Antibacterial Properties of Citric Acid/β-Alanine Carbon Dots against Gram-Negative Bacteria
Abstract
:1. Introduction
2. Materials and Methods
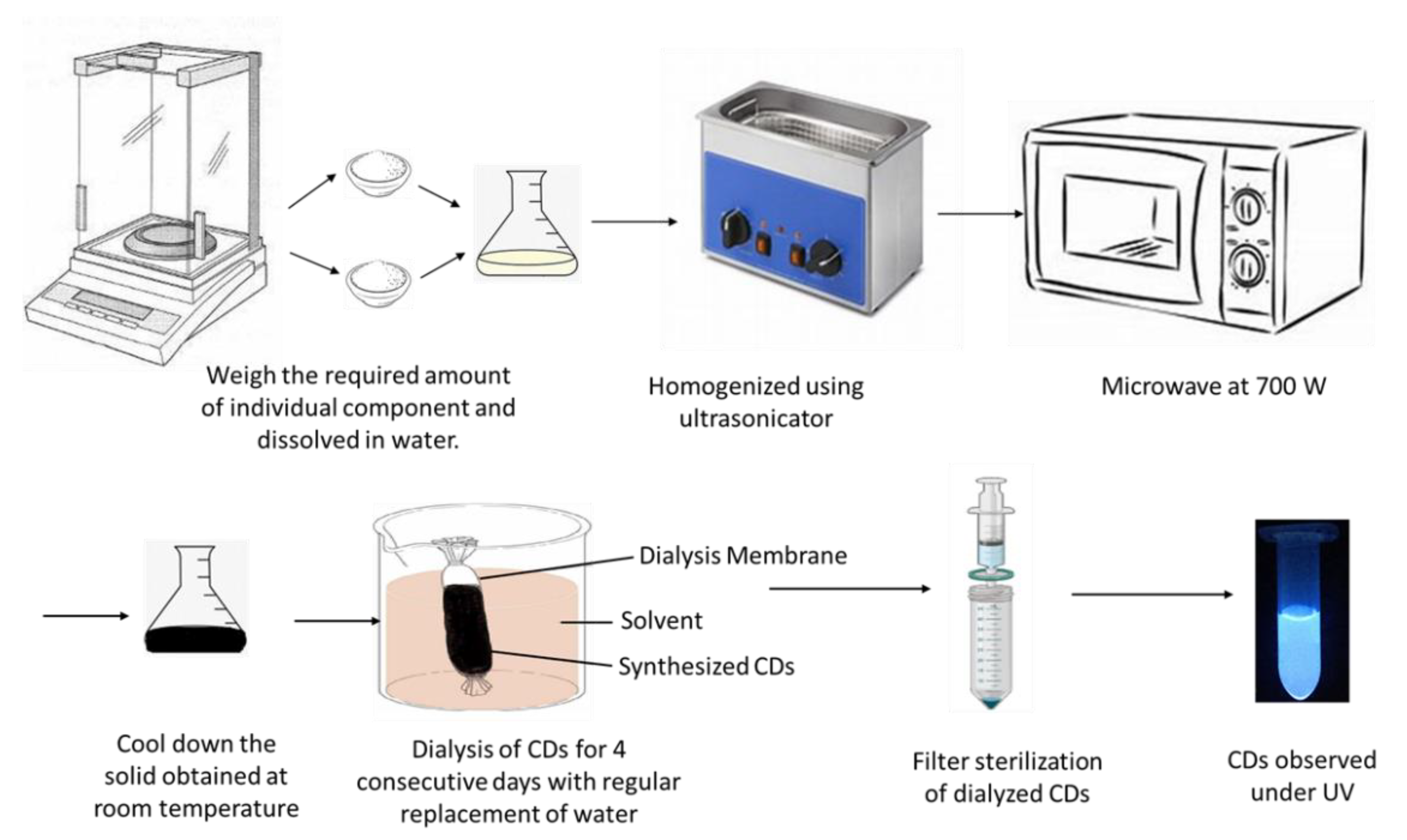
2.1. Synthesis of Carbon Dots (CDs)
2.2. Purification of CDs
2.3. Preparation of Bacteria Strains
2.4. Characterization of CDs
2.5. Antimicrobial Study
2.6. Effect of Light on Antimicrobial Properties of CDs
2.7. Statistical Analysis
3. Results and Discussion
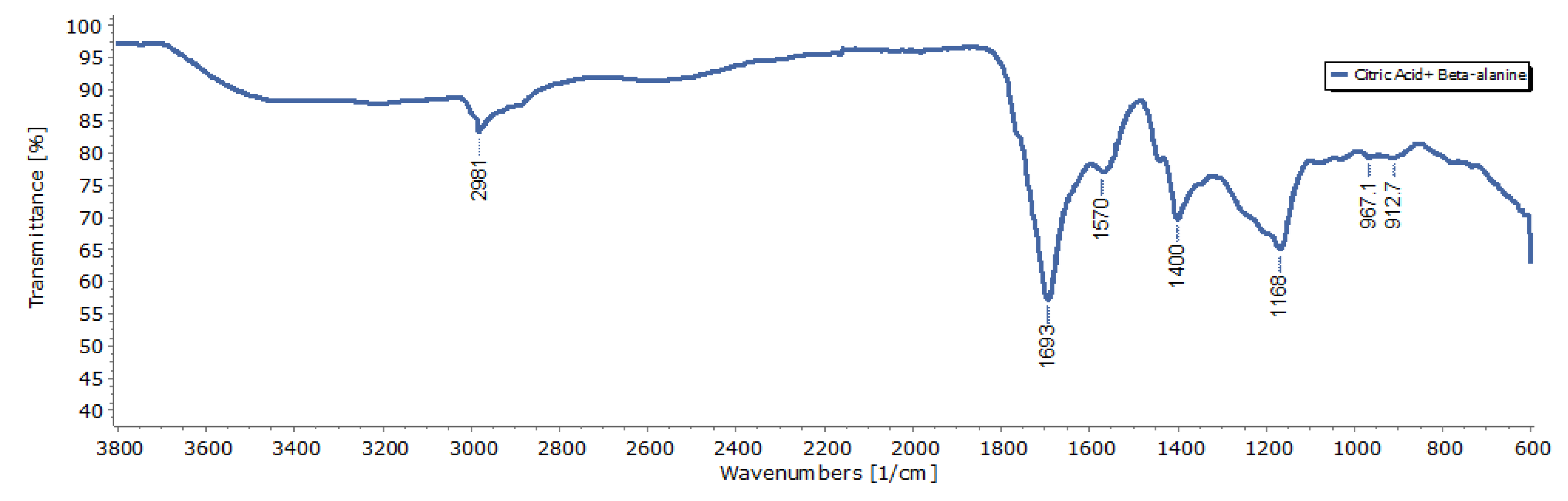
3.1. Characterization of CDs
3.2. Interaction between CDs and Bacterial Cells
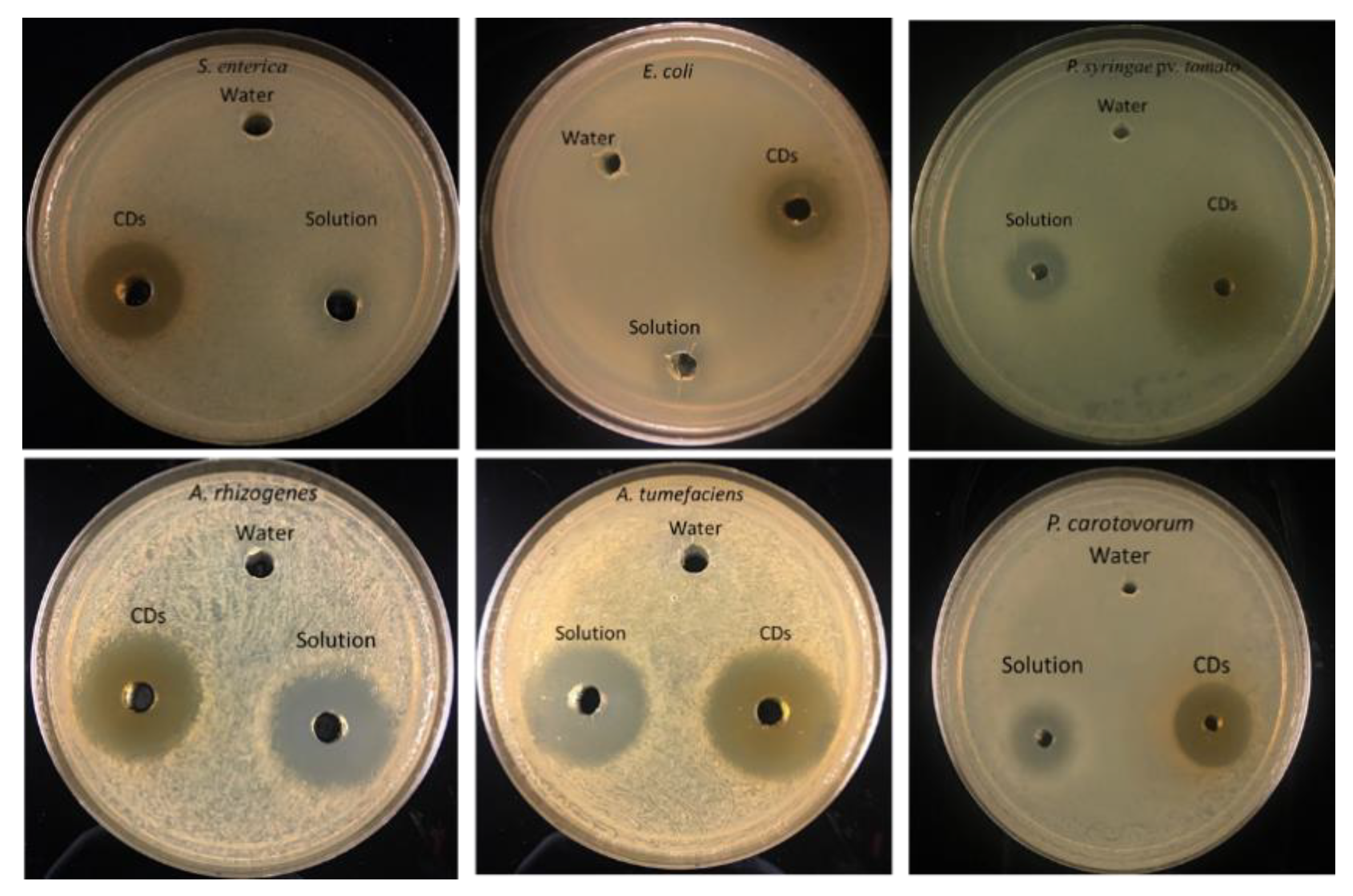
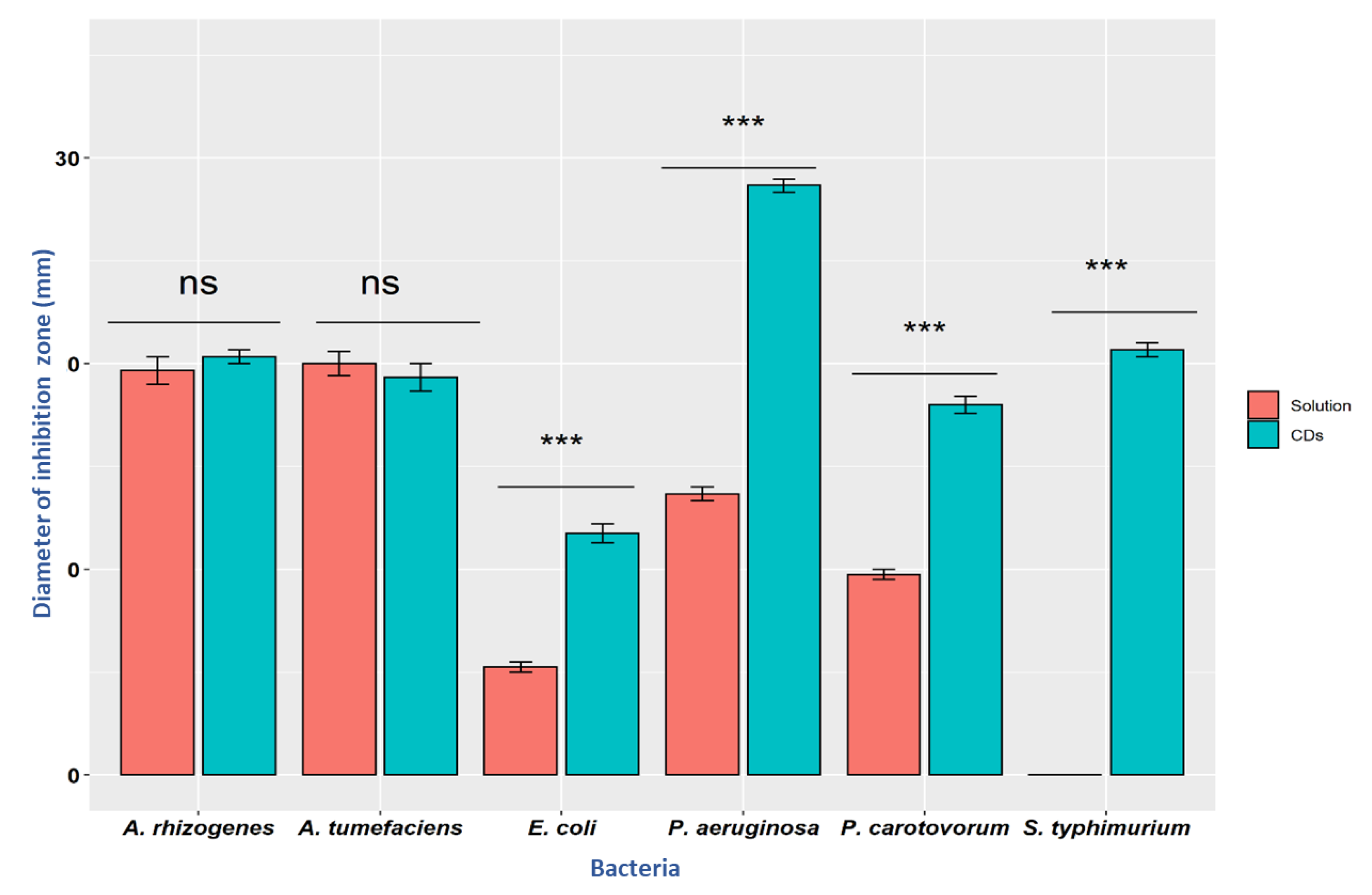
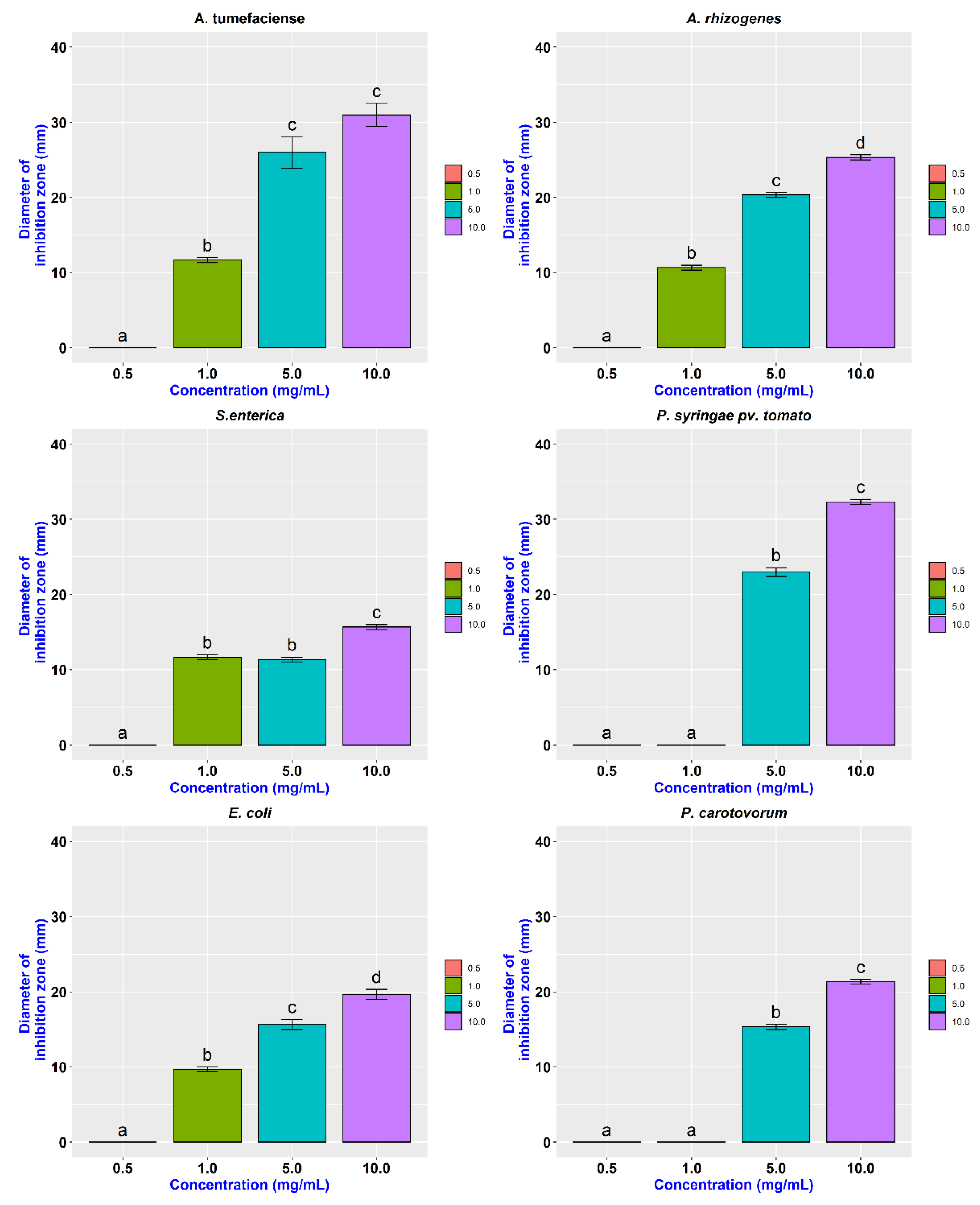
3.3. Antibacterial Activity
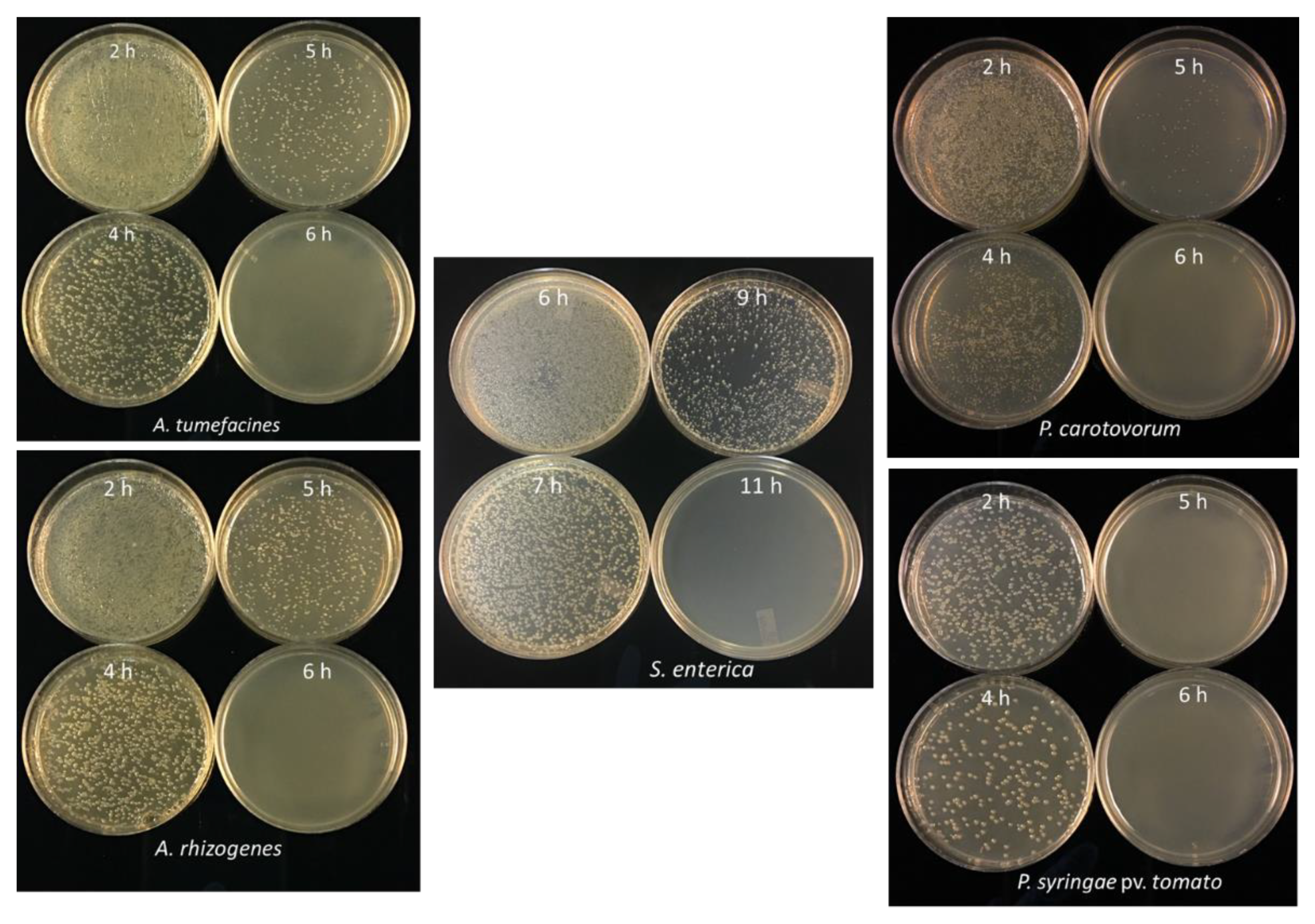
3.4. Minimum Incubation Time for Complete Inhibition of Bacterial Growth
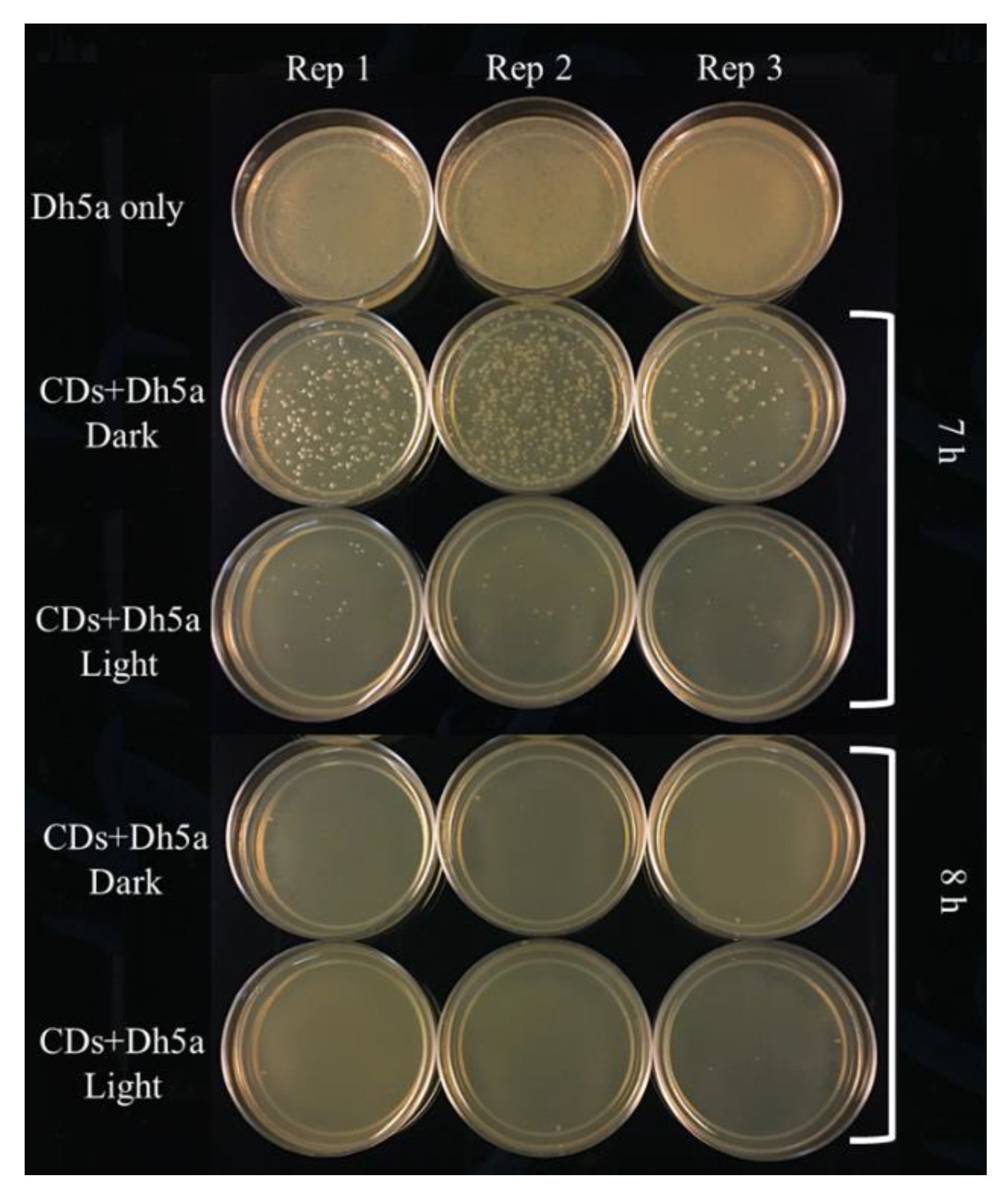
3.5. Effect of Light on Antimicrobial Activity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruppé, É.; Woerther, P.-L.; Barbier, F. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann. Intensive Care 2015, 5, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules 2020, 25, 1340. [Google Scholar] [CrossRef] [Green Version]
- Delcour, A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta Proteins Proteom. 2009, 1794, 808–816. [Google Scholar] [CrossRef] [Green Version]
- Hawkey, P.M.; Warren, R.E.; Livermore, D.M.; McNulty, C.A.M.; Enoch, D.A.; Otter, J.A.; Wilson, A.P.R. Treatment of infections caused by multidrug-resistant gram-negative bacteria: Report of the British society for antimicrobial chemotherapy/healthcare infection society/british infection association joint working party. J. Antimicrob. Chemother. 2018, 73, iii2–iii78. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Ray, R.; Gu, Y.; Ploehn, H.J.; Gearheart, L.; Raker, K.; Scrivens, W.A. Electrophoretic analysis and purification of fluorescent single-walled carbon nanotube fragments. J. Am. Chem. Soc. 2004, 126, 12736–12737. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, P.; Zhai, X.; Tian, F.; Li, W.; Yang, J.; Liu, Y.; Wang, H.; Wang, W.; Liu, W. Nano-carrier for gene delivery and bioimaging based on carbon dots with PEI-passivation enhanced fluorescence. Biomaterials 2012, 33, 3604–3613. [Google Scholar] [CrossRef]
- Wei, C.; Li, J.; Xiao, X.; Yue, T.; Zhao, D. The one-step preparation of green-emission carbon dots based on the deactivator-reducing reagent synergistic effect and the study on their luminescence mechanism. RSC Adv. 2018, 8, 20016–20024. [Google Scholar] [CrossRef] [Green Version]
- Jung, Y.K.K.; Shin, E.; Kim, B.S.S. Cell Nucleus-Targeting Zwitterionic Carbon Dots. Sci. Rep. 2015, 5, 18807. [Google Scholar] [CrossRef] [Green Version]
- Hasanzadeh, A.; Radmanesh, F.; Kiani, J.; Bayandori, M.; Fatahi, Y.; Aref, A.R.; Karimi, M. Photoluminescent functionalized carbon dots for CRISPR delivery: Synthesis, optimization and cellular investigation. Nanotechnology 2019, 30, 135101. [Google Scholar] [CrossRef]
- Cao, X.; Wang, J.; Deng, W.; Chen, J.; Wang, Y.; Zhou, J.; Du, P.; Xu, W.; Wang, Q.; Wang, Q.; et al. Photoluminescent Cationic Carbon Dots as efficient Non-Viral Delivery of Plasmid SOX9 and Chondrogenesis of Fibroblasts. Sci. Rep. 2018, 8, 7057. [Google Scholar] [CrossRef]
- Iravani, S.; Varma, S.R. Green synthesis, biomedical and biotechnological applications of carbon and graphene quantum dots. A review. Environ. Chem. Lett. 2020, 18, 703–727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saravanan, A.; Maruthapandi, M.; Das, P.; Ganguly, S.; Margel, S.; Luong, J.H.T.; Gedanken, A. Applications of N-Doped Carbon Dots as Antimicrobial Agents, Antibiotic Carriers, and Selective Fluorescent Probes for Nitro Explosives. ACS Appl. Bio Mater. 2020, 3, 8023–8031. [Google Scholar] [CrossRef]
- Dong, X.; Liang, W.; Meziani, M.J.; Sun, Y.P.; Yang, L. Carbon dots as potent antimicrobial agents. Theranostics 2020, 10, 671–686. [Google Scholar] [CrossRef]
- Dong, X.; al Awak, M.; Tomlinson, N.; Tang, Y.; Sun, Y.-P.; Yang, L. Antibacterial effects of carbon dots in combination with other antimicrobial reagents. PLoS ONE 2017, 12, e0185324. [Google Scholar] [CrossRef] [Green Version]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Choi, Y.; Kwon, O.-H.; Kim, B.-S. Carbon Dots: Bottom-Up Syntheses, Properties, and Light-Harvesting Applications. Chem. Asian J. 2018, 13, 586–598. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Krysmann, M.J.; Kelarakis, A.; Koutnik, P.; Anzenbacher, P., Jr.; Roland, P.J.; Ellingson, R.; Sun, L. Understanding the Photoluminescence Mechanism of Carbon Dots. MRS Adv. 2017, 2, 2927–2934. [Google Scholar] [CrossRef] [Green Version]
- Carbonaro, C.M.; Chiriu, D.; Stagi, L.; Casula, M.F.; Thakkar, S.V.; Malfatti, L.; Suzuki, K.; Ricci, P.C.; Corpino, R. Carbon Dots in Water and Mesoporous Matrix: Chasing the Origin of their Photoluminescence. J. Phys. Chem. C 2018, 122, 25638–25650. [Google Scholar] [CrossRef]
- Peng, H.; Travas-Sejdic, J. Simple Aqueous Solution Route to Luminescent Carbogenic Dots from Carbohydrates. Chem. Mater. 2009, 21, 5563. [Google Scholar] [CrossRef]
- Choi, Y.; Kim, S.; Choi, M.-H.; Ryoo, S.-R.; Park, J.; Min, D.-H.; Kim, B.-S. Highly Biocompatible Carbon Nanodots for Simultaneous Bioimaging and Targeted Photodynamic Therapy In Vitro and In Vivo. Adv. Funct. Mater. 2014, 24, 5781–5789. [Google Scholar] [CrossRef]
- Mendes, C.A.C.; Burdmann, E.A. Polymyxins—A review focusing on their nephrotoxicity. Rev. Assoc. Med. Bras. 2009, 55, 752–759. [Google Scholar] [CrossRef] [Green Version]
- Dixon, R.A.; Chopra, I. Leakage of periplasmic proteins from Escherichia coli mediated by polymyxin B nonapeptide. Antimicrob. Agents Chemother. 1986, 29, 781–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halder, S.; Yadav, K.K.; Sarkar, R.; Mukherjee, S.; Saha, P.; Haldar, S.; Karmakar, S.; Sen, T. Alteration of Zeta potential and membrane permeability in bacteria: A study with cationic agents. SpringerPlus 2015, 4, 672. [Google Scholar] [CrossRef] [Green Version]
- Jhonsi, M.A.; Ananth, D.A.; Nambirajan, G.; Sivasudha, T.; Yamini, R.; Bera, S.; Kathiravan, A. Antimicrobial activity, cytotoxicity and DNA binding studies of carbon dots. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 196, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Huang, J.; Song, Y.; Zhang, M.; Wang, H.; Lu, F.; Huang, H.; Liu, Y.; Dai, X.; Gu, Z.; et al. Degradable Carbon Dots with Broad-Spectrum Antibacterial Activity. ACS Appl. Mater. Interfaces 2018, 10, 26936–26946. [Google Scholar] [CrossRef]
- Li, Y.J.; Harroun, S.G.; Su, Y.C.; Huang, C.F.; Unnikrishnan, B.; Lin, H.J.; Lin, C.H.; Huang, C.C. Synthesis of Self-Assembled Spermidine-Carbon Quantum Dots Effective against Multidrug-Resistant Bacteria. Adv. Healthc. Mater. 2016, 5, 2545–2554. [Google Scholar] [CrossRef]
- Ge, J.; Jia, Q.; Liu, W.; Lan, M.; Zhou, B.; Guo, L.; Zhou, H.; Zhang, H.; Wang, Y.; Gu, Y.; et al. Carbon Dots with Intrinsic Theranostic Properties for Bioimaging, Red-Light-Triggered Photodynamic/Photothermal Simultaneous Therapy In Vitro and In Vivo. Adv. Healthc. Mater. 2016, 5, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Yu, N.Y.; di Fang, W.; Tan, Q.G.; Ji, R.; Yang, L.Y.; Wei, S.; Zhang, X.W.; Miao, A.J. Photodegradation of carbon dots cause cytotoxicity. Nat. Commun. 2021, 12, 812. [Google Scholar] [CrossRef] [PubMed]
- Travlou, N.A.; Giannakoudakis, D.A.; Algarra, M.; Labella, A.M.; Rodríguez-Castellón, E.; Bandosz, T.J. S- and N-doped carbon quantum dots: Surface chemistry dependent antibacterial activity. Carbon 2018, 135, 104–111. [Google Scholar] [CrossRef]




| Bacteria | Inhibition Zone Diameter (mm) | ||
|---|---|---|---|
| Water | Solution | Carbon Dots | |
| P. carotovorum | 0.00 ± 0.00 | 9.75 ± 0.50 | 18.00 ± 0.82 |
| E. coli | 0.00 ± 0.00 | 5.25 ± 0.50 | 11.75 ± 0.96 |
| P. syringae pv. Tomato | 0.00 ± 0.00 | 13.67 ± 0.58 | 28.67 ± 0.58 |
| A. tumefaciens | 0.00 ± 0.00 | 20.00 ± 1.00 | 19.33 ± 1.15 |
| A. rhizogenes | 0.00 ± 0.00 | 19.67 ± 1.15 | 20.33 ± 0.58 |
| S. enterica subsp enterica serovar Typhimurium | 0.00 ± 0.00 | 0.00 ± 0.00 | 20.67 ± s0.58 |
| Bacteria | Concentration of CDs | |||
|---|---|---|---|---|
| 0.5 mg/mL | 1 mg/mL | 5 mg/mL | 10 mg/mL | |
| A. tumefaciens | 0.00 ± 0.00 | 11.67 ± 0.47 | 26.00 ± 2.94 | 31.00 ± 2.16 |
| A. rhizogenes | 0.00 ± 0.00 | 10.67 ± 0.47 | 20.33 ± 0.47 | 25.33 ± 0.47 |
| S. enterica | 0.00 ± 0.00 | 11.67 ± 0.47 | 11.33 ± 0.47 | 15.67 ± 0.47 |
| P. syringae pv tomato | 0.00 ± 0.00 | 0.00 ± 0.00 | 23.00 ± 0.82 | 32.33 ± 0.47 |
| E. coli | 0.00 ± 0.00 | 9.67 ± 0.47 | 15.67 ± 0.94 | 19.67 ± 0.94 |
| P. carotovorum | 0.00 ± 0.00 | 0.00 ± 0.00 | 15.33 ± 0.47 | 21.33 ± 0.47 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandey, A.; Devkota, A.; Yadegari, Z.; Dumenyo, K.; Taheri, A. Antibacterial Properties of Citric Acid/β-Alanine Carbon Dots against Gram-Negative Bacteria. Nanomaterials 2021, 11, 2012. https://doi.org/10.3390/nano11082012
Pandey A, Devkota A, Yadegari Z, Dumenyo K, Taheri A. Antibacterial Properties of Citric Acid/β-Alanine Carbon Dots against Gram-Negative Bacteria. Nanomaterials. 2021; 11(8):2012. https://doi.org/10.3390/nano11082012
Chicago/Turabian StylePandey, Anju, Asmita Devkota, Zeinab Yadegari, Korsi Dumenyo, and Ali Taheri. 2021. "Antibacterial Properties of Citric Acid/β-Alanine Carbon Dots against Gram-Negative Bacteria" Nanomaterials 11, no. 8: 2012. https://doi.org/10.3390/nano11082012
APA StylePandey, A., Devkota, A., Yadegari, Z., Dumenyo, K., & Taheri, A. (2021). Antibacterial Properties of Citric Acid/β-Alanine Carbon Dots against Gram-Negative Bacteria. Nanomaterials, 11(8), 2012. https://doi.org/10.3390/nano11082012








