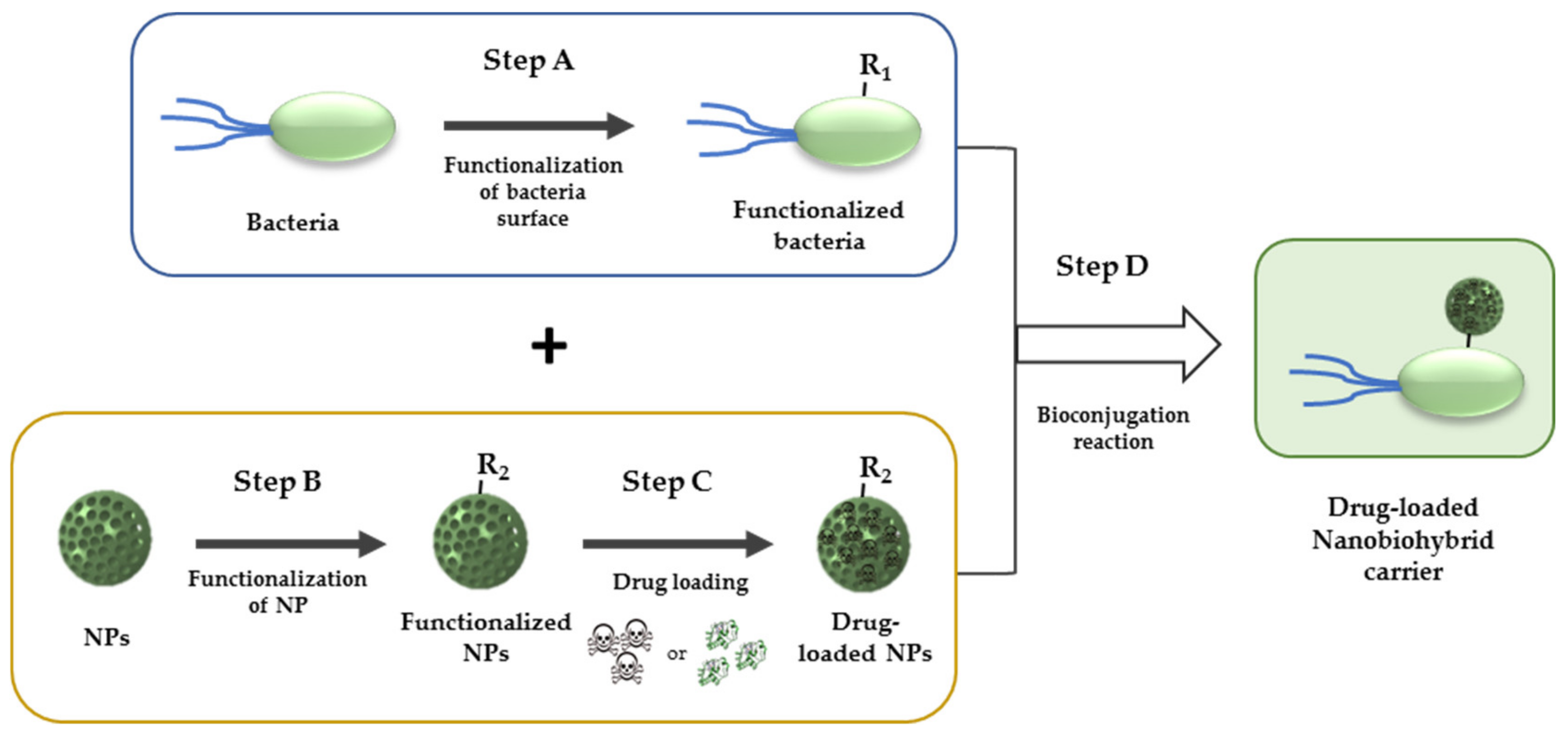
4.3. Bioconjugation in Living Organisms
- Copper-catalyzed [3+2] azide-alkyne cycloaddition: Azides are 1,3-dipoles, thus can undergo reactions with dipolarophiles as activated alkynes (H in Table 1). The reaction is thermodynamically favourable since the dipolarophile is activated but requires Cu (I) catalyst for an efficient reaction [115]. However, present a major disadvantage since the metal catalyst exhibit cellular toxicity to bacteria.
- Strain-promoted [3+2] azide-alkyne cycloaddition: Represent a catalyst-free alternative that employ as complementary group a highly strained cyclooctyne ring (I in Table 1). The reaction between azide and strained alkyne is thermodynamically favoured at room temperature and no toxic effects are observable [114].
5. Nanobiohybrid Bacterial Carriers
5.1. Polymeric Nanoparticles
5.2. Silica Nanoparticles (SiNPs)
5.3. Carbon Nanoparticles
5.4. Metallic Nanoparticles
5.5. Liposomal Nanoparticles
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chabner, B.A.; Roberts, T.G., Jr. Chemotherapy and the war on cancer. Nat. Rev. Cancer 2005, 5, 65–72. [Google Scholar] [CrossRef]
- Tannock, I.F. Conventional cancer therapy: Promise broken or promise delayed? Lancet 1998, 351, 9–16. [Google Scholar] [CrossRef]
- Palumbo, M.O.; Kavan, P.; Miller, W.H.; Panasci, L.; Assouline, S.; Johnson, N.; Cohen, V.; Patenaude, F.; Pollak, M.; Jagoe, R.T.; et al. Systemic cancer therapy: Achievements and challenges that lie ahead. Front. Pharmacol. 2013, 4, 57. [Google Scholar] [CrossRef] [Green Version]
- Matsumura, Y.; Maeda, H. A new concept for macromolecular therapeutics in cancer chemotherapy: Mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986, 46, 6387–6392. [Google Scholar]
- Wicki, A.; Witzigmann, D.; Balasubramanian, V.; Huwyler, J. Nanomedicine in cancer therapy: Challenges, opportunities, and clinical applications. J. Control. Release 2015, 200, 138–157. [Google Scholar] [CrossRef]
- Vallet-Regí, M.; Balas, F.; Arcos, D. Mesoporous materials for drug delivery. Angew. Chem. Int. Ed. 2007, 46, 7548–7558. [Google Scholar] [CrossRef]
- Riedinger, A.; Guardia, P.; Curcio, A.; Garcia, M.A.; Cingolani, R.; Manna, L.; Pellegrino, T. Subnanometer local temperature probing and remotely controlled drug release based on Azo-functionalized iron oxide nanoparticles. Nano Lett. 2013, 13, 2399–2406. [Google Scholar] [CrossRef]
- Martínez-Carmona, M.; Izquierdo-Barba, I.; Colilla, M.; Vallet-Regí, M. Concanavalin A-targeted mesoporous silica nanoparticles for infection treatment. Acta Biomater. 2019, 96, 547–556. [Google Scholar] [CrossRef]
- Cao-Milán, R.; Liz-Marzán, L.M. Gold nanoparticle conjugates: Recent advances toward clinical applications. Expert Opin. Drug Deliv. 2014, 11, 741–752. [Google Scholar] [CrossRef]
- Talelli, M.; Barz, M.; Rijcken, C.J.F.; Kiessling, F.; Hennink, W.E.; Lammers, T. Core-crosslinked polymeric micelles: Principles, preparation, biomedical applications and clinical translation. Nano Today 2015, 10, 93–117. [Google Scholar] [CrossRef] [Green Version]
- Deshpande, P.P.; Biswas, S.; Torchilin, V.P. Current trends in the use of liposomes for tumor targeting. Nanomedicine 2013, 8, 1509–1528. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, H.; Pérez-Andrés, E.; Thevenot, J.; Sandre, O.; Berra, E.; Lecommandoux, S. Magnetic field triggered drug release from polymersomes for cancer therapeutics. J. Control. Release 2013, 169, 165–170. [Google Scholar] [CrossRef] [Green Version]
- Villegas, M.R.; Baeza, A.; Vallet-Regí, M. Nanotechnological Strategies for Protein Delivery. Molecules 2018, 23, 1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, C.; Lu, J.; Lin, W. Hybrid nanoparticles for combination therapy of cancer. J. Control. Release 2015, 219, 224–236. [Google Scholar] [CrossRef] [Green Version]
- García, A.; González, B.; Harvey, C.; Izquierdo-Barba, I.; Vallet-Regí, M. Effective reduction of biofilm through photothermal therapy by gold core@shell based mesoporous silica nanoparticles. Microporous Mesoporous Mater. 2021, 328, 111489. [Google Scholar] [CrossRef]
- Manzano, M.; Vallet-Regí, M. Mesoporous silica nanoparticles in nanomedicine applications. J. Mater. Sci. Mater. Med. 2018, 29, 65. [Google Scholar] [CrossRef]
- Vallet-Regi, M.; Rámila, A.; Del Real, R.P.; Pérez-Pariente, J. A new property of MCM-41: Drug delivery system. Chem. Mater. 2001, 13, 308–311. [Google Scholar] [CrossRef]
- Aguilar-Colomer, A.; Colilla, M.; Izquierdo-Barba, I.; Jiménez-Jiménez, C.; Mahillo, I.; Esteban, J.; Vallet-Regí, M. Impact of the antibiotic-cargo from MSNs on gram-positive and gram-negative bacterial biofilms. Microporous Mesoporous Mater. 2021, 311, 110681. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Carmona, M.; Gun’ko, Y.K.; Vallet-Regí, M. Mesoporous silica materials as drug delivery: “The nightmare” of bacterial infection. Pharmaceutics 2018, 10, 279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallet-Regí, M.; González, B.; Izquierdo-Barba, I. Nanomaterials as promising alternative in the infection treatment. Int. J. Mol. Sci. 2019, 20, 3806. [Google Scholar] [CrossRef] [Green Version]
- Heras, C.; Jiménez-Holguín, J.; Doadrio, A.L.; Vallet-Regí, M.; Sánchez-Salcedo, S.; Salinas, A.J. Multifunctional antibiotic- and zinc-containing mesoporous bioactive glass scaffolds to fight bone infection. Acta Biomater. 2020, 114, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Vallet-Regí, M.; Lozano, D.; González, B.; Izquierdo-Barba, I. Biomaterials against Bone Infection. Adv. Healthc. Mater. 2020, 9, 2000310. [Google Scholar] [CrossRef] [PubMed]
- Colilla, M.; Vallet-Regí, M. Targeted stimuli-responsive mesoporous silica nanoparticles for bacterial infection treatment. Int. J. Mol. Sci. 2020, 21, 8605. [Google Scholar] [CrossRef]
- Blanco, E.; Shen, H.; Ferrari, M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015, 33, 941–951. [Google Scholar] [CrossRef]
- Felgner, S.; Kocijancic, D.; Frahm, M.; Weiss, S. Bacteria in cancer therapy: Renaissance of an old concept. Int. J. Microbiol. 2016, 2016, 8451728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busch, W. Aus der Sitzung der medicinischen Section vom 13 November 1867. Berl. Klin. Wochenschr. 1868, 5, 137. [Google Scholar]
- Coley, W.B. The treatment of inoperable sarcoma with the ’mixed toxins of erysipelas and bacillus prodigiosus. J. Am. Med. Assoc. 1898, XXXI, 456–465. [Google Scholar] [CrossRef]
- Nauts, H.C.; Swift, W.E.; Coley, B.L. The Treatment of Malignant Tumors by Bacterial Toxins as Developed by the Late William B. Coley, M.D., Reviewed in the Light of Modern Research. Cancer Res. 1946, 6, 205–216. [Google Scholar]
- McCarthy, E.F. The toxins of William B. Coley and the treatment of bone and soft-tissue sarcomas. Iowa Orthop. J. 2006, 26, 154–158. [Google Scholar] [PubMed]
- Kienle, G.S. Fever in Cancer Treatment: Coley’s Therapy and Epidemiologic Observations. Glob. Adv. Health Med. 2012, 1, 92–100. [Google Scholar] [CrossRef] [Green Version]
- Cann, S.A.H.; Van Netten, J.P.; Van Netten, C. Dr William Coley and tumour regression: A place in history or in the future. Postgrad. Med. J. 2003, 79, 672–680. [Google Scholar]
- Pylaeva, E.; Lang, S.; Jablonska, J. The essential role of type I interferons in differentiation and activation of tumor-associated neutrophils. Front. Immunol. 2016, 7, 629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baron, S. Medical Microbiology, 4th ed.; Medical Microbiology; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- Yulo, P.R.J.; Hendrickson, H.L. The evolution of spherical cell shape; progress and perspective. Biochem. Soc. Trans. 2019, 47, 1621–1634. [Google Scholar] [CrossRef]
- Matica, M.A.; Aachmann, F.L.; Tøndervik, A.; Sletta, H.; Ostafe, V. Chitosan as a wound dressing starting material: Antimicrobial properties and mode of action. Int. J. Mol. Sci. 2019, 20, 5889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, D.C.; Blair, K.M.; Salama, N.R. Staying in Shape: The Impact of Cell Shape on Bacterial Survival in Diverse Environments. Microbiol. Mol. Biol. Rev. 2016, 80, 187–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zapun, A.; Vernet, T.; Pinho, M.G. The different shapes of cocci. FEMS Microbiol. Rev. 2008, 32, 345–360. [Google Scholar] [CrossRef] [Green Version]
- Strong, D.R. Food Chains and Food Webs. In Encyclopedia of Ecology; Jørgensen, S.E., Fath, B.D., Eds.; Academic Press: Cambridge, MA, USA, 2008; pp. 1627–1636. [Google Scholar] [CrossRef]
- Finke, N.; Hoehler, T.M.; Polerecky, L.; Buehring, B.; Thamdrup, B. Competition for inorganic carbon between oxygenic and anoxygenic phototrophs in a hypersaline microbial mat, Guerrero Negro, Mexico. Environ. Microbiol. 2013, 15, 1532–1550. [Google Scholar] [CrossRef]
- Hamer, G. Microbiology of Treatment Processes. Pr. Biotech. Spec. Prod. Serv. Act 1985, 4, 819–834. [Google Scholar]
- Gram, C. Über die isolierte Färbung der Schizomyceten in Schnitt-und Trockenpräparaten. Fortschr. Med. 1884, 2, 185–189. [Google Scholar]
- Mitchell, P. Approaches to the analysis of specific membrane transport. Biol. Struct. Funct. 1961, 2, 581–599. [Google Scholar]
- Pajerski, W.; Ochonska, D.; Brzychczy-Wloch, M.; Indyka, P.; Jarosz, M.; Golda-Cepa, M.; Sojka, Z.; Kotarba, A. Attachment efficiency of gold nanoparticles by Gram-positive and Gram-negative bacterial strains governed by surface charges. J. Nanopart. Res. 2019, 21, 186. [Google Scholar] [CrossRef] [Green Version]
- Opal, S.M.; Scannon, P.J.; Vincent, J.L.; White, M.; Carroll, S.F.; Palardy, J.E.; Parejo, N.A.; Pribble, J.P.; Lemke, J.H. Relationship between plasma levels of lipopolysaccharide (LPS) and LPS-binding protein in patients with severe sepsis and septic shock. J. Infect. Dis. 1999, 180, 1584–1589. [Google Scholar] [CrossRef] [Green Version]
- Vandeputte-Rutten, L.; Kramer, R.A.; Kroon, J.; Dekker, N.; Egmond, M.R.; Gros, P. Crystal structure of the outer membrane protease Ompt from Escherichia coli suggests a novel catalytic site. EMBO J. 2001, 20, 5033–5039. [Google Scholar] [CrossRef] [Green Version]
- Hwang, P.M.; Choy, W.Y.; Lo, E.I.; Chen, L.; Forman-Kay, J.D.; Raetz, C.R.H.; Privé, G.G.; Bishop, R.E.; Kay, L.E. Solution structure and dynamics of the outer membrane enzyme PagP by NMR. Proc. Natl. Acad. Sci. USA 2002, 99, 13560–13565. [Google Scholar] [CrossRef] [Green Version]
- Snijder, H.J.; Ubarretxena-Belandia, I.; Blaauw, M.; Kalk, K.H.; Verheij, H.M.; Egmond, M.R.; Dekker, N.; Dijkstra, B.W. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature 1999, 401, 717–721. [Google Scholar] [CrossRef] [PubMed]
- De Tejada, G.M.; Heinbockel, L.; Ferrer-Espada, R.; Heine, H.; Alexander, C.; Bárcena-Varela, S.; Goldmann, T.; Correa, W.; Wiesmüller, K.H.; Gisch, N.; et al. Lipoproteins/peptides are sepsis-inducing toxins from bacteria that can be neutralized by synthetic anti-endotoxin peptides. Sci. Rep. 2015, 5, 14292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurtado, S.R.; Iregui, C.A. El Lipopolisacárido. Rev. Med. Vet. 2010, 37–45. [Google Scholar] [CrossRef]
- Braun, V. Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim. Biophys. Acta BBA-Rev. Biomembr. 1975, 415, 335–377. [Google Scholar] [CrossRef]
- Yem, D.W.; Wu, H.C. Physiological characterization of an Escherichia coli mutant altered in the structure of murein lipoprotein. J. Bacteriol. 1978, 133, 1419–1426. [Google Scholar] [CrossRef] [Green Version]
- Mullineaux, C.W.; Nenninger, A.; Ray, N.; Robinson, C. Diffusion of green fluorescent protein in three cell environments in Escherichia coli. J. Bacteriol. 2006, 188, 3442–3448. [Google Scholar] [CrossRef] [Green Version]
- Krojer, T.; Sawa, J.; Schäfer, E.; Saibil, H.R.; Ehrmann, M.; Clausen, T. Structural basis for the regulated protease and chaperone function of DegP. Nature 2008, 453, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.I.; Salama, N.R. The gram-negative bacterial periplasm: Size matters. PLoS Biol. 2018, 16, e2004935. [Google Scholar] [CrossRef]
- Raetz, C.R.; Dowhan, W. Biosynthesis and function of phospholipids in Escherichia coli. J. Biol. Chem. 1990, 265, 1235–1238. [Google Scholar] [CrossRef]
- Neuhaus, F.C.; Baddiley, J. A Continuum of Anionic Charge: Structures and Functions of d -Alanyl-Teichoic Acids in Gram-Positive Bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 686–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, J.R.; Barnett, T.C. Surface proteins of gram-positive bacteria and how they get there. Annu. Rev. Microbiol. 2006, 60, 397–423. [Google Scholar] [CrossRef]
- Vollmer, W.; Blanot, D.; De Pedro, M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef] [Green Version]
- Rogers, H.J.; Perkins, H.R.; Ward, J.B. Microbial Cell Walls and Membranes; Chapman & Hall: London, UK, 1980; ISBN 0-412-12030-5. [Google Scholar]
- Davis, K.M.; Weiser, J.N. Modifications to the peptidoglycan backbone help bacteria to establish infection. Infect. Immun. 2011, 79, 562–570. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Sussman, M.; Liu, D.; Poxton, I.; Schwartzman, J. Chapter 6—Peptidoglycan. In Molecular Medical Microbiology; Academic Press: Cambridge, MA, USA, 2015; pp. 105–124. [Google Scholar]
- Barreteau, H.; Kovač, A.; Boniface, A.; Sova, M.; Gobec, S.; Blanot, D. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 168–207. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, H.; Watanabe, R.; Choyke, P.L. Improving conventional enhanced permeability and retention (EPR) effects; What is the appropriate target? Theranostics 2014, 4, 81–89. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Folkman, J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell 1996, 86, 353–364. [Google Scholar] [CrossRef] [Green Version]
- Fang, J.; Nakamura, H.; Maeda, H. The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Adv. Drug Deliv. Rev. 2011, 63, 136–151. [Google Scholar] [CrossRef]
- Gillies, R.J.; Schomack, P.A.; Secomb, T.W.; Raghunand, N. Causes and Effects of Heterogeneous Perfusion in Tumors. Neoplasia 1999, 1, 197–207. [Google Scholar] [CrossRef] [Green Version]
- Heldin, C.H.; Rubin, K.; Pietras, K.; Östman, A. High interstitial fluid pressure—An obstacle in cancer therapy. Nat. Rev. Cancer 2004, 4, 806–813. [Google Scholar] [CrossRef] [PubMed]
- Horsman, M.R.; Vaupel, P. Pathophysiological basis for the formation of the tumor microenvironment. Front. Oncol. 2016, 6, 66. [Google Scholar] [CrossRef] [Green Version]
- Swartz, M.A.; Lund, A.W. Lymphatic and interstitial flow in the tumour microenvironment: Linking mechanobiology with immunity. Nat. Rev. Cancer 2012, 12, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.M.; Wilson, W.R. Exploiting tumour hypoxia in cancer treatment. Cancer Res. 2004, 4, 437–447. [Google Scholar] [CrossRef]
- Werb, Z.; Lu, P. The Role of Stroma in Tumor Development. Cancer J. 2015, 21, 250–253. [Google Scholar] [CrossRef] [Green Version]
- Jin, M.Z.; Jin, W.L. The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct. Target. Ther. 2020, 5, 166. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, B.A.; El-Deiry, W.S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020, 17, 395–417. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Gravekamp, C.; Bermudes, D.; Liu, K. Tumour-targeting bacteria engineered to fight cancer. Nat. Rev. Cancer 2018, 18, 727–743. [Google Scholar] [CrossRef]
- Lambin, P.; Theys, J.; Landuyt, W.; Rijken, P.; Kogel, A.V.D.; Schueren, E.V.D.; Hodgkiss, R.; Fowler, J.; Nuyts, S.; Bruijn, E.D.; et al. Colonisation of Clostridium in the body is restricted to hypoxic and necrotic areas of tumours. Anaerobe 1998, 4, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.Q.; Ellem, K.A.O.; Dunn, P.; West, M.J.; Bai, C.X.; Vogelstein, B. Facultative or obligate anaerobic bacteria have the potential for multimodality therapy of solid tumours. Eur. J. Cancer 2007, 43, 490–496. [Google Scholar] [CrossRef]
- Roberts, N.J.; Zhang, L.; Janku, F.; Collins, A.; Bai, R.Y.; Staedtke, V.; Rusk, A.W.; Tung, D.; Miller, M.; Roix, J.; et al. Intratumoral injection of Clostridium novyi-NT spores induces antitumor responses. Sci. Transl. Med. 2014, 6, 249ra111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staedtke, V.; Bai, R.Y.; Sun, W.; Huang, J.; Kibler, K.K.; Tyler, B.M.; Gallia, G.L.; Kinzler, K.; Vogelstein, B.; Zhou, S.; et al. Clostridium novyi-NT can cause regression of orthotopically implanted glioblastomas in rats. Oncotarget 2015, 6, 5536–5546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yazawa, K.; Fujimori, M.; Nakamura, T.; Sasaki, T.; Amano, J.; Kano, Y.; Taniguchi, S. Bifidobacterium longum as a delivery system for gene therapy of chemically induced rat mammary tumors. Breast Cancer Res. Treat. 2001, 66, 165–170. [Google Scholar] [CrossRef]
- Malmgren, R.A.; Flanigan, C.C. Localization of the Vegetative Form of Clostridium tetani in Mouse Tumors Following Intravenous Spore Administration. Cancer Res. 1955, 15, 473–478. [Google Scholar]
- He, L.; Yang, H.; Liu, F.; Chen, Y.; Tang, S.; Ji, W.; Tang, J.; Liu, Z.; Sun, Y.; Hu, S.; et al. Escherichia coli Nissle 1917 engineered to express Tum-5 can restrain murine melanoma growth. Oncotarget 2017, 8, 85772–85782. [Google Scholar] [CrossRef] [Green Version]
- Momiyama, M.; Zhao, M.; Kimura, H.; Tran, B.; Chishima, T.; Bouvet, M.; Endo, I.; Hoffman, R.M. Inhibition and eradication of human glioma with tumor-targeting Salmonella typhimurium in an orthotopic nude-mouse model. Cell Cycle 2012, 11, 628–632. [Google Scholar] [CrossRef] [Green Version]
- Miwa, S.; Yano, S.; Zhang, Y.; Matsumoto, Y.; Uehara, F.; Yamamoto, M.; Hiroshima, Y.; Kimura, H.; Hayashi, K.; Yamamoto, N.; et al. Tumor-targeting Salmonella typhimurium A1-R prevents experimental human breast cancer bone metastasis in nude mice. Oncotarget 2014, 5, 7119–7125. [Google Scholar] [CrossRef] [Green Version]
- Keenan, B.P.; Saenger, Y.; Kafrouni, M.I.; Leubner, A.; Lauer, P.; Maitra, A.; Rucki, A.A.; Gunderson, A.J.; Coussens, L.M.; Brockstedt, D.G.; et al. A listeria vaccine and depletion of t-regulatory cells activate immunity against early stage pancreatic intraepithelial neoplasms and prolong survival of mice. Gastroenterology 2014, 146, 1784–1794. [Google Scholar] [CrossRef] [Green Version]
- Lizotte, P.H.; Baird, J.R.; Stevens, C.A.; Lauer, P.; Green, W.R.; Brockstedt, D.G.; Fiering, S.N. Attenuated Listeria monocytogenes reprograms M2-polarized tumor-associated macrophages in ovarian cancer leading to iNOS-mediated tumor cell lysis. Oncoimmunology 2014, 3, e28926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, M.; Yang, M.; Li, X.M.; Jiang, P.; Baranov, E.; Li, S.; Xu, M.; Penman, S.; Hoffman, R.M. Tumor-targeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella typhimurium. Urol. Oncol. Semin. Orig. Investig. 2005, 23, 380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leschner, S.; Westphal, K.; Dietrich, N.; Viegas, N.; Jablonska, J.; Lyszkiewicz, M.; Lienenklaus, S.; Falk, W.; Gekara, N.; Loessner, H.; et al. Tumor invasion of Salmonella enterica serovar Typhimurium is accompanied by strong hemorrhage promoted by TNF-α. PLoS ONE 2009, 4, e6692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasinskas, R.W.; Forbes, N.S. Salmonella typhimurium lacking ribose chemoreceptors localize in tumor quiescence and induce apoptosis. Cancer Res. 2007, 67, 3201–3209. [Google Scholar] [CrossRef] [Green Version]
- Sznol, M.; Lin, S.L.; Bermudes, D.; Zheng, L.M.; King, I. Use of preferentially replicating bacteria for the treatment of cancer. J. Clin. Investig. 2000, 105, 1027–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duong, M.T.Q.; Qin, Y.; You, S.H.; Min, J.J. Bacteria-cancer interactions: Bacteria-based cancer therapy. Exp. Mol. Med. 2019, 51, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zargar, A.; Chang, S.; Kothari, A.; Snijders, A.M.; Mao, J.-H.; Wang, J.; Hernández, A.C.; Keasling, J.D.; Bivona, T.G. Overcoming the challenges of cancer drug resistance through bacterial-mediated therapy. Chronic Dis. Transl. Med. 2019, 5, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Li, Z.; Lin, S.; Le, T.; Ittensohn, M.; Bermudes, D.; Runyab, J.D.; Shen, S.Y.; Chen, J.; King, I.C.; et al. Antitumor effect of VNP20009, an attenuated Salmonella, in murine tumor models. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2001, 12, 501–508. [Google Scholar] [CrossRef] [Green Version]
- Low, K.B.; Ittensohn, M.; Le, T.; Platt, J.; Sodi, S.; Amoss, M.; Ash, O.; Carmichael, E.; Chakraborty, A.; Fischer, J.; et al. Lipid a mutant Salmonella with suppressed virulence and TNFα induction retain tumor-targeting in vivo. Nat. Biotechnol. 1999, 17, 37–41. [Google Scholar] [CrossRef]
- Loeffler, M.; Le’Negrate, G.; Krajewska, M.; Reed, J.C. Salmonella typhimurium engineered to produce CCL21 inhibit tumor growth. Cancer Immunol. Immunother. 2009, 58, 769–775. [Google Scholar] [CrossRef]
- Xu, Y.F.; Zhu, L.P.; Hu, B.; Fu, G.F.; Zhang, H.Y.; Wang, J.J.; Xu, G.X. A new expression plasmid in Bifidobacterium longum as a delivery system of endostatin for cancer gene therapy. Cancer Gene Ther. 2007, 14, 151–157. [Google Scholar] [CrossRef] [Green Version]
- Baban, C.K.; Cronin, M.; O’Hanlon, D.; O’Sullivan, G.C.; Tangney, M. Bacteria as vectors for gene therapy of cancer. Bioeng. Bugs 2010, 1, 385–394. [Google Scholar] [CrossRef] [Green Version]
- Akin, D.; Sturgis, J.; Ragheb, K.; Sherman, D.; Burkholder, K.; Robinson, J.P.; Bhunia, A.K.; Mohammed, S.; Bashir, R. Bacteria-mediated delivery of nanoparticles and cargo into cells. Nat. Nanotechnol. 2007, 2, 441–449. [Google Scholar] [CrossRef] [Green Version]
- Hosseinidoust, Z.; Mostaghaci, B.; Yasa, O.; Park, B.-W.; Singh, A.V.; Sitti, M. Bioengineered and biohybrid bacteria-based systems for drug delivery. Adv. Drug Deliv. Rev. 2016, 106, 27–44. [Google Scholar] [CrossRef] [PubMed]
- Florence, A.T. “Targeting” nanoparticles: The constraints of physical laws and physical barriers. J. Control. Release 2012, 164, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.K.; Stylianopoulos, T. Delivering nanomedicine to solid tumors. Nat. Rev. Clin. Oncol. 2010, 7, 653–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taherkhani, S.; Mohammadi, M.; Daoud, J.; Martel, S.; Tabrizian, M. Covalent Binding of Nanoliposomes to the Surface of Magnetotactic Bacteria for the Synthesis of Self-Propelled Therapeutic Agents. ACS Nano 2014, 8, 5049–5060. [Google Scholar] [CrossRef] [PubMed]
- Forbes, N.S. Engineering the perfect (bacterial) cancer therapy. Nat. Rev. Cancer 2010, 10, 785–794. [Google Scholar] [CrossRef] [Green Version]
- Nichols, J.W.; Bae, Y.H. Odyssey of a cancer nanoparticle: From injection site to site of action. Nano Today 2012, 7, 606–618. [Google Scholar] [CrossRef] [Green Version]
- Khawar, I.A.; Kim, J.H.; Kuh, H.J. Improving drug delivery to solid tumors: Priming the tumor microenvironment. J. Control. Release 2015, 201, 78–89. [Google Scholar] [CrossRef]
- Haider, T.; Pandey, V.; Banjare, N.; Gupta, P.N.; Soni, V. Drug resistance in cancer: Mechanisms and tackling strategies. Pharmacol. Rep. 2020, 72, 1125–1151. [Google Scholar] [CrossRef]
- Hu, Q.; Wu, M.; Fang, C.; Cheng, C.; Zhao, M.; Fang, W.; Chu, P.K.; Ping, Y.; Tang, G. Engineering Nanoparticle-Coated Bacteria as Oral DNA Vaccines for Cancer Immunotherapy. Nano Lett. 2015, 15, 2732–2739. [Google Scholar] [CrossRef]
- Zheng, D.W.; Chen, Y.; Li, Z.H.; Xu, L.; Li, C.X.; Li, B.; Fan, J.X.; Cheng, S.X.; Zhang, X.Z. Optically-controlled bacterial metabolite for cancer therapy. Nat. Commun. 2018, 9, 1680. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Yang, Q.; Lv, F.; Liu, L.; Wang, S. Conjugated polymer-coated bacteria for multimodal intracellular and extracellular anticancer activity. Adv. Mater. 2013, 25, 1203–1208. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Chen, M.; Song, X.; Zhang, Z.; Zhang, Z.; Chen, Z.; Li, X. Bacterial microbots for acid-labile release of hybrid micelles to promote the synergistic antitumor efficacy. Acta Biomater. 2018, 78, 198–210. [Google Scholar] [CrossRef]
- Suh, S.; Jo, A.; Traore, M.A.; Zhan, Y.; Coutermarsh-Ott, S.L.; Ringel-Scaia, V.M.; Allen, I.C.; Davis, R.M.; Behkam, B. Nanoscale Bacteria-Enabled Autonomous Drug Delivery System (NanoBEADS) Enhances Intratumoral Transport of Nanomedicine. Adv. Sci. 2019, 6, 1801309. [Google Scholar] [CrossRef] [Green Version]
- Park, S.J.; Park, S.H.; Cho, S.; Kim, D.M.; Lee, Y.; Ko, S.Y.; Hong, Y.; Choy, H.E.; Min, J.J.; Park, J.O.; et al. New paradigm for tumor theranostic methodology using bacteria-based microrobot. Sci. Rep. 2013, 3, 3394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno, V.M.; Álvarez, E.; Izquierdo-Barba, I.; Baeza, A.; Serrano-López, J.; Vallet-Regí, M. Bacteria as Nanoparticles Carrier for Enhancing Penetration in a Tumoral Matrix Model. Adv. Mater. Interfaces 2020, 7, 1901942. [Google Scholar] [CrossRef]
- Zoaby, N.; Shainsky-Roitman, J.; Badarneh, S.; Abumanhal, H.; Leshansky, A.; Yaron, S.; Schroeder, A. Autonomous bacterial nanoswimmers target cancer. J. Control. Release 2017, 257, 68–75. [Google Scholar] [CrossRef]
- Prescher, J.A.; Bertozzi, C.R. Chemistry in Living Systems. Nat. Chem. Biol. 2005, 1, 13–21. [Google Scholar] [CrossRef]
- Sletten, E.M.; Bertozzi, C.R. From mechanism to mouse: A tale of two bioorthogonal reactions. Acc. Chem. Res. 2011, 44, 666–676. [Google Scholar] [CrossRef]
- Borrmann, A.; Van Hest, J.C.M. Bioorthogonal chemistry in living organisms. Chem. Sci. 2014, 5, 2123–2134. [Google Scholar] [CrossRef]
- Sletten, E.M.; Bertozzi, C.R. Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angew. Chem. Int. Ed. 2009, 48, 6974–6998. [Google Scholar] [CrossRef] [Green Version]
- Carell, T.; Vrabel, M. Bioorthogonal Chemistry—Introduction and Overview. Top. Curr. Chem. 2016, 374, 5–25. [Google Scholar] [CrossRef]
- Cañeque, T.; Müller, S.; Rodriguez, R. Visualizing biologically active small molecules in cells using click chemistry. Nat. Rev. Chem. 2018, 2, 202–215. [Google Scholar] [CrossRef]
- Nguyen, S.S.; Prescher, J.A. Developing bioorthogonal probes to span a spectrum of reactivities. Nat. Rev. Chem. 2020, 4, 476–489. [Google Scholar] [CrossRef]
- Liu, B. Bio-orthogonal Click Chemistry for In Vivo Bioimaging. Trends Chem. 2019, 1, 763–778. [Google Scholar] [CrossRef]
- Algar, W.R.; Prasuhn, D.E.; Stewart, M.H.; Jennings, T.L.; Blanco-Canosa, J.B.; Dawson, P.E.; Medintz, I.L. The Controlled Display of Biomolecules on Nanoparticles: A Challenge Suited to Bioorthogonal Chemistry. Bioconjug. Chem. 2011, 22, 825–858. [Google Scholar] [CrossRef] [PubMed]
- Heuer-Jungemann, A.; Feliu, N.; Bakaimi, I.; Hamaly, M.; Alkilany, A.; Chakraborty, I.; Masood, A.; Casula, M.F.; Kostopoulou, A.; Oh, E.; et al. The role of ligands in the chemical synthesis and applications of inorganic nanoparticles. Chem. Rev. 2019, 119, 4819–4880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biju, V. Chemical modifications and bioconjugate reactions of nanomaterials for sensing, imaging, drug delivery and therapy. Chem. Soc. Rev. 2014, 43, 744–764. [Google Scholar] [CrossRef]
- Stauber, R.H.; Siemer, S.; Becker, S.; Ding, G.B.; Strieth, S.; Knauer, S.K. Small Meets Smaller: Effects of Nanomaterials on Microbial Biology, Pathology, and Ecology. ACS Nano 2018, 12, 6351–6359. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, R.; Zuniga, M.; Sassine, F.R.; Karakoy, M.; Gracias, D.H. Enabling cargo-carrying bacteria via surface attachment and triggered release. Small 2011, 7, 588–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haensch, C.; Hoeppener, S.; Schubert, U.S. Chemical modification of self-assembled silane based monolayers by surface reactions. Chem. Soc. Rev. 2010, 39, 2323–2334. [Google Scholar] [CrossRef] [PubMed]
- Kuru, E.; Hughes, H.V.; Brown, P.J.; Hall, E.; Tekkam, S.; Cava, F.; Pedro, M.A.D.; Brun, Y.V.; VanNieuwenhze, M.S. In Situ Probing of Newly Synthesized Peptidoglycan in Live Bacteria with Fluorescent D-amino Acids. Angew. Chem. Int. Ed. 2012, 124, 12687–12691. [Google Scholar] [CrossRef] [Green Version]
- Patterson, D.M.; Nazarova, L.A.; Prescher, J.A. Finding the Right (Bioorthogonal) Chemistry. ACS Chem. Biol. 2014, 9, 592–605. [Google Scholar] [CrossRef]
- Stephanopoulos, N.; Francis, M.B. Choosing an effective protein bioconjugation strategy. Nat. Chem. Biol. 2011, 7, 876–884. [Google Scholar] [CrossRef]
- Devaraj, N.K. The Future of Bioorthogonal Chemistry. ACS Cent. Sci. 2018, 4, 952–959. [Google Scholar] [CrossRef] [Green Version]
- Schäfer, R.J.B.; Monaco, M.R.; Li, M.; Tirla, A.; Rivera-Fuentes, P.; Wennemers, H. The Bioorthogonal Isonitrile—Chlorooxime Ligation. J. Am. Chem. Soc. 2019, 141, 18644–18648. [Google Scholar] [CrossRef]
- Leriche, G.; Chisholm, L.; Wagner, A. Cleavable linkers in chemical biology. Bioorganic Med. Chem. 2012, 20, 571–582. [Google Scholar] [CrossRef]
- Stanton, M.M.; Simmchen, J.; Ma, X.; Miguel-López, A.; Sánchez, S. Biohybrid Janus Motors Driven by Escherichia coli. Adv. Mater. Interfaces 2016, 3, 1500505. [Google Scholar] [CrossRef]
- Huang, Z.; Chen, P.; Zhu, G.; Yang, Y.; Xu, Z.; Yan, L.T. Bacteria-Activated Janus Particles Driven by Chemotaxis. ACS Nano 2018, 12, 6725–6733. [Google Scholar] [CrossRef]
- Xie, S.; Zhao, L.; Song, X.; Tang, M.; Mo, C.; Li, X. Doxorubicin-conjugated Escherichia coli Nissle 1917 swimmers to achieve tumor targeting and responsive drug release. J. Control. Release 2017, 268, 390–399. [Google Scholar] [CrossRef]
- Lin, D.; Feng, X.; Mai, B.; Li, X.; Wang, F.; Liu, J.; Liu, X.; Zhang, K.; Wang, X. Bacterial-based cancer therapy: An emerging toolbox for targeted drug/gene delivery. Biomaterials 2021, 277, 121124. [Google Scholar] [CrossRef]
- Li, Q.; Chen, H.; Feng, X.; Yu, C.; Feng, F.; Chai, Y.; Lu, P.; Song, T.; Wang, X.; Yao, L. Nanoparticle-Regulated Semiartificial Magnetotactic Bacteria with Tunable Magnetic Moment and Magnetic Sensitivity. Small 2019, 15, 1900427. [Google Scholar] [CrossRef]
- Xing, J.; Yin, T.; Li, S.; Xu, T.; Ma, A.; Chen, Z.; Luo, Y.; Lai, Z.; Lv, Y.; Pan, H.; et al. Sequential Magneto-Actuated and Optics-Triggered Biomicrorobots for Targeted Cancer Therapy. Adv. Funct. Mater. 2021, 31, 2008262. [Google Scholar] [CrossRef]
- Chen, W.; Wang, Y.; Qin, M.; Zhang, X.; Zhang, Z.; Sun, X.; Gu, Z. Bacteria-Driven Hypoxia Targeting for Combined Biotherapy and Photothermal Therapy. ACS Nano 2018, 12, 5995–6005. [Google Scholar] [CrossRef] [PubMed]
- Park, B.W.; Zhuang, J.; Yasa, O.; Sitti, M. Multifunctional Bacteria-Driven Microswimmers for Targeted Active Drug Delivery. ACS Nano 2017, 11, 8910–8923. [Google Scholar] [CrossRef] [PubMed]
- Baeza, A.; Manzano, M.; Colilla, M.; Vallet-Regí, M. Recent advances in mesoporous silica nanoparticles for antitumor therapy: Our contribution. Biomater. Sci. 2016, 4, 803–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paris, J.L.; Colilla, M.; Izquierdo-Barba, I.; Manzano, M.; Vallet-Regí, M. Tuning mesoporous silica dissolution in physiological environments: A review. J. Mater. Sci. 2017, 52, 8761–8771. [Google Scholar] [CrossRef] [Green Version]
- Vallet-Regí, M. Our contributions to applications of mesoporous silica nanoparticles. Acta Biomater. 2021, 137, 44–52. [Google Scholar] [CrossRef]
- Tang, F.; Li, L.; Chen, D. Mesoporous silica nanoparticles: Synthesis, biocompatibility and drug delivery. Adv. Mater. 2012, 24, 1504–1534. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, H.; Shi, J. In vivo bio-safety evaluations and diagnostic/therapeutic applications of chemically designed mesoporous silica nanoparticles. Adv. Mater. 2013, 25, 3144–3176. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Liong, M.; Li, Z.; Zink, J.I.; Tamanoi, F. Biocompatibility, biodistribution, and drug-delivery efficiency of mesoporous silica nanoparticles for cancer therapy in animals. Small 2010, 6, 1794–1805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Encinas, N.; Angulo, M.; Astorga, C.; Colilla, M.; Izquierdo-Barba, I.; Vallet-Regí, M. Mixed-charge pseudo-zwitterionic mesoporous silica nanoparticles with low-fouling and reduced cell uptake properties. Acta Biomater. 2019, 84, 317–327. [Google Scholar] [CrossRef]
- Tang, J.; Chu, B.; Wang, J.; Song, B.; Su, Y.; Wang, H.; He, Y. Multifunctional nanoagents for ultrasensitive imaging and photoactive killing of Gram-negative and Gram-positive bacteria. Nat. Commun. 2019, 10, 4057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.F.; Li, S.; Liang, L.; Huang, Q.; Yuwen, L.; Yang, W.; Wang, R.; Wang, L.H. Highly Biocompatible Chlorin e6-Loaded Chitosan Nanoparticles for Improved Photodynamic Cancer Therapy. ACS Appl. Mater. Interfaces 2018, 10, 9980–9987. [Google Scholar] [CrossRef]
- Park, W.; Cho, S.; Huang, X.; Larson, A.C.; Kim, D.H. Branched Gold Nanoparticle Coating of Clostridium novyi-NT Spores for CT-Guided Intratumoral Injection. Small 2017, 13, 1602722. [Google Scholar] [CrossRef] [Green Version]
- Luo, C.H.; Huang, C.T.; Su, C.H.; Yeh, C.S. Bacteria-Mediated Hypoxia-Specific Delivery of Nanoparticles for Tumors Imaging and Therapy. Nano Lett. 2016, 16, 3493–3499. [Google Scholar] [CrossRef]
- Dong, H.; Sarkes, D.A.; Rice, J.J.; Hurley, M.M.; Fu, A.J.; Stratis-Cullum, D.N. Living Bacteria—Nanoparticle Hybrids Mediated through Surface-Displayed Peptides. Langmuir 2018, 34, 5837–5848. [Google Scholar] [CrossRef]
- Fan, J.X.; Peng, M.Y.; Wang, H.; Zheng, H.R.; Liu, Z.L.; Li, C.X.; Wang, X.N.; Liu, X.H.; Cheng, S.X.; Zhang, X.Z. Engineered Bacterial Bioreactor for Tumor Therapy via Fenton-Like Reaction with Localized H2O2 Generation. Adv. Mater. 2019, 31, 1808278. [Google Scholar] [CrossRef]
- Peer, D.; Karp, J.M.; Hong, S.; Farokhzad, O.C.; Margalit, R.; Langer, R. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotechnol. 2007, 2, 751–760. [Google Scholar] [CrossRef]
- Dogra, N.; Izadi, H.; Vanderlick, T.K. Micro-motors: A motile bacteria based system for liposome cargo transport. Sci. Rep. 2016, 6, 29396. [Google Scholar] [CrossRef] [Green Version]
- Felfoul, O.; Mohammadi, M.; Taherkhani, S.; De Lanauze, D.; Xu, Y.Z.; Loghin, D.; Essa, S.; Jancik, S.; Houle, D.; Lafleur, M.; et al. Magneto-aerotactic bacteria deliver drug-containing nanoliposomes to tumour hypoxic regions. Nat. Nanotechnol. 2016, 11, 941–947. [Google Scholar] [CrossRef]

| Chemical Group (R1 or R2) | Partner Group (R2 or R1) | Conjugation Product | Ref. | |||
|---|---|---|---|---|---|---|
| A | 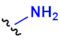 |  | NHS-ester | 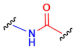 | Amide derivate | [117] |
| B | Amine |  | Isocyanate or isothiocyanate |  | (Thio)Urea derivate | |
| C |  | 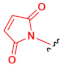 | Maleimide |  | Thioether derivate | [117] |
| D | Thiol |  | Iodoacetamide |  | Thioether derivate | |
| E |  Ketone or aldehyde |  | Hydrazide | 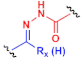 | Hydrazone derivate | [114,117] |
| F |  | Aminooxy |  | Oxime derivate | ||
| G |  | Amine | 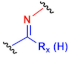 | Imine derivate | ||
| H | 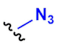 Azide |  | Alkyne | 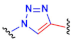 | Triazole derivate | [114,115] |
| I |  | Cyclooctyne |  | Triazole derivate | ||
| J | 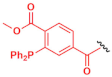 | Staudinger phosphine | 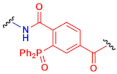 | Amide derivate | ||
| Bacteria Type | NP-Bacteria Interaction | Bioconjugation Method | Nanomaterial | Therapeutic Strategy | Ref. |
|---|---|---|---|---|---|
| L. monocytogenes | Attached | Antigen/antibody and Avidin/neutravidin | Polystyrene NPs | Gene delivery and protein expression in tumoral cells | [97] |
| E. coli | Attached | Acid-labile linker | Free drug | Sustained release of drug | [136] |
| Salmonella | Adsorbed | Electrostatic interactions | PEI NPs | Cancer immunotherapy | [106] |
| E. coli | Attached | Tetrazine/norbornene click reaction | Polymeric pro-micelles | On-demand release of two drugs | [109] |
| Salmonella | Attached | Biotin/Streptavidin | PLGA NPs | - | [110] |
| Magnetospirillum magneticum | Attached | Michael addition to maleimide | Indocyanine green PLGA NPs | Photothermal therapy | [138] |
| Salmonella | Attached | Oxidation and self-polymerization | Polydopamine NPs | Photothermal therapy | [140] |
| E. coli | Adsorbed | Electrostatic interactions | Polyelectrolyte multilayer microparticles | Drug delivery with magnetic guidance | [141] |
| E. coli | Attached | Azide/DBCO click chemistry | MSNs | Transport of high amounts of drug | [112] |
| E. coli | Adsorbed | Electrostatic interactions | Carbon nitride NPs | Photoinduced in situ generation of cytotoxic species | [107] |
| Clostridium novyi-NT spores | Adsorbed | Electrostatic interactions | Branched Au NPs | Theragnostic combination therapy | [151] |
| Bifidobacterium and Clostridium difficile | Adsorbed/Attached | Electrostatic interactions and antigen/antibody | Au nanorods | Photothermal ablation | [152] |
| E. coli | Adsorbed | Metal-peptide affinity | Au NPs | - | [153] |
| E. coli | Attached | Carbodiimide chemistry | Fe3O4 NPs | Chemodynamic therapy | [154] |
| E. coli and Salmonella | Engulfed | Incubation and electroporation | Liposomes | Enhanced drug delivery | [113] |
| E. coli | Attached | Bacterial affinity with glycolipids | SUVs, LUVs, and GUVs | - | [156] |
| Magnetococcus marinus | Attached | Carbodiimide chemistry | Liposomes | Enhanced drug delivery | [157] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiménez-Jiménez, C.; Moreno, V.M.; Vallet-Regí, M. Bacteria-Assisted Transport of Nanomaterials to Improve Drug Delivery in Cancer Therapy. Nanomaterials 2022, 12, 288. https://doi.org/10.3390/nano12020288
Jiménez-Jiménez C, Moreno VM, Vallet-Regí M. Bacteria-Assisted Transport of Nanomaterials to Improve Drug Delivery in Cancer Therapy. Nanomaterials. 2022; 12(2):288. https://doi.org/10.3390/nano12020288
Chicago/Turabian StyleJiménez-Jiménez, Carla, Víctor M. Moreno, and María Vallet-Regí. 2022. "Bacteria-Assisted Transport of Nanomaterials to Improve Drug Delivery in Cancer Therapy" Nanomaterials 12, no. 2: 288. https://doi.org/10.3390/nano12020288
APA StyleJiménez-Jiménez, C., Moreno, V. M., & Vallet-Regí, M. (2022). Bacteria-Assisted Transport of Nanomaterials to Improve Drug Delivery in Cancer Therapy. Nanomaterials, 12(2), 288. https://doi.org/10.3390/nano12020288







