Methylglyoxal Adducts Levels in Blood Measured on Dried Spot by Portable Near-Infrared Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Control
2.2. Blood-Derived Cell Culture (BDC)
2.3. Secretome Collection and Characterisation
2.4. SeOECT Chip Operation
2.5. Analysis of Variance of Biochip Data
2.6. NIR Sample Preparation and Chemometric Calibration Procedures
2.7. NIR Spectra Acquisition
2.8. Spectral Pretreatments
2.9. Regression Model
2.10. Data Analysis
2.11. Theory of the Machine Learning Algorithm (ML)
3. Results
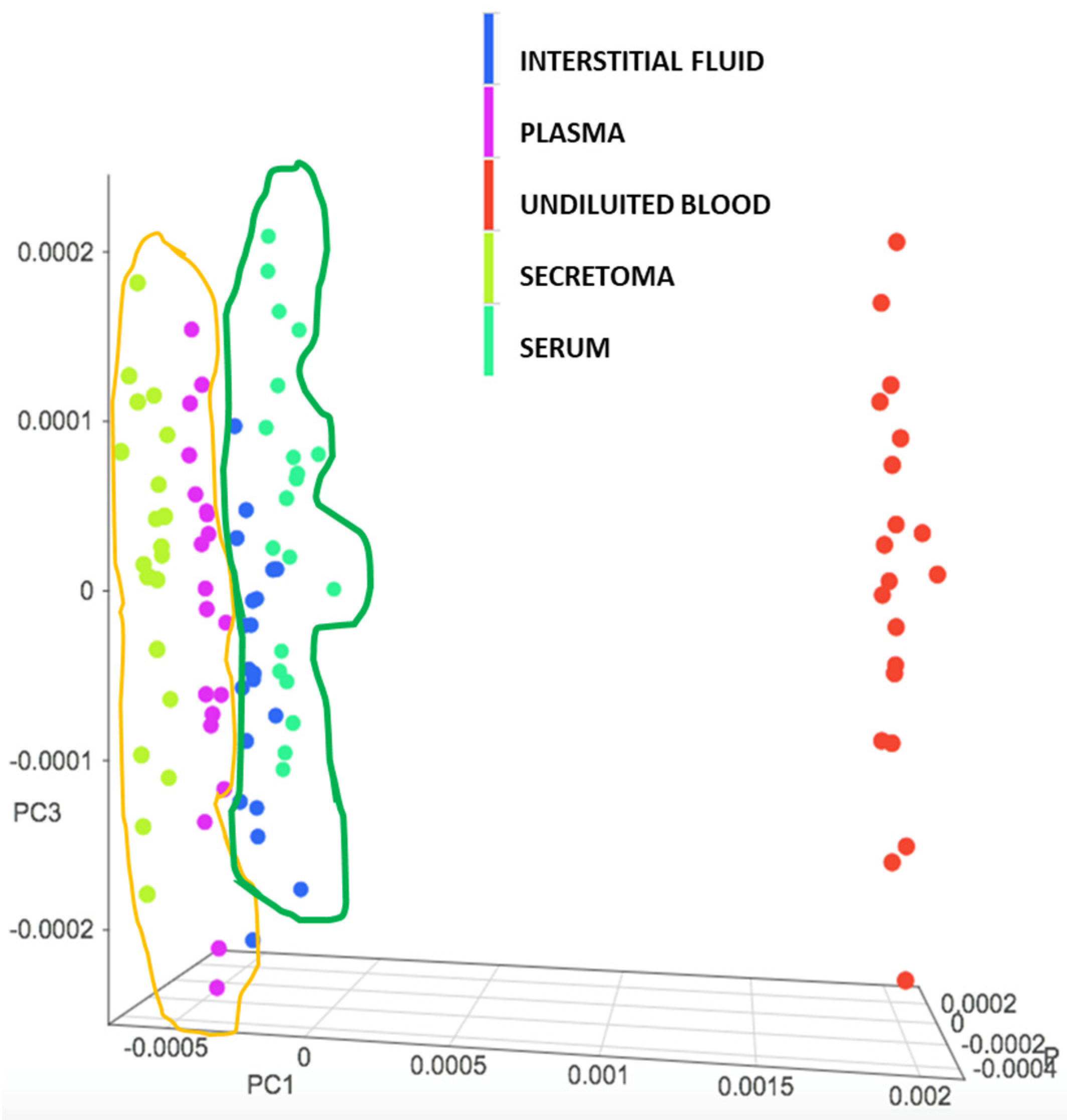
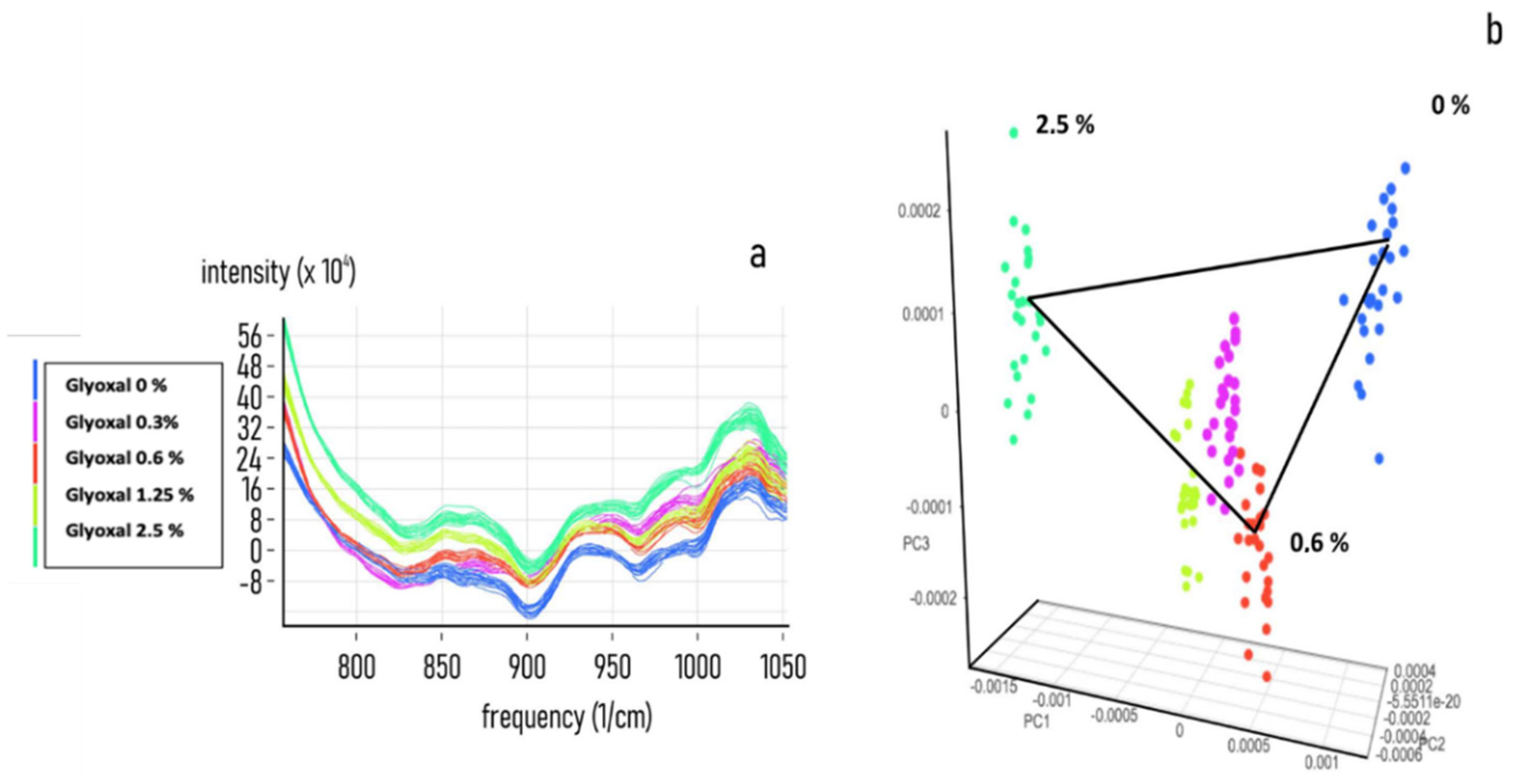
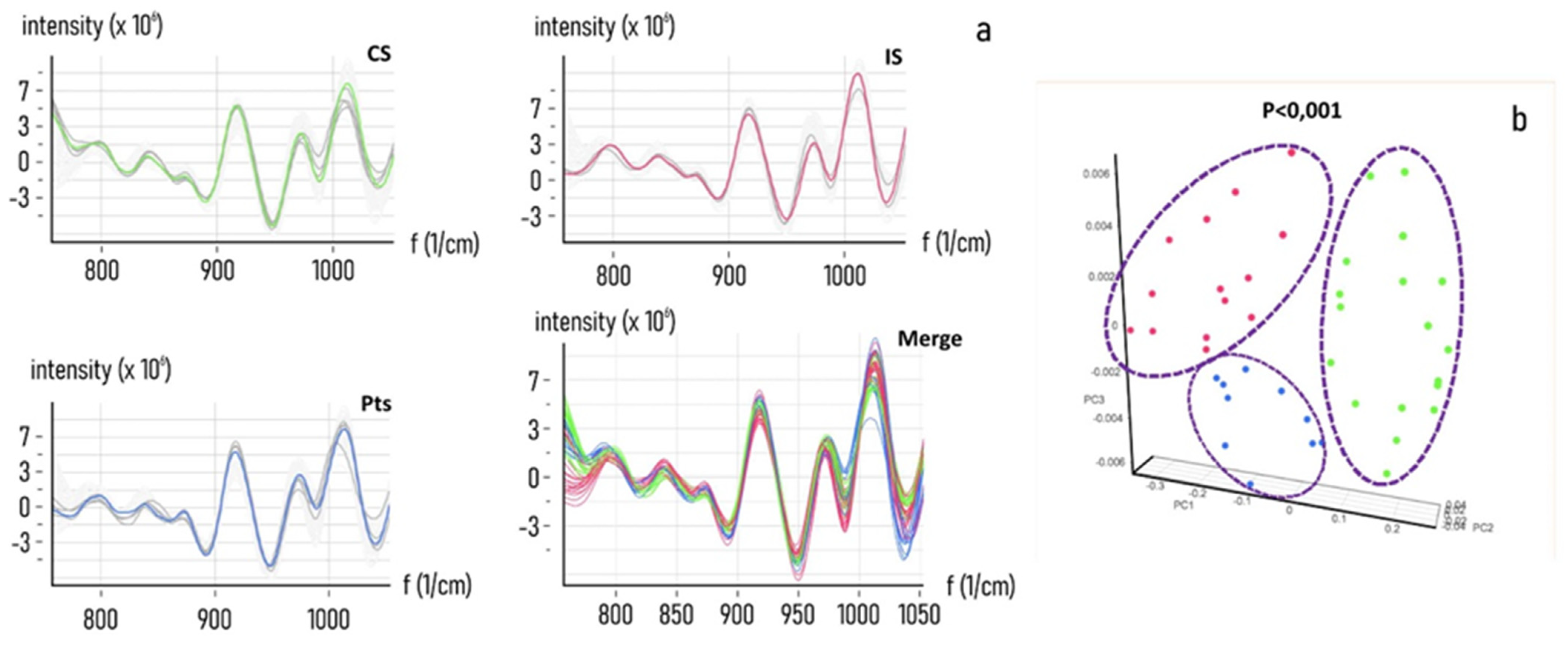
3.1. Evaluation of the NIR Spectroscopic Response of Different Biological Matrices
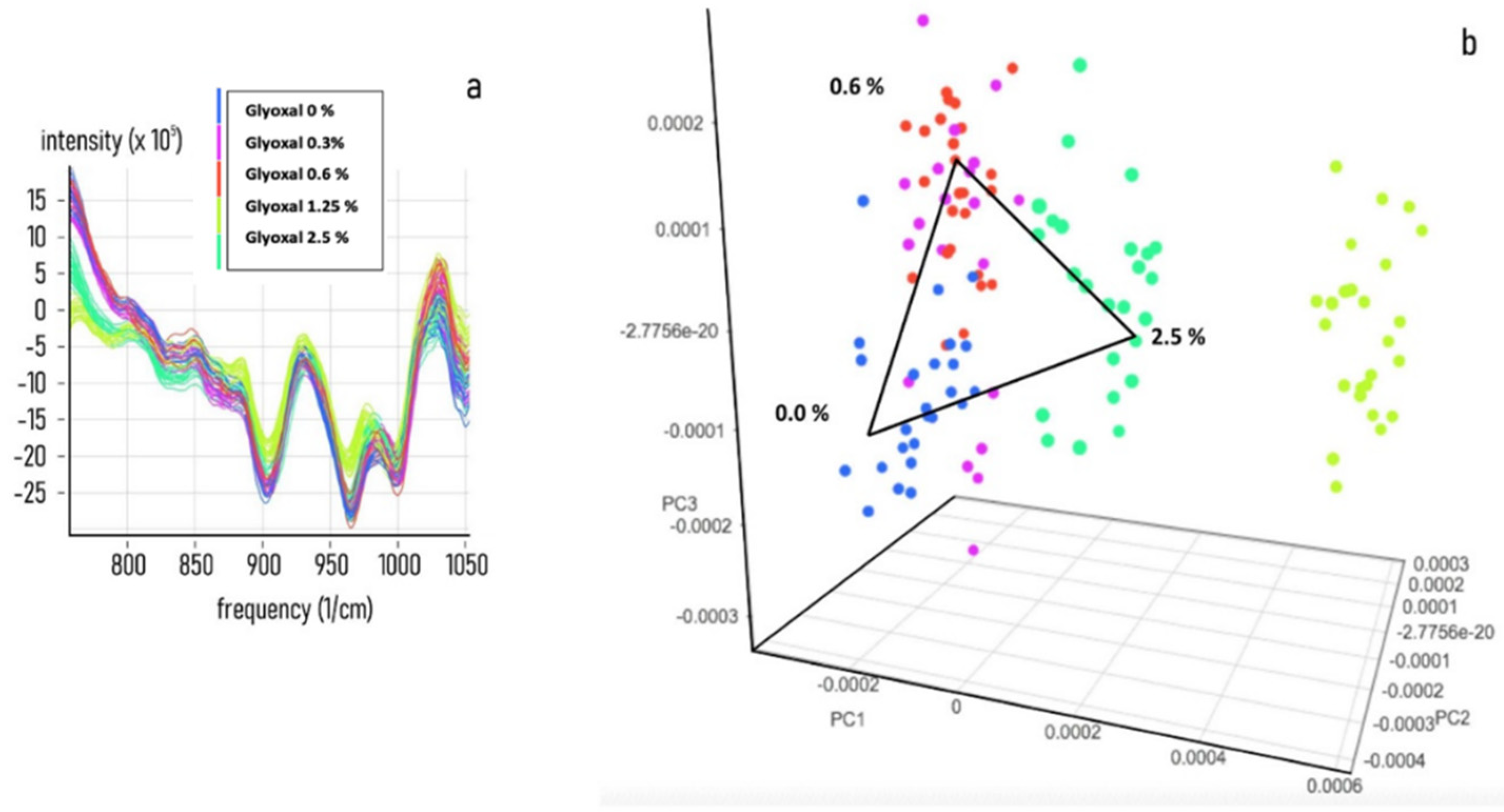
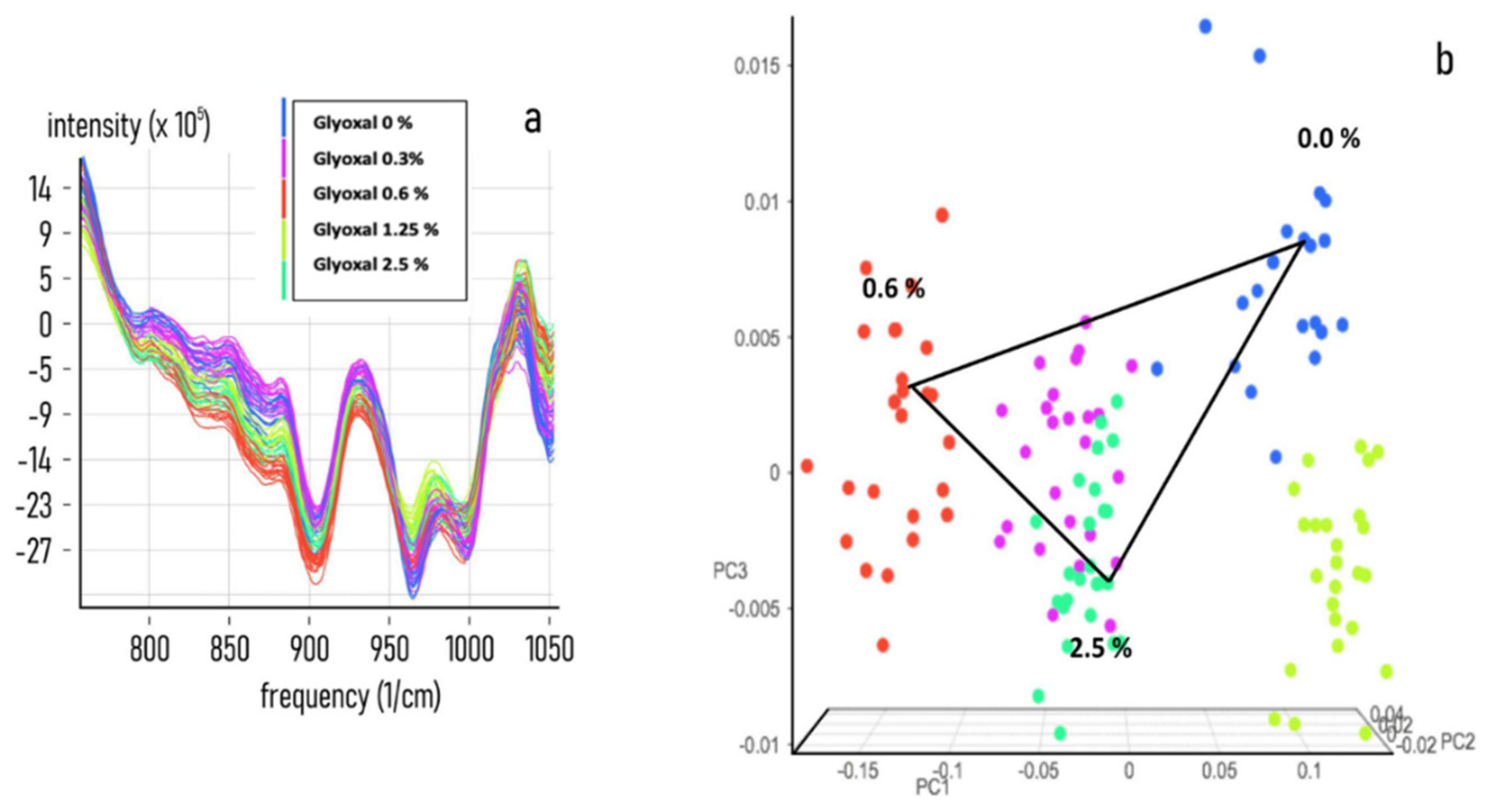
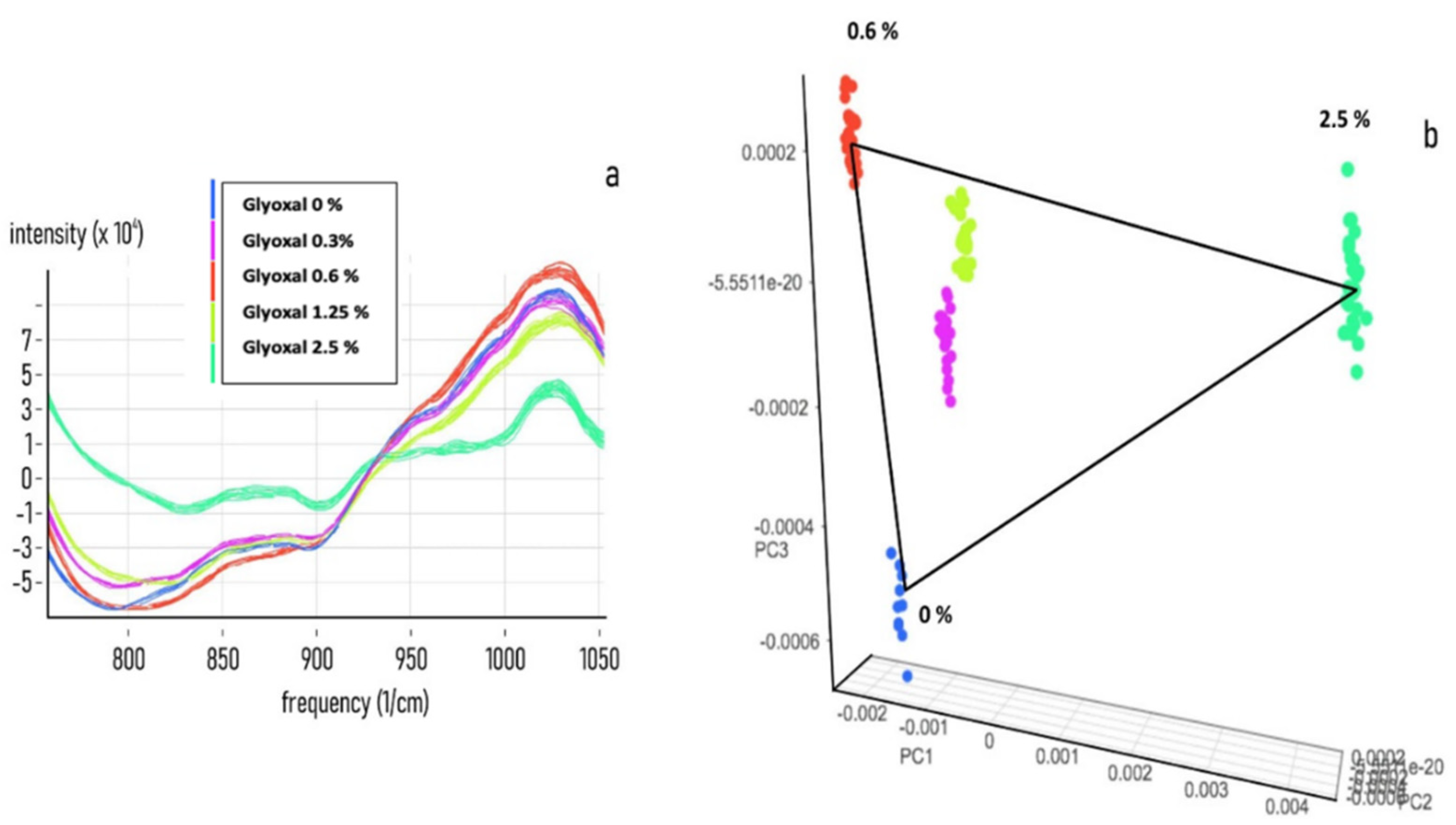
3.2. Chemometric Calibration
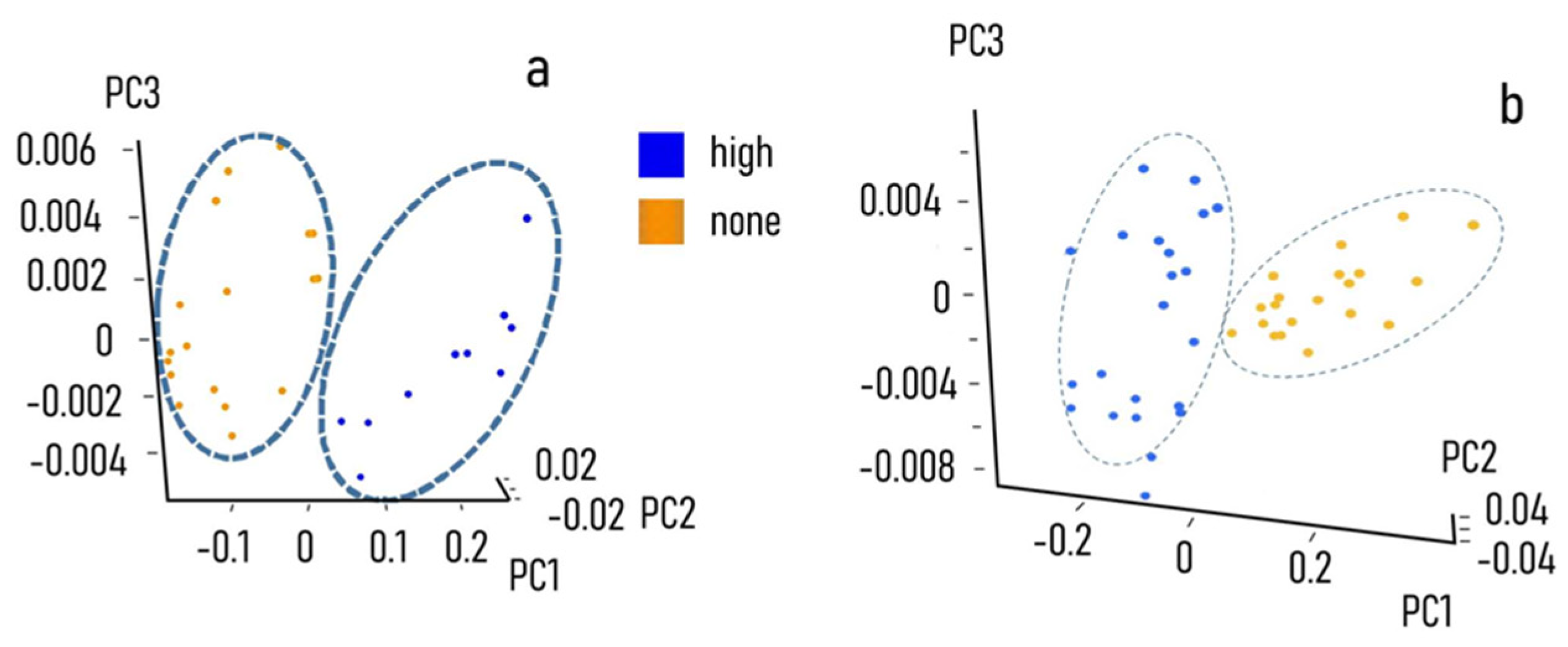
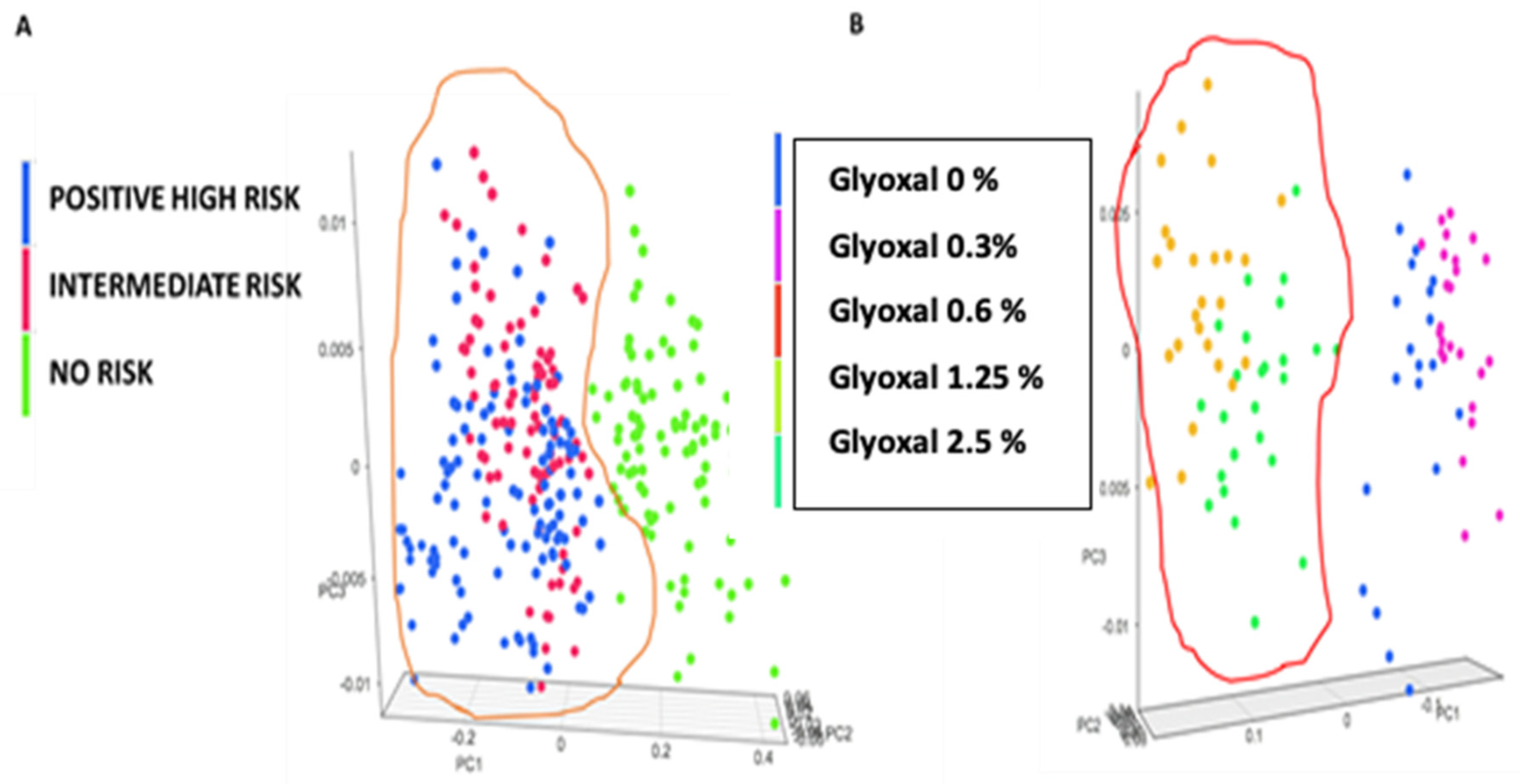
3.3. Chemometric Model Construction
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Garabedian, A.; Benigni, P.; Ramirez, C.E.; Baker, E.S.; Liu, T.; Smith, R.D.; Fernandez-Lima, F. Towards Discovery and Targeted Peptide Biomarker Detection Using nanoESI-TIMS-TOF MS. J. Am. Soc. Mass Spectrom. 2018, 29, 817–826. [Google Scholar] [CrossRef]
- Yang, Z.; Sweedler, J.V. Application of capillary electrophoresis for the early diagnosis of cancer. Anal. Bioanal. Chem. 2014, 406, 4013–4031. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, Y.H.; Kim, K.H.; Cho, J.Y.; Woo, S.M.; Yoo, B.C.; Kim, S.C. Profiling of Serum Metabolites Using MALDI-TOF and Triple-TOF Mass Spectrometry to Develop a Screen for Ovarian Cancer. Cancer Res. Treat. 2018, 50, 883–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugumaran, S.; Jamlos, M.F.; Ahmad, M.N.; Bellan, C.S.; Schreurs, D. Nanostructured materials with plasmonic nanobiosensors for early cancer detection: A past and future prospect. Biosens. Bioelectron. 2018, 100, 361–373. [Google Scholar] [CrossRef]
- Zhang, R.R.; Schroeder, A.B.; Grudzinski, J.J.; Rosenthal, E.L.; Warram, J.M.; Pinchuk, A.N.; Eliceiri, K.W.; Kuo, J.S.; Weichert, J.P. Beyond the margins: Real-time detection of cancer using targeted fluorophores. Nat. Rev. Clin. Oncol. 2017, 14, 347–364. [Google Scholar] [CrossRef] [Green Version]
- Rong, W.; Yong, W. Fourier Transform Infrared Spectroscopy in Oral Cancer Diagnosis. J. Mol. Sci. 2021, 22, 1206–1227. [Google Scholar]
- Jiale, D.; Yanping, Y.; Lei, Y.; Xue, Z.; Yi, T.; Shuxian, Z.; Hanjuan, G.; Ming, X.; Ming, L.; Qingshu, L.; et al. Preliminary Study on the Identification of BRAFV600E Mutation in Colorectal Cancer by Near-Infrared Spectroscopy. OncoTargets Ther. 2020, 13, 13077–13085. [Google Scholar]
- Xiaozhou, L.; Xuchao, M.; Weijin, X.; Qin, Y.; Sisi, W.; Juqiang, L.; Chao, L.; Zufang, H. Filter-Membrane-Based Ultrafiltration Coupled with Surface-Enhanced Raman Spectroscopy for Potential Differentiation of Benign and Malignant Thyroid Tumors from Blood Plasma. Int. J. Nanomed. 2020, 15, 2303–2314. [Google Scholar]
- Hackshaw, K.V.; Miller, J.S.; Aykas, D.P.; Rodriguez-Saona, L. Vibrational Spectroscopy for Identification of Metabolites in Biologic Samples. Molecules 2020, 25, 4725. [Google Scholar] [CrossRef]
- Hayashi, M.; Yoshizawa, N.; Ueda, Y.; Mimura, T.; Ohmae, E.; Yoshimoto, K.; Wada, H.; Nasu, H.; Ogura, H.; Sakahara, H. Effect of Source-Detector Distance on the Measurement of Hemoglobin Using Near-Infrared Spectroscopy in Breast Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819830411. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Wen, Q.; Nie, Q.; Chang, F.; Zhou, Y.; Wen, Z. Miniaturized NIR Spectrometer Based on Novel MOEMS Scanning Tilted Grating. Micromachines 2018, 20, 478. [Google Scholar] [CrossRef] [Green Version]
- Manley, M. Near-infrared spectroscopy and hyperspectral imaging: Non-destructive analysis of biological materials. Chem. Soc. Rev. 2014, 43, 8200–8214. [Google Scholar] [CrossRef] [Green Version]
- Malara, N.; Gentile, F.; Coppedè, N.; Coluccio, M.; Candeloro, P.; Perozziello, G.; Ferrara, L.; Giannetto, M.; Careri, M.; Castellini, A.; et al. Superhydrophobic lab-on-chip measures secretome protonation state and provides a personalized risk assessment of sporadic tumour. NPJ Precis. Oncol. 2018, 2, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coluccio, M.L.; Gentile, F.; Presta, I.; Donato, G.; Coppedè, N.; Valprapuram, I.; Mignogna, C.; Lavecchia, A.; Figuccia, F.; Garo, V.M.; et al. Tailoring chemometric models on blood-derived cultures secretome to assess personalized cancer risk score. Cancers 2020, 12, 1362. [Google Scholar] [CrossRef]
- Malara, N.; Trunzo, V.; Foresta, U.; Amodio, N.; De Vitis, S.; Roveda, L.; Fava, M.; Coluccio, M.L.; Macrì, R.; Di Vito, A.; et al. Ex-vivo characterization of circulating colon cancer cells distinguished in stem and differentiated subset provides useful biomarker for personalized metastatic risk assessment. J. Transl. Med. 2016, 14, 133–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malara, N.; Gentile, F.; Ferrara, L.; Villani, M.; Iannotta, S.; Zappettini, A.; Di Fabrizio, E.; Trunzo, V.; Mollace, V.; Coppedé, N. Tailoring super-hydrophobic properties of electrochemical biosensor for early cancer detection. MRS Adv. 2016, 1, 3545–3552. [Google Scholar] [CrossRef] [Green Version]
- Coluccio, M.L.; Presta, I.; Greco, M.; Gervasi, R.; La Torre, D.; Renne, M.; Voci, C.P.; Lunelli, L.; Donato, G.; Malara, N. Microenvironment Molecular Profile Combining Glycation Adducts and Cytokines Patterns on Secretome of Short-term Blood-derived Cultures during Tumour Progression. Int. J. Mol. Sci. 2020, 21, 4711. [Google Scholar] [CrossRef] [PubMed]
- Perozziello, G.; Simone, G.; Candeloro, P.; Gentile, F.; Malara, N.; Larocca, R.; Coluccio, M.; Andrea Pullano, S.; Tirinato, L.; Geschke, O.; et al. A Fluidic Motherboard for Multiplexed Simultaneous and Modular Detection in Microfluidic Systems for Biological Application. Micro Nanosyst. 2012, 2, 227–238. [Google Scholar] [CrossRef] [Green Version]
- Coluccio, M.L.; Pullano, S.A.; Vismara, M.F.M.; Coppedè, N.; Perozziello, G.; Candeloro, P.; Gentile, F.; Malara, N. Emerging Designs of Electronic Devices in Biomedicine. Micromachines 2020, 11, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinnan, A.; Berg, F.V.D.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Workman, J.J. NIR spectroscopy calibration basics. In Handbook of Near-Infrared Analysis, 3rd ed.; Burns, D.A., Ciurczak, E.W., Eds.; CRC Press: Boca Raton, FL, USA, 2007; pp. 123–150. [Google Scholar]
- Osisanwo, F.Y.; Akinsola, J.E.T.; Awodele, O.; Hinmikaiye, J.O.; Olakanmi, O.; Akinjobi, J. Supervised Machine Learning Algorithms: Classification and Comparison. Int. J. Comput. Trends Technol. (IJCTT) 2007, 48, 128–138. [Google Scholar]
- Taiwo, O.A. Types of Machine Learning Algorithms. In New Advances in Machine Learning; Zhang, Y., Ed.; InTech: London, UK, 2010; ISBN 978-953-307-034-6. Available online: http://www.intechopen.com/books/new-advances-in-machine-learning/types-of-machine-learning-algorithms (accessed on 15 July 2021).
- Elsohaby, I.; Burns, J.B.; Riley, C.B.; Shaw, R.A.; McClure, J.T. Application of laboratory and portable attenuated total reflectance infrared spectroscopic approaches for rapid quantification of alpaca serum immunoglobulin G. PLoS ONE 2017, 26, e0179644. [Google Scholar] [CrossRef] [Green Version]
- Perozziello, G.; Catalano, R.; Francardi, M.; Rondanina, E.; Pardeo, F.; De Angelis, F.; Malara, N.; Candeloro, P.; Morrone, G.; Di Fabrizio, E. A microfluidic device integrating plasmonic nanodevices for Raman spectroscopy analysis on trapped single living cells. Microelectron. Eng. 2013, 111, 314–319. [Google Scholar] [CrossRef]
- Hasegawa, K.; Funatsu, K. Advanced PLS Techniques in Chemometrics and Their Applications to Molecular Design. In Chemoinformatics and Advanced Machine Learning Perspectives: Complex Computational Methods and Collaborative Techniques; Lodhi, H., Yamanishi, Y., Eds.; IGI Global: Hershey, PA, USA, 2011; pp. 145–168. [Google Scholar]
- Bonapace, G.; Marasco, O.; Scozzafava, G.; Michael, A.; Pittelli, M.; Greto, T.; Moricca, M.T.; Vismara, S.A.; Valentini, A.; Vismara, M.F.; et al. Phenylalanine measurements in human blood using NIR spectroscopy and DBS, a preliminary study. Arch. Dis. Child. 2019, 104, 35–36. [Google Scholar]
| Classified Known Class | High | Intermediate | None |
|---|---|---|---|
| High | 99% | 0% | 0% |
| Intermediate | 3% | 96% | 0% |
| None | 0% | 0% | 100% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonapace, G.; Gentile, F.; Coppedé, N.; Coluccio, M.L.; Garo, V.; Vismara, M.F.M.; Candeloro, P.; Donato, G.; Malara, N. Methylglyoxal Adducts Levels in Blood Measured on Dried Spot by Portable Near-Infrared Spectroscopy. Nanomaterials 2021, 11, 2432. https://doi.org/10.3390/nano11092432
Bonapace G, Gentile F, Coppedé N, Coluccio ML, Garo V, Vismara MFM, Candeloro P, Donato G, Malara N. Methylglyoxal Adducts Levels in Blood Measured on Dried Spot by Portable Near-Infrared Spectroscopy. Nanomaterials. 2021; 11(9):2432. https://doi.org/10.3390/nano11092432
Chicago/Turabian StyleBonapace, Giuseppe, Francesco Gentile, Nicola Coppedé, Maria Laura Coluccio, Virginia Garo, Marco Flavio Michele Vismara, Patrizio Candeloro, Giuseppe Donato, and Natalia Malara. 2021. "Methylglyoxal Adducts Levels in Blood Measured on Dried Spot by Portable Near-Infrared Spectroscopy" Nanomaterials 11, no. 9: 2432. https://doi.org/10.3390/nano11092432
APA StyleBonapace, G., Gentile, F., Coppedé, N., Coluccio, M. L., Garo, V., Vismara, M. F. M., Candeloro, P., Donato, G., & Malara, N. (2021). Methylglyoxal Adducts Levels in Blood Measured on Dried Spot by Portable Near-Infrared Spectroscopy. Nanomaterials, 11(9), 2432. https://doi.org/10.3390/nano11092432









