Novel Perspectives towards RNA-Based Nano-Theranostic Approaches for Cancer Management
Abstract
1. Introduction
2. RNA Nanotechnology for Diagnosis of Cancers
2.1. Benefits of RNA Nanotechnology in Targeting Cancer Treatment
2.2. Nano-Biosensors as Developing Trend in Cancer Diagnostics
2.3. RNA Nano-Biosensors
| RNA-Based Nanoparticles | Key Feature |
|---|---|
| Immune-Magnetic Exosome RNA (iMER) | Exosomal analysis of glioblastoma multiforme (GBM). |
| Anti-RNA aptamer | Initial detection and analysis of residual GBM. |
| RNA tetrahedrons | Target triple-negative breast cancer cells. |
| Oligonucleotide-treated Au-NPs | Analyzing circulating tumor cells (CTCs) of the prostate. |
| miR-122 mimicked using cationic lipid NPs | Theranostic agent against hepatocellular carcinoma. |
| Superparamagnetic iron oxide NPs (PEG-g-PEI-SPION) | Initial detection and analysis of gastric cancer. |
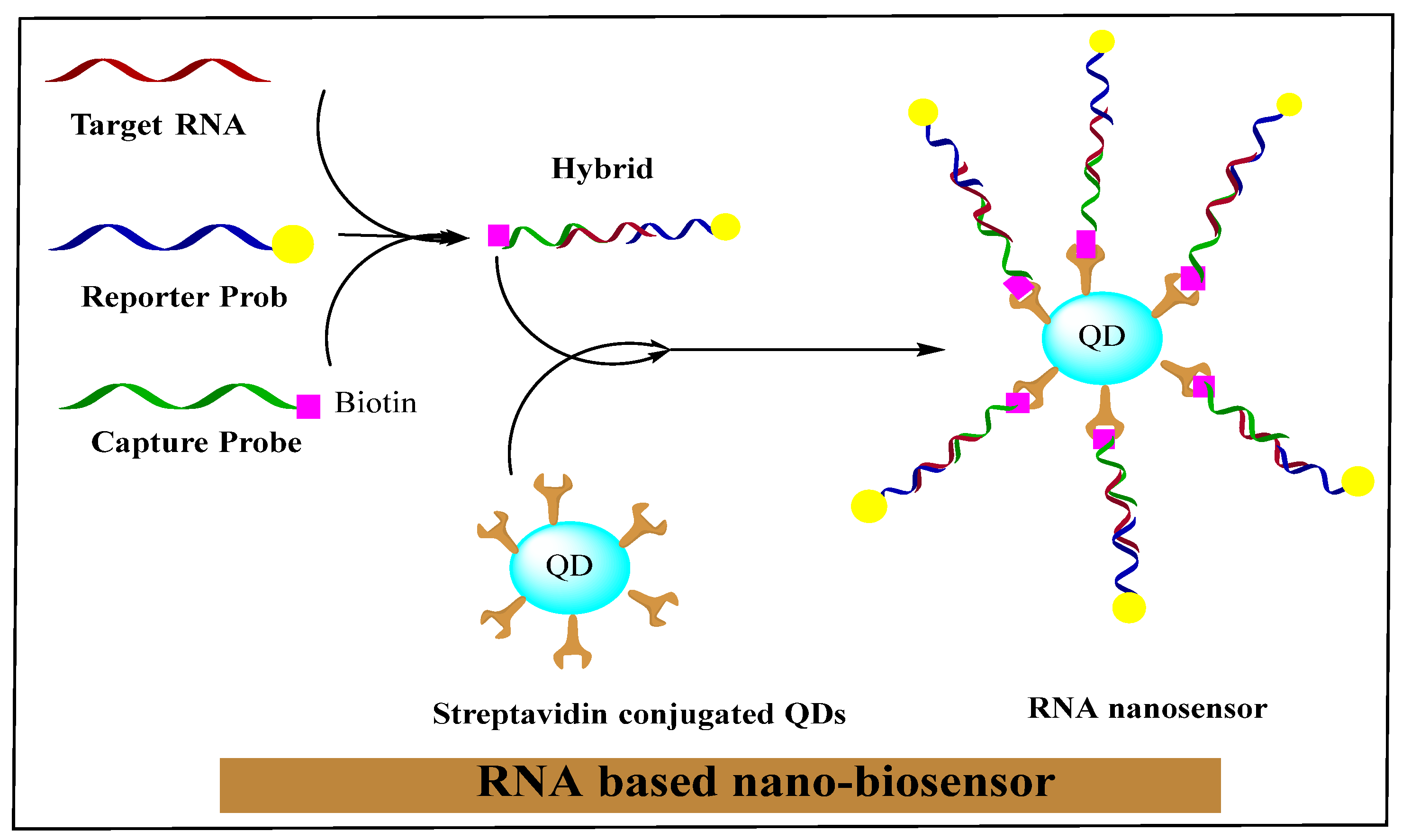
2.4. RNA Nanotechnology in Diagnosis of Different Cancers
2.4.1. GBM
2.4.2. Breast Cancer
2.4.3. Prostate Cancer
2.4.4. Liver Cancer
2.4.5. Gastric Cancer
3. RNA-Nanomaterials for Targeted Therapy of Different Cancers
3.1. RNA NPs
3.2. Nanotechnology for Transfer of Therapeutic RNAs
3.3. Small Interfering RNA-Selenium NPs
3.4. siRNA-Polymeric NPs
3.5. siRNA-Superparamagnetic Iron Oxide NPs
3.6. RNA-Mesoporous Silica NPs
4. Advantages and Limitations of RNA-Based Nano-Theranostic Systems
5. Conclusions and Future Prospective
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| NPs | Nanoparticles |
| siRNAs | Short interfering RNAs |
| PTX | Paclitaxel |
| SPR | Surface plasmon resonance |
| PRRs | Pattern recognition receptors |
| EPR | Enhanced permeability and retention |
| AuNPs | Gold nanoparticles |
| QDs | Quantum dots |
| ssDNA | Single-stranded DNA |
| SELEX | Systemic Evolution of Ligands by EXponential enrichment |
| NIRF | Near-infrared fluorescent |
| MBS | m-maleimidobenzoyl N-hydroxysuccinimde ester |
| GBM | Glioblastoma multiforme |
| iMER | Immune-Magnetic Exosome RNA |
| EGFR | Epidermal growth factor receptor |
| GSCs | GBM stem cells |
| TICc | Tumor-initiating cells |
| TNBC | Triple-negative breast cancer |
| CTCs | Circulating tumor cells |
| PSA | Prostate-specific antigen |
| CLs | Cationic lipids |
| PEG | Polyethylene glycol |
| ECM | Extracellular matrix |
| PD-L1 | Programmed death-ligand 1 |
| TEM | Transmission electron microscopy |
| FTIR | Fourier transform infrared spectroscopy |
| EDS | Energy dispersive spectroscopy |
| ROS | Reactive oxygen species |
| PHB1 | Prohibitin 1 |
| FA | Folic acid |
| MRI | Magnetic resonance imaging |
| HCC | Hepatocellular carcinoma |
| CRC | Colorectal cancer |
| PLL | Poly-L-lysine |
| PBS | Phosphate-buffered saline |
| RT-PCR | Real-time-polymerase chain reaction |
| MSNs | Mesoporous silica nanoparticles |
| PDA | Polymeric dopamine |
References
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Moumaris, M.; Bretagne, J.-M.; Abuaf, N. Nanomedical Devices and Cancer Theranostics. Open Nanomed. Nanotechnol. J. 2020, 61, 69–90. [Google Scholar] [CrossRef]
- Barani, M.; Nematollahi, M.H.; Zaboli, M.; Mirzaei, M.; Torkzadeh-Mahani, M.; Pardakhty, A.; Karam, G.A. In silico and in vitro study of magnetic niosomes for gene delivery: The effect of ergosterol and cholesterol. Mater. Sci. Eng. C 2019, 94, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Mora-Huertas, C.E.; Fessi, H.; Elaissari, A. Polymer-based nanocapsules for drug delivery. Int. J. Pharm. 2010, 385, 113–142. [Google Scholar] [CrossRef] [PubMed]
- Mashayekhi, S.; Rasoulpoor, S.; Shabani, S.; Esmaeilizadeh, N.; Serati-Nouri, H.; Sheervalilou, R.; Pilehvar-Soltanahmadi, Y. Curcumin-loaded mesoporous silica nanoparticles/nanofiber composites for supporting long-term proliferation and stemness preservation of adipose-derived stem cells. Int. J. Pharm. 2020, 587, 119656. [Google Scholar] [CrossRef] [PubMed]
- Sheervalilou, R.; Shirvaliloo, M.; Sargazi, S.; Ghaznavi, H. Recent advances in iron oxide nanoparticles for brain cancer theranostics: From in vitro to clinical applications. Expert Opin. Drug Deliv. 2021, 31, 1–29. [Google Scholar] [CrossRef]
- Kafshdooz, L.; Pourfathi, H.; Akbarzadeh, A.; Kafshdooz, T.; Razban, Z.; Sheervalilou, R.; Ebrahimi Sadr, N.; Khalilov, R.; Saghfi, S.; Kavetskyy, T. The role of microRNAs and nanoparticles in ovarian cancer: A review. Artif. Cells Nanomed. Biotechnol. 2018, 46, 241–247. [Google Scholar] [CrossRef]
- Irajirad, R.; Ahmadi, A.; Najafabad, B.K.; Abed, Z.; Sheervalilou, R.; Khoei, S.; Shiran, M.B.; Ghaznavi, H.; Shakeri-Zadeh, A. Combined thermo-chemotherapy of cancer using 1 MHz ultrasound waves and a cisplatin-loaded sonosensitizing nanoplatform: An in vivo study. Cancer Chemother. Pharmacol. 2019, 84, 1315–1321. [Google Scholar] [CrossRef]
- Raghubir, M.; Rahman, C.N.; Fang, J.; Matsui, H.; Mahajan, S.S. Osteosarcoma growth suppression by riluzole delivery via iron oxide nanocage in nude mice. Oncol. Rep. 2020, 43, 169–176. [Google Scholar] [CrossRef]
- Mishra, S.; Sharma, S.; Javed, M.N.; Pottoo, F.H.; Barkat, M.A.; Alam, M.S.; Amir, M.; Sarafroz, M. Bioinspired nanocomposites: Applications in disease diagnosis and treatment. Pharm. Nanotechnol. 2019, 7, 206–219. [Google Scholar] [CrossRef]
- Haghiralsadat, F.; Amoabediny, G.; Naderinezhad, S.; Nazmi, K.; De Boer, J.P.; Zandieh-Doulabi, B.; Forouzanfar, T.; Helder, M.N. EphA2 targeted doxorubicin-nanoliposomes for osteosarcoma treatment. Pharm. Res. 2017, 34, 2891–2900. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; He, C.; Cheng, Y.; Li, D.; Gong, Y.; Liu, J.; Tian, H.; Chen, X. PLK1shRNA and doxorubicin co-loaded thermosensitive PLGA-PEG-PLGA hydrogels for osteosarcoma treatment. Biomaterials 2014, 35, 8723–8734. [Google Scholar] [CrossRef]
- Shakeri-Zadeh, A.; Zareyi, H.; Sheervalilou, R.; Laurent, S.; Ghaznavi, H.; Samadian, H. Gold nanoparticle-mediated bubbles in cancer nanotechnology. J. Control. Release 2020, 330, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Rauf, A.; Tabish, T.A.; Ibrahim, I.M.; ul Hassan, M.R.; Tahseen, S.; Sandhu, M.A.; Shahnaz, G.; Rahdar, A.; Cucchiarini, M.; Pandey, S. Design of Mannose-Coated Rifampicin nanoparticles modulating the immune response and Rifampicin induced hepatotoxicity with improved oral drug delivery. Arab. J. Chem. 2021, 14, 103321. [Google Scholar] [CrossRef]
- Niazi, M.; Zakeri-Milani, P.; Najafi Hajivar, S.; Soleymani Goloujeh, M.; Ghobakhlou, N.; Shahbazi Mojarrad, J.; Valizadeh, H. Nano-based strategies to overcome p-glycoprotein-mediated drug resistance. Expert Opin. Drug Metab. Toxicol. 2016, 12, 1021–1033. [Google Scholar] [CrossRef]
- Rahdar, A.; Hasanein, P.; Bilal, M.; Beyzaei, H.; Kyzas, G.Z. Quercetin-loaded F127 nanomicelles: Antioxidant activity and protection against renal injury induced by gentamicin in rats. Life Sci. 2021, 276, 119420. [Google Scholar] [CrossRef]
- Javad Farhangi, M.; Es-Haghi, A.; Taghavizadeh Yazdi, M.E.; Rahdar, A.; Baino, F. MOF-Mediated Synthesis of CuO/CeO2 Composite Nanoparticles: Characterization and Estimation of the Cellular Toxicity against Breast Cancer Cell Line (MCF-7). J. Funct. Biomater. 2021, 12, 53. [Google Scholar] [CrossRef]
- Arshad, R.; Tabish, T.A.; Kiani, M.H.; Ibrahim, I.M.; Shahnaz, G.; Rahdar, A.; Kang, M.; Pandey, S. A Hyaluronic Acid Functionalized Self-Nano-Emulsifying Drug Delivery System (SNEDDS) for Enhancement in Ciprofloxacin Targeted Delivery against Intracellular Infection. Nanomaterials 2021, 11, 1086. [Google Scholar] [CrossRef]
- Bilal, M.; Qindeel, M.; Raza, A.; Mehmood, S.; Rahdar, A. Stimuli-responsive nanoliposomes as prospective nanocarriers for targeted drug delivery. J. Drug Deliv. Sci. Technol. 2021, 102916. [Google Scholar] [CrossRef]
- Rahdar, A.; Taboada, P.; Aliahmad, M.; Hajinezhad, M.R.; Sadeghfar, F. Iron oxide nanoparticles: Synthesis, physical characterization, and intraperitoneal biochemical studies in Rattus norvegicus. J. Mol. Struct. 2018, 1173, 240–245. [Google Scholar] [CrossRef]
- Sivasankarapillai, V.S.; Somakumar, A.K.; Joseph, J.; Nikazar, S.; Rahdar, A.; Kyzas, G.Z. Cancer theranostic applications of MXene nanomaterials: Recent updates. Nano-Struct. Nano-Objects 2020, 22, 100457. [Google Scholar] [CrossRef]
- Bertrand, N.; Wu, J.; Xu, X.; Kamaly, N.; Farokhzad, O.C. Cancer nanotechnology: The impact of passive and active targeting in the era of modern cancer biology. Adv. Drug Deliv. Rev. 2014, 66, 2–25. [Google Scholar] [CrossRef] [PubMed]
- Paris, J.L.; Vallet-Regí, M. Mesoporous Silica Nanoparticles for Co-Delivery of Drugs and Nucleic Acids in Oncology: A Review. Pharmaceutics 2020, 12, 526. [Google Scholar] [CrossRef]
- Chaturvedi, V.K.; Singh, A.; Singh, V.K.; Singh, M.P. Cancer nanotechnology: A new revolution for cancer diagnosis and therapy. Curr. Drug Metab. 2019, 20, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Eivazzadeh-Keihan, R.; Pashazadeh-Panahi, P.; Baradaran, B.; Maleki, A.; Hejazi, M.; Mokhtarzadeh, A.; de la Guardia, M. Recent advances on nanomaterial based electrochemical and optical aptasensors for detection of cancer biomarkers. TrAC Trends Anal. Chem. 2018, 100, 103–115. [Google Scholar] [CrossRef]
- Datta, L.P.; Manchineella, S.; Govindaraju, T. Biomolecules-derived biomaterials. Biomaterials 2020, 230, 119633. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y.; Nie, G. Multifunctional biomolecule nanostructures for cancer therapy. Nat. Rev. Mater. 2021, 19, 1–18. [Google Scholar] [CrossRef]
- Guo, P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 2010, 5, 833–842. [Google Scholar] [CrossRef]
- Mansouri, S.; Cuie, Y.; Winnik, F.; Shi, Q.; Lavigne, P.; Benderdour, M.; Beaumont, E.; Fernandes, J.C. Characterization of folate-chitosan-DNA nanoparticles for gene therapy. Biomaterials 2006, 27, 2060–2065. [Google Scholar] [CrossRef]
- Mao, H.-Q.; Roy, K.; Troung-Le, V.L.; Janes, K.A.; Lin, K.Y.; Wang, Y.; August, J.T.; Leong, K.W. Chitosan-DNA nanoparticles as gene carriers: Synthesis, characterization and transfection efficiency. J. Control. Release 2001, 70, 399–421. [Google Scholar] [CrossRef]
- Liu, G.; Li, D.; Pasumarthy, M.K.; Kowalczyk, T.H.; Gedeon, C.R.; Hyatt, S.L.; Payne, J.M.; Miller, T.J.; Brunovskis, P.; Fink, T.L. Nanoparticles of compacted DNA transfect postmitotic cells. J. Biol. Chem. 2003, 278, 32578–32586. [Google Scholar] [CrossRef]
- Mastorakos, P.; Da Silva, A.L.; Chisholm, J.; Song, E.; Choi, W.K.; Boyle, M.P.; Morales, M.M.; Hanes, J.; Suk, J.S. Highly compacted biodegradable DNA nanoparticles capable of overcoming the mucus barrier for inhaled lung gene therapy. Proc. Natl. Acad. Sci. USA 2015, 112, 8720–8725. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, A.L.; Oliveira, G.P.; Kitoko, J.Z.; Blanco, N.G.; Suk, J.S.; Hanes, J.; Olsen, P.C.; Morales, M.M.; Rocco, P.R. Thymulin Gene Therapy Delivered By A New Biodegradable Dna Nanoparticle In Experimental Chronic Allergic Asthma. In D21. Asthma Treatment: Glucocorticoids, Biologicals and Beyond; American Thoracic Society: New York, NY, USA, 2016; p. A6490. [Google Scholar]
- Haque, F.; Pi, F.; Zhao, Z.; Gu, S.; Hu, H.; Yu, H.; Guo, P. RNA versatility, flexibility, and thermostability for practice in RNA nanotechnology and biomedical applications. Wiley Interdiscip. Rev. RNA 2018, 9, e1452. [Google Scholar] [CrossRef]
- Guo, P. The Emerging Field of RNA Nanotechnology. In Nano-Enabled Medical Applications; Jenny Stanford Publishing: Dubai, United Arab Emirates, 2020; pp. 131–157. [Google Scholar]
- Arshad, R.; Barani, M.; Rahdar, A.; Sargazi, S.; Cucchiarini, M.; Pandey, S.; Kang, M. Multi-Functionalized Nanomaterials and Nanoparticles for Diagnosis and Treatment of Retinoblastoma. Biosensors 2021, 11, 97. [Google Scholar] [CrossRef] [PubMed]
- Barani, M.; Sargazi, S.; Mohammadzadeh, V.; Rahdar, A.; Pandey, S.; Jha, N.K.; Gupta, P.K.; Thakur, V.K. Theranostic Advances of Bionanomaterials against Gestational Diabetes Mellitus: A Preliminary Review. J. Funct. Biomater. 2021, 12, 54. [Google Scholar] [CrossRef] [PubMed]
- Barani, M.; Zeeshan, M.; Kalantar-Neyestanaki, D.; Farooq, M.A.; Rahdar, A.; Jha, N.K.; Sargazi, S.; Gupta, P.K.; Thakur, V.K. Nanomaterials in the Management of Gram-Negative Bacterial Infections. Nanomaterials 2021, 11, 2535. [Google Scholar] [CrossRef] [PubMed]
- Fiume, E.; Magnaterra, G.; Rahdar, A.; Verné, E.; Baino, F. Hydroxyapatite for Biomedical Applications: A Short Overview. Ceramics 2021, 4, 542–563. [Google Scholar] [CrossRef]
- Hashemi, B.; Firouzi-amandi, A.; Amirazad, H.; Dadashpour, M.; Shirvaliloo, M.; Nasrabadi, D.; Ahmadi, M.; Sheervalilou, R.; Reza, M.A.S.; Ghazi, F. Emerging importance of nanotechnology-based approaches to control the COVID-19 pandemic; focus on nanomedicine iterance in diagnosis and treatment of COVID-19 patients. J. Drug Deliv. Sci. Technol. 2021, 102967. [Google Scholar] [CrossRef] [PubMed]
- Hassanisaadi, M.; Bonjar, G.H.S.; Rahdar, A.; Pandey, S.; Hosseinipour, A.; Abdolshahi, R. Environmentally Safe Biosynthesis of Gold Nanoparticles Using Plant Water Extracts. Nanomaterials 2021, 11, 2033. [Google Scholar] [CrossRef]
- Mukhtar, M.; Sargazi, S.; Barani, M.; Madry, H.; Rahdar, A.; Cucchiarini, M. Application of Nanotechnology for Sensitive Detection of Low-Abundance Single-Nucleotide Variations in Genomic DNA: A Review. Nanomaterials 2021, 11, 1384. [Google Scholar] [CrossRef] [PubMed]
- Norouzi, M.; Yasamineh, S.; Montazeri, M.; Dadashpour, M.; Sheervalilou, R.; Abasi, M.; Pilehvar-Soltanahmadi, Y. Recent advances on nanomaterials-based fluorimetric approaches for microRNAs detection. Mater. Sci. Eng. C 2019, 104, 110007. [Google Scholar] [CrossRef]
- Rahdar, A.; Hajinezhad, M.R.; Bilal, M.; Askari, F.; Kyzas, G.Z. Behavioral effects of zinc oxide nanoparticles on the brain of rats. Inorg. Chem. Commun. 2020, 119, 108131. [Google Scholar] [CrossRef]
- Sargazi, S.; Mukhtar, M.; Rahdar, A.; Barani, M.; Pandey, S.; Díez-Pascual, A.M. Active targeted nanoparticles for delivery of poly (ADP-ribose) polymerase (PARP) inhibitors: A preliminary review. Int. J. Mol. Sci. 2021, 22, 10319. [Google Scholar] [CrossRef] [PubMed]
- Shirvalilou, S.; Khoei, S.; Esfahani, A.J.; Kamali, M.; Shirvaliloo, M.; Sheervalilou, R.; Mirzaghavami, P. Magnetic Hyperthermia as an adjuvant cancer therapy in combination with radiotherapy versus radiotherapy alone for recurrent/progressive glioblastoma: A systematic review. J. Neuro-Oncol. 2021, 13, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Nangreave, J.; Liu, Y.; Yan, H. Structural DNA nanotechnology: State of the art and future perspective. J. Am. Chem. Soc. 2014, 136, 11198–11211. [Google Scholar] [CrossRef]
- Ohno, H.; Akamine, S.; Saito, H. RNA nanostructures and scaffolds for biotechnology applications. Curr. Opin. Biotechnol. 2019, 58, 53–61. [Google Scholar] [CrossRef]
- Jasinski, D.; Haque, F.; Binzel, D.W.; Guo, P. Advancement of the emerging field of RNA nanotechnology. ACS Nano 2017, 11, 1142–1164. [Google Scholar] [CrossRef]
- Shu, Y.; Pi, F.; Sharma, A.; Rajabi, M.; Haque, F.; Shu, D.; Leggas, M.; Evers, B.M.; Guo, P. Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Adv. Drug Deliv. Rev. 2014, 66, 74–89. [Google Scholar] [CrossRef]
- Guo, S.; Xu, C.; Yin, H.; Hill, J.; Pi, F.; Guo, P. Tuning the size, shape and structure of RNA nanoparticles for favorable cancer targeting and immunostimulation. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, 12, e1582. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zheng, M.; Wu, S.; Tian, C.; Liu, D.; Weizmann, Y.; Jiang, W.; Wang, G.; Mao, C. In vivo production of RNA nanostructures via programmed folding of single-stranded RNAs. Nat. Commun. 2018, 9, 1–9. [Google Scholar] [CrossRef]
- Farokhzad, O.C.; Jon, S.; Khademhosseini, A.; Tran, T.-N.T.; LaVan, D.A.; Langer, R. Nanoparticle-aptamer bioconjugates: A new approach for targeting prostate cancer cells. Cancer Res. 2004, 64, 7668–7672. [Google Scholar] [CrossRef]
- Germer, K.; Leonard, M.; Zhang, X. RNA aptamers and their therapeutic and diagnostic applications. Int. J. Biochem. Mol. Biol. 2013, 4, 27. [Google Scholar]
- Shu, Y.; Shu, D.; Diao, Z.; Shen, G.; Guo, P. Fabrication of polyvalent therapeutic RNA nanoparticles for specific delivery of siRNA, ribozyme and drugs to targeted cells for cancer therapy. In Proceedings of the 2009 IEEE/NIH Life Science Systems and Applications Workshop, Bethesda, MD, USA, 8–10 April 2009; pp. 9–12. [Google Scholar]
- Yin, H.; Xiong, G.; Guo, S.; Xu, C.; Xu, R.; Guo, P.; Shu, D. Delivery of anti-miRNA for triple-negative breast cancer therapy using RNA nanoparticles targeting stem cell marker CD133. Mol. Ther. 2019, 27, 1252–1261. [Google Scholar] [CrossRef] [PubMed]
- Ghimire, C.; Wang, H.; Li, H.; Vieweger, M.; Xu, C.; Guo, P. RNA nanoparticles as rubber for compelling vessel extravasation to enhance tumor targeting and for fast renal excretion to reduce toxicity. ACS Nano 2020, 14, 13180–13191. [Google Scholar] [CrossRef]
- Wang, H.; Guo, P. Radiolabeled RNA Nanoparticles for Highly Specific Targeting and Efficient Tumor Accumulation with Favorable In Vivo Biodistribution. Mol. Pharm. 2021, 18, 2924–2934. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Hyun, H.N.; Heo, R.; Nam, K.; Yang, K.; Kim, Y.M.; Lee, Y.S.; An, J.Y.; Park, J.H.; Choi, K.Y. Dual-targeting RNA nanoparticles for efficient delivery of polymeric siRNA to cancer cells. Chem. Commun. 2020, 56, 6624–6627. [Google Scholar] [CrossRef]
- Xu, Y.; Pang, L.; Wang, H.; Xu, C.; Shah, H.; Guo, P.; Shu, D.; Qian, S.Y. Specific delivery of delta-5-desaturase siRNA via RNA nanoparticles supplemented with dihomo-γ-linolenic acid for colon cancer suppression. Redox Biol. 2019, 21, 101085. [Google Scholar] [CrossRef]
- Haque, F.; Shu, D.; Shu, Y.; Shlyakhtenko, L.S.; Rychahou, P.G.; Evers, B.M.; Guo, P. Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today 2012, 7, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Metkar, S.K.; Girigoswami, K. Diagnostic biosensors in medicine—A review. Biocatal. Agric. Biotechnol. 2019, 17, 271–283. [Google Scholar] [CrossRef]
- El Harrad, L.; Bourais, I.; Mohammadi, H.; Amine, A. Recent advances in electrochemical biosensors based on enzyme inhibition for clinical and pharmaceutical applications. Sensors 2018, 18, 164. [Google Scholar] [CrossRef]
- Bagherzade, G.; Tavakoli, M.M.; Namaei, M.H. Green synthesis of silver nanoparticles using aqueous extract of saffron (Crocus sativus L.) wastages and its antibacterial activity against six bacteria. Asian Pac. J. Trop. Biomed. 2017, 7, 227–233. [Google Scholar] [CrossRef]
- Usman, A.I.; Aziz, A.A.; Noqta, O.A. Application of green synthesis of gold nanoparticles: A review. J. Teknol. 2019, 81. [Google Scholar] [CrossRef]
- Shanaa, O.A.; Rumyantsev, A.; Sambuk, E.; Padkina, M. In Vivo Production of RNA Aptamers and Nanoparticles: Problems and Prospects. Molecules 2021, 26, 1422. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Wang, H.; Li, Z.; Shu, D.; Guo, P. RNA micelles for the systemic delivery of anti-miRNA for cancer targeting and inhibition without ligand. ACS Nano 2018, 13, 706–717. [Google Scholar] [CrossRef]
- Zhang, Y.; Leonard, M.; Shu, Y.; Yang, Y.; Shu, D.; Guo, P.; Zhang, X. Overcoming tamoxifen resistance of human breast cancer by targeted gene silencing using multifunctional pRNA nanoparticles. ACS Nano 2017, 11, 335–346. [Google Scholar] [CrossRef]
- Xu, C.; Haque, F.; Jasinski, D.L.; Binzel, D.W.; Shu, D.; Guo, P. Favorable biodistribution, specific targeting and conditional endosomal escape of RNA nanoparticles in cancer therapy. Cancer Lett. 2018, 414, 57–70. [Google Scholar] [CrossRef]
- Shi, J.; Kantoff, P.W.; Wooster, R.; Farokhzad, O.C. Cancer nanomedicine: Progress, challenges and opportunities. Nat. Rev. Cancer 2017, 17, 20–37. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Li, H.; Ma, M.; Fu, J.; Dong, Y.; Guo, P. Size, shape, and sequence-dependent immunogenicity of RNA nanoparticles. Mol. Ther. Nucleic Acids 2017, 9, 399–408. [Google Scholar] [CrossRef]
- Salvati, E.; Stellacci, F.; Krol, S. Nanosensors for early cancer detection and for therapeutic drug monitoring. Nanomedicine 2015, 10, 3495–3512. [Google Scholar] [CrossRef]
- Rossetti, M.; Del Grosso, E.; Ranallo, S.; Mariottini, D.; Idili, A.; Bertucci, A.; Porchetta, A. Programmable RNA-based systems for sensing and diagnostic applications. Anal. Bioanal. Chem. 2019, 411, 4293–4302. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, A.R. DNA nanobiosensors: An outlook on signal readout strategies. J. Nanomater. 2017, 2017, 2820619. [Google Scholar] [CrossRef]
- Shandilya, R.; Bhargava, A.; Bunkar, N.; Tiwari, R.; Goryacheva, I.Y.; Mishra, P.K. Nanobiosensors: Point-of-care approaches for cancer diagnostics. Biosens. Bioelectron. 2019, 130, 147–165. [Google Scholar] [CrossRef] [PubMed]
- Bartosik, K.; Debiec, K.; Czarnecka, A.; Sochacka, E.; Leszczynska, G. Synthesis of Nucleobase-Modified RNA Oligonucleotides by Post-Synthetic Approach. Molecules 2020, 25, 3344. [Google Scholar] [CrossRef] [PubMed]
- Binzel, D.W.; Li, X.; Burns, N.; Khan, E.; Lee, W.-J.; Chen, L.-C.; Ellipilli, S.; Miles, W.; Ho, Y.S.; Guo, P. Thermostability, Tunability, and Tenacity of RNA as Rubbery Anionic Polymeric Materials in Nanotechnology and Nanomedicine—Specific Cancer Targeting with Undetectable Toxicity. Chem. Rev. 2021, 121, 1322–1335. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Piñeiro, I.; Badiola, I.; Sanchez, A. Nanocarriers for microRNA delivery in cancer medicine. Biotechnol. Adv. 2017, 35, 350–360. [Google Scholar] [CrossRef] [PubMed]
- Tommasini-Ghelfi, S.; Lee, A.; Mirkin, C.A.; Stegh, A.H. Synthesis, physicochemical, and biological evaluation of spherical nucleic acids for RNAi-based therapy in glioblastoma. In RNA Interference and Cancer Therapy; Springer: Berlin/Heidelberg, Germany, 2019; pp. 371–391. [Google Scholar]
- Hasan, M.R.; Hassan, N.; Khan, R.; Kim, Y.-T.; Iqbal, S.M. Classification of cancer cells using computational analysis of dynamic morphology. Comput. Methods Programs Biomed. 2018, 156, 105–112. [Google Scholar] [CrossRef]
- Nuzzo, S.; Brancato, V.; Affinito, A.; Salvatore, M.; Cavaliere, C.; Condorelli, G. The role of RNA and dna aptamers in glioblastoma diagnosis and therapy: A systematic review of the literature. Cancers 2020, 12, 2173. [Google Scholar] [CrossRef]
- Fechter, P.; Da Silva, E.C.; Mercier, M.-C.; Noulet, F.; Etienne-Seloum, N.; Guenot, D.; Lehmann, M.; Vauchelles, R.; Martin, S.; Lelong-Rebel, I. RNA Aptamers Targeting Integrin α5β1 as Probes for Cyto-and Histofluorescence in Glioblastoma. Mol. Ther. Nucleic Acids 2019, 17, 63–77. [Google Scholar] [CrossRef]
- Affinito, A.; Quintavalle, C.; Esposito, C.L.; Roscigno, G.; Vilardo, C.; Nuzzo, S.; Ricci-Vitiani, L.; De Luca, G.; Pallini, R.; Kichkailo, A.S. The discovery of RNA aptamers that selectively bind glioblastoma stem cells. Mol. Ther. Nucleic Acids 2019, 18, 99–109. [Google Scholar] [CrossRef]
- Hadjidemetriou, M.; Kostarelos, K. Evolution of the nanoparticle corona. Nat. Nanotechnol. 2017, 12, 288–290. [Google Scholar] [CrossRef]
- Ganju, A.; Khan, S.; Hafeez, B.B.; Behrman, S.W.; Yallapu, M.M.; Chauhan, S.C.; Jaggi, M. miRNA nanotherapeutics for cancer. Drug Discov. Today 2017, 22, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Liu, Y.; Yung, B.; Xiong, Y.; Chen, X. Nanotechnology-enhanced no-wash biosensors for in vitro diagnostics of cancer. ACS Nano 2017, 11, 5238–5292. [Google Scholar] [CrossRef]
- Huang, X.; O’Connor, R.; Kwizera, E.A. Gold nanoparticle based platforms for circulating cancer marker detection. Nanotheranostics 2017, 1, 80. [Google Scholar] [CrossRef]
- Matsuda, M.; Ishikawa, E.; Yamamoto, T.; Hatano, K.; Joraku, A.; Iizumi, Y.; Masuda, Y.; Nishiyama, H.; Matsumura, A. Potential use of prostate specific membrane antigen (PSMA) for detecting the tumor neovasculature of brain tumors by PET imaging with 89 Zr-Df-IAB2M anti-PSMA minibody. J. Neuro-Oncol. 2018, 138, 581–589. [Google Scholar] [CrossRef]
- Tanjore Ramanathan, J.; Lehtipuro, S.; Sihto, H.; Tóvári, J.; Reiniger, L.; Téglási, V.; Moldvay, J.; Nykter, M.; Haapasalo, H.; Le Joncour, V. Prostate-specific membrane antigen expression in the vasculature of primary lung carcinomas associates with faster metastatic dissemination to the brain. J. Cell. Mol. Med. 2020, 24, 6916–6927. [Google Scholar] [CrossRef]
- Engur, C.O.; Turoglu, H.T.; Ozguven, S.; Tanidir, Y.; Erdil, T.Y. 68Ga-Prostate-Specific Membrane Antigen PET-Positive Paget Bone Disease With Metastatic Prostatic Carcinoma. Clin. Nucl. Med. 2020, 45, e425–e426. [Google Scholar] [CrossRef]
- Anwer, K.; Meaney, C.; Kao, G.; Hussain, N.; Shelvin, R.; Earls, R.M.; Leonard, P.; Quezada, A.; Rolland, A.P.; Sullivan, S.M. Cationic lipid-based delivery system for systemic cancer gene therapy. Cancer Gene Ther. 2000, 7, 1156–1164. [Google Scholar] [CrossRef][Green Version]
- Chaudhary, V.; Jangra, S.; Yadav, N.R. Nanotechnology based approaches for detection and delivery of microRNA in healthcare and crop protection. J. Nanobiotechnol. 2018, 16, 1–18. [Google Scholar] [CrossRef]
- Li, R.; Liu, B.; Gao, J. The application of nanoparticles in diagnosis and theranostics of gastric cancer. Cancer Lett. 2017, 386, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Amorim, S.; Reis, C.A.; Reis, R.L.; Pires, R.A. Extracellular matrix mimics using hyaluronan-based biomaterials. Trends Biotechnol. 2021, 39, 90–104. [Google Scholar] [CrossRef] [PubMed]
- Jaakonmäki, R.; Simons, A.; Müller, O.; vom Brocke, J. ECM implementations in practice: Objectives, processes, and technologies. J. Enterp. Inf. Manag. 2018, 5, 704–723. [Google Scholar] [CrossRef]
- Marchandet, L.; Lallier, M.; Charrier, C.; Baud’huin, M.; Ory, B.; Lamoureux, F. Mechanisms of Resistance to Conventional Therapies for Osteosarcoma. Cancers 2021, 13, 683. [Google Scholar] [CrossRef] [PubMed]
- Arnett, A.B.; Wang, T.; Eichler, E.E.; Bernier, R.A. Reflections on the genetics-first approach to advancements in molecular genetic and neurobiological research on neurodevelopmental disorders. J. Neurodev. Disord. 2021, 13, 1–10. [Google Scholar] [CrossRef]
- Falese, J.P.; Donlic, A.; Hargrove, A.E. Targeting RNA with small molecules: From fundamental principles towards the clinic. Chem. Soc. Rev. 2021, 50, 2224–2243. [Google Scholar] [CrossRef]
- Saraswathy, M.; Gong, S. Recent developments in the co-delivery of siRNA and small molecule anticancer drugs for cancer treatment. Mater. Today 2014, 17, 298–306. [Google Scholar] [CrossRef]
- Bajan, S.; Hutvagner, G. RNA-based therapeutics: From antisense oligonucleotides to miRNAs. Cells 2020, 9, 137. [Google Scholar] [CrossRef]
- Elcheva, I.A.; Spiegelman, V.S. Targeting RNA-binding proteins in acute and chronic leukemia. Leukemia 2021, 35, 360–376. [Google Scholar] [CrossRef]
- Guo, S.; Vieweger, M.; Zhang, K.; Yin, H.; Wang, H.; Li, X.; Li, S.; Hu, S.; Sparreboom, A.; Evers, B.M.; et al. Ultra-thermostable RNA nanoparticles for solubilizing and high-yield loading of paclitaxel for breast cancer therapy. Nat. Commun. 2020, 11, 972. [Google Scholar] [CrossRef]
- Xia, Y.; Tang, G.; Wang, C.; Zhong, J.; Chen, Y.; Hua, L.; Li, Y.; Liu, H.; Zhu, B. Functionalized selenium nanoparticles for targeted siRNA delivery silence Derlin1 and promote antitumor efficacy against cervical cancer. Drug Deliv. 2020, 27, 15–25. [Google Scholar] [CrossRef]
- Li, Y.; Duo, Y.; Bi, J.; Zeng, X.; Mei, L.; Bao, S.; He, L.; Shan, A.; Zhang, Y.; Yu, X. Targeted delivery of anti-miR-155 by functionalized mesoporous silica nanoparticles for colorectal cancer therapy. Int. J. Nanomed. 2018, 13, 1241. [Google Scholar] [CrossRef]
- Wu, Z.; Xu, X.-L.; Zhang, J.-Z.; Mao, X.-H.; Xie, M.-W.; Cheng, Z.-L.; Lu, L.-J.; Duan, X.-H.; Zhang, L.-M.; Shen, J. Magnetic cationic amylose nanoparticles used to deliver survivin-small interfering RNA for gene therapy of hepatocellular carcinoma in vitro. Nanomaterials 2017, 7, 110. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wu, J.; Liu, Y.; Saw, P.E.; Tao, W.; Yu, M.; Zope, H.; Si, M.; Victorious, A.; Rasmussen, J. Multifunctional envelope-type siRNA delivery nanoparticle platform for prostate cancer therapy. ACS Nano 2017, 11, 2618–2627. [Google Scholar] [CrossRef]
- Luo, X.; Peng, X.; Hou, J.; Wu, S.; Shen, J.; Wang, L. Folic acid-functionalized polyethylenimine superparamagnetic iron oxide nanoparticles as theranostic agents for magnetic resonance imaging and PD-L1 siRNA delivery for gastric cancer. Int. J. Nanomed. 2017, 12, 5331. [Google Scholar] [CrossRef] [PubMed]
- Lio, D.C.S.; Liu, C.; Oo, M.M.S.; Wiraja, C.; Teo, M.H.Y.; Zheng, M.; Chew, S.W.T.; Wang, X.; Xu, C. Transdermal delivery of small interfering RNAs with topically applied mesoporous silica nanoparticles for facile skin cancer treatment. Nanoscale 2019, 11, 17041–17051. [Google Scholar] [CrossRef]
- Batool, A.; Arshad, R.; Razzaq, S.; Nousheen, K.; Kiani, M.H.; Shahnaz, G. Formulation and evaluation of hyaluronic acid-based mucoadhesive self nanoemulsifying drug delivery system (SNEDDS) of tamoxifen for targeting breast cancer. Int. J. Biol. Macromol. 2020, 152, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Marta, G.; Ramiah, D.; Kaidar-Person, O.; Kirby, A.; Coles, C.; Jagsi, R.; Hijal, T.; Sancho, G.; Zissiadis, Y.; Pignol, J.-P. The financial impact on reimbursement of moderately hypofractionated postoperative radiation therapy for breast cancer: An international consortium report. Clin. Oncol. 2021, 33, 322–330. [Google Scholar] [CrossRef]
- Sato, Y.; Hashiba, K.; Sasaki, K.; Maeki, M.; Tokeshi, M.; Harashima, H. Understanding structure-activity relationships of pH-sensitive cationic lipids facilitates the rational identification of promising lipid nanoparticles for delivering siRNAs in vivo. J. Control. Release 2019, 295, 140–152. [Google Scholar] [CrossRef]
- Lin, Y.-X.; Wang, Y.; Blake, S.; Yu, M.; Mei, L.; Wang, H.; Shi, J. RNA nanotechnology-mediated cancer immunotherapy. Theranostics 2020, 10, 281. [Google Scholar] [CrossRef]
- Jasinski, D.L.; Yin, H.; Li, Z.; Guo, P. Hydrophobic effect from conjugated chemicals or drugs on in vivo biodistribution of RNA nanoparticles. Hum. Gene Ther. 2018, 29, 77–86. [Google Scholar] [CrossRef]
- Gangopadhyay, S.; Nikam, R.R.; Gore, K.R. Folate Receptor-Mediated Small Interfering RNA Delivery: Recent Developments and Future Directions for RNA Interference Therapeutics. Nucleic Acid Ther. 2021, 31, 245–270. [Google Scholar] [CrossRef]
- Mendonça, M.C.; Kont, A.; Aburto, M.R.; Cryan, J.F.; O’Driscoll, C.M. Advances in the Design of (Nano) Formulations for Delivery of Antisense Oligonucleotides and Small Interfering RNA: Focus on the Central Nervous System. Mol. Pharm. 2021, 18, 1491–1506. [Google Scholar] [CrossRef] [PubMed]
- Arbyn, M.; Gultekin, M.; Morice, P.; Nieminen, P.; Cruickshank, M.; Poortmans, P.; Kelly, D.; Poljak, M.; Bergeron, C.; Ritchie, D. The European response to the WHO call to eliminate cervical cancer as a public health problem. Int. J. Cancer 2021, 148, 277–284. [Google Scholar] [CrossRef]
- Barani, M.; Sabir, F.; Rahdar, A.; Arshad, R.; Kyzas, G.Z. Nanotreatment and Nanodiagnosis of Prostate Cancer: Recent Updates. Nanomaterials 2020, 10, 1696. [Google Scholar] [CrossRef]
- Chiang, T.-H.; Chang, W.-J.; Chen, S.L.-S.; Yen, A.M.-F.; Fann, J.C.-Y.; Chiu, S.Y.-H.; Chen, Y.-R.; Chuang, S.-L.; Shieh, C.-F.; Liu, C.-Y. Mass eradication of Helicobacter pylori to reduce gastric cancer incidence and mortality: A long-term cohort study on Matsu Islands. Gut 2021, 70, 243–250. [Google Scholar]
- Lei, M.; Siemers, N.O.; Pandya, D.; Chang, H.; Sanchez, T.; Harbison, C.; Szabo, P.M.; Janjigian, Y.; Ott, P.A.; Sharma, P. Analyses of PD-L1 and Inflammatory Gene Expression Association with Efficacy of Nivolumab±Ipilimumab in Gastric Cancer/Gastroesophageal Junction Cancer. Clin. Cancer Res. 2021, 27. [Google Scholar] [CrossRef]
- Wu, L.; Cai, S.; Deng, Y.; Zhang, Z.; Zhou, X.; Su, Y.; Xu, D. PD-1/PD-L1 enhanced cisplatin resistance in gastric cancer through PI3K/AKT mediated P-gp expression. Int. Immunopharmacol. 2021, 94, 107443. [Google Scholar] [CrossRef]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Chew, X.H.; Sultana, R.; Mathew, E.N.; Ng, D.C.E.; Lo, R.H.; Toh, H.C.; Tai, D.; Choo, S.P.; Goh, B.K.P.; Yan, S.X. Real-World Data on Clinical Outcomes of Patients with Liver Cancer: A Prospective Validation of the National Cancer Centre Singapore Consensus Guidelines for the Management of Hepatocellular Carcinoma. Liver Cancer 2021, 7, 1–16. [Google Scholar] [CrossRef]
- uz Zaman, S.; Arshad, R.; Tabish, T.A.; Naseem, A.A.; Shahnaz, G. Mapping the potential of thiolated pluronic based nanomicelles for the safe and targeted delivery of vancomycin against staphylococcal blepharitis. J. Drug Deliv. Sci. Technol. 2021, 61, 102220. [Google Scholar]
- Nguyen, T.V.; Damiani, G.; Orenstein, L.A.; Hamzavi, I.; Jemec, G. Hidradenitis suppurativa: An update on epidemiology, phenotypes, diagnosis, pathogenesis, comorbidities and quality of life. J. Eur. Acad. Dermatol. Venereol. 2021, 35, 50–61. [Google Scholar] [CrossRef]
- Arshad, R.; Tabish, T.A.; Naseem, A.A.; Hassan, M.R.u.; Hussain, I.; Hussain, S.S.; Shahnaz, G. Development of poly-L-lysine multi-functionalized muco-penetrating self- emulsifying drug delivery system (SEDDS) for improved solubilization and targeted delivery of ciprofloxacin against intracellular Salmonella typhi. J. Mol. Liq. 2021, 333, 115972. [Google Scholar] [CrossRef]
- Olivieri, F.; Castaldo, R.; Cocca, M.; Gentile, G.; Lavorgna, M. Mesoporous silica nanoparticles as carriers of active agents for smart anticorrosion organic coatings. A critical review. Nanoscale 2021, 13, 9091–9111. [Google Scholar] [CrossRef] [PubMed]
- López, V.; Villegas, M.R.; Rodríguez, V.; Villaverde, G.; Lozano, D.; Baeza, A.; Vallet-Regí, M. Janus mesoporous silica nanoparticles for dual targeting of tumor cells and mitochondria. ACS Appl. Mater. Interfaces 2017, 9, 26697–26706. [Google Scholar] [CrossRef]
- Arshad, R.; Pal, K.; Sabir, F.; Rahdar, A.; Bilal, M.; Shahnaz, G.; Kyzas, G.Z. A review of the nanomaterials use for the diagnosis and therapy of salmonella typhi. J. Mol. Struct. 2021, 1230, 129928. [Google Scholar] [CrossRef]
- Aagaard, L.; Rossi, J.J. RNAi therapeutics: Principles, prospects and challenges. Adv. Drug Deliv. Rev. 2007, 59, 75–86. [Google Scholar] [CrossRef]
- Pecot, C.V.; Calin, G.A.; Coleman, R.L.; Lopez-Berestein, G.; Sood, A.K. RNA interference in the clinic: Challenges and future directions. Nat. Rev. Cancer 2011, 11, 59–67. [Google Scholar] [CrossRef]
- Mura, S.; Nicolas, J.; Couvreur, P. Stimuli-responsive nanocarriers for drug delivery. Nat. Mater. 2013, 12, 991–1003. [Google Scholar] [CrossRef]
- Xiong, Q.; Lee, G.Y.; Ding, J.; Li, W.; Shi, J. Biomedical applications of mRNA nanomedicine. Nano Res. 2018, 11, 5281–5309. [Google Scholar] [CrossRef]
- Guo, P.; Haque, F.; Hallahan, B.; Reif, R.; Li, H. Uniqueness, advantages, challenges, solutions, and perspectives in therapeutics applying RNA nanotechnology. Nucleic Acid Ther. 2012, 22, 226–245. [Google Scholar] [CrossRef]
- Abdelmawla, S.; Guo, S.; Zhang, L.; Pulukuri, S.M.; Patankar, P.; Conley, P.; Trebley, J.; Guo, P.; Li, Q.-X. Pharmacological characterization of chemically synthesized monomeric phi29 pRNA nanoparticles for systemic delivery. Mol. Ther. 2011, 19, 1312–1322. [Google Scholar] [CrossRef] [PubMed]
- de Fougerolles, A.; Vornlocher, H.-P.; Maraganore, J.; Lieberman, J. Interfering with disease: A progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007, 6, 443–453. [Google Scholar] [CrossRef]
- Kim, J.; Eygeris, Y.; Gupta, M.; Sahay, G. Self-assembled mRNA vaccines. Adv. Drug Deliv. Rev. 2021, 170, 83–112. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Breton, B.; Zheng, W.; McFadyen, I.; Hopson, K.; Frederick, J.; Meehan, R.S.; Zaks, T. Bioinformatics algorithm of mRNA-4157 identifies neoantigens with pre-existing TIL reactivities in colorectal tumors. In Proceedings of the AACR Annual Meeting, Philadelphia, PA, USA, 22–24 June 2020. [Google Scholar]
- Paquet-Fifield, S.; Koh, S.L.; Cheng, L.; Beyit, L.M.; Shembrey, C.; Mølck, C.; Behrenbruch, C.; Papin, M.; Gironella, M.; Guelfi, S. Tight junction protein claudin-2 promotes self-renewal of human colorectal cancer stem-like cells. Cancer Res. 2018, 78, 2925–2938. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, P.S.; Rudra, A.; Miao, L.; Anderson, D.G. Delivering the messenger: Advances in technologies for therapeutic mRNA delivery. Mol. Ther. 2019, 27, 710–728. [Google Scholar] [CrossRef]
- Lim, S.A.; Cox, A.; Tung, M.; Chung, E.J. Clinical progress of nanomedicine-based RNA therapies. Bioact. Mater. 2021, in press. [Google Scholar] [CrossRef]





| Nanostructure | Key Feature | Ref |
|---|---|---|
| Ultra-thermostable RNA NPs | RNA ligand proved to dramatically inhibit the growth of breast cancer with non-detectable toxicity and immune responses in mice. | [102] |
| Selenium-siRNA NPs | Small interfering RNA (siRNA) showed great potential in advanced therapeutics because of its highly sequential ability for silencing HeLa genes for cervical cancer | [103] |
| MSN-anti-miR-155 NPs | miR-155 was highly over-expressed in colorectal tissues and cell lines as compared to the control groups and showed enhanced therapeutic efficacy. | [104] |
| Survivin-siRNA NPs | The novel nanocarrier system was able to initiate a specified and safe cellular uptake with increased transfection efficacy, promoting the downregulation of HCC cells. | [105] |
| Enveloped siRNA NPs | siRNA multi-functionalized nano-enveloped carriers can strongly silence target genes expressions as well as strongly pre-dominant genes, such as prohibitin 1 (PHB1), resulting in significantly culminating prostate tumor growth | [106] |
| FA-PEI-Fe3O4-siRNA NPs | Effective targeted PD-L1-knockdown therapy as well as a diagnosis in gastric cancers, thus favoring towards the best theranostic approach | [107] |
| PLL-siRNA-MSN NPs | MSNPs-PLL proved to be an accomplished candidate for non-invasive transdermal drug delivery in alleviating skin cancer cells division | [108] |
| RNA Based Nanomaterials | Clinical Trials | Ref. |
|---|---|---|
| The self-delivering RNA (sd RNA) | Combination of immunotherapy and chemotherapy for cancer treatment in pre-clinical trials. | [136] |
| Single mRNA-4157vaccine | Preclinical phase 2 against melanoma. | [137] |
| Adjuvant claudin mRNA cells | Pre-clinical stages against metastatic breast cancer. | [138] |
| mRNA 5671 based NPs | Pre-clinical stages against colorectal cancer, lungs cancer, and pancreatic cancer. | [139] |
| mRNA 2416 based NPs | Pre-clinical stages against solid tumors in ovarian cancer. | [140] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arshad, R.; Fatima, I.; Sargazi, S.; Rahdar, A.; Karamzadeh-Jahromi, M.; Pandey, S.; Díez-Pascual, A.M.; Bilal, M. Novel Perspectives towards RNA-Based Nano-Theranostic Approaches for Cancer Management. Nanomaterials 2021, 11, 3330. https://doi.org/10.3390/nano11123330
Arshad R, Fatima I, Sargazi S, Rahdar A, Karamzadeh-Jahromi M, Pandey S, Díez-Pascual AM, Bilal M. Novel Perspectives towards RNA-Based Nano-Theranostic Approaches for Cancer Management. Nanomaterials. 2021; 11(12):3330. https://doi.org/10.3390/nano11123330
Chicago/Turabian StyleArshad, Rabia, Iqra Fatima, Saman Sargazi, Abbas Rahdar, Milad Karamzadeh-Jahromi, Sadanand Pandey, Ana M. Díez-Pascual, and Muhammad Bilal. 2021. "Novel Perspectives towards RNA-Based Nano-Theranostic Approaches for Cancer Management" Nanomaterials 11, no. 12: 3330. https://doi.org/10.3390/nano11123330
APA StyleArshad, R., Fatima, I., Sargazi, S., Rahdar, A., Karamzadeh-Jahromi, M., Pandey, S., Díez-Pascual, A. M., & Bilal, M. (2021). Novel Perspectives towards RNA-Based Nano-Theranostic Approaches for Cancer Management. Nanomaterials, 11(12), 3330. https://doi.org/10.3390/nano11123330









